We narrowed to 45,592 results for: cha;
-
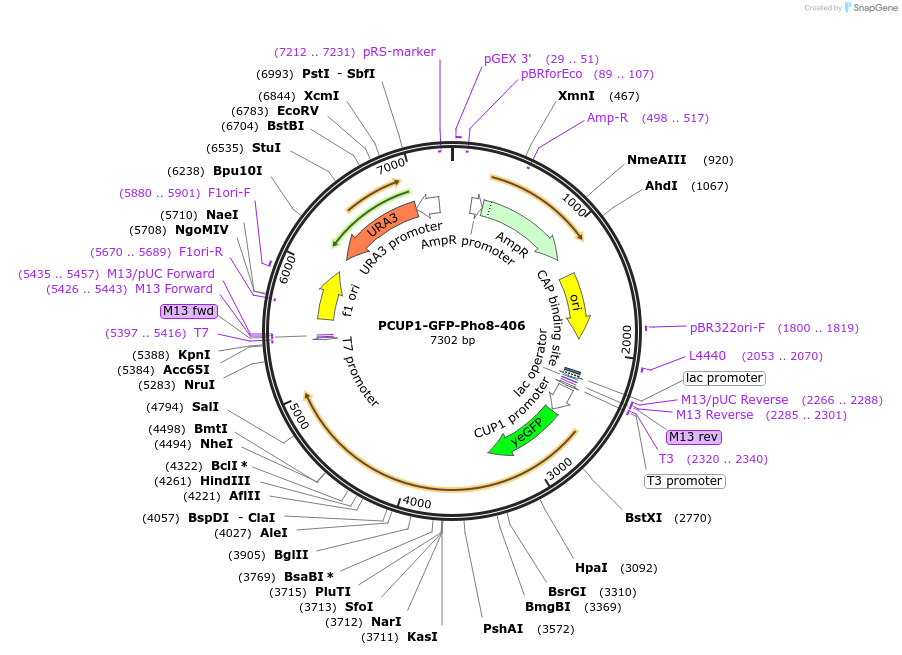
Plasmid#207006PurposeIntegration of GFP-Pho8 as an additional copy in genome, expressed under CUP1 promoter. Uses auxotrophic marker URA3 (Saccharomyces cerevisiae) for selection. Marker for yeast vacuolar membrane.DepositorInsertPHO8
TagsGFPExpressionYeastAvailable SinceJune 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
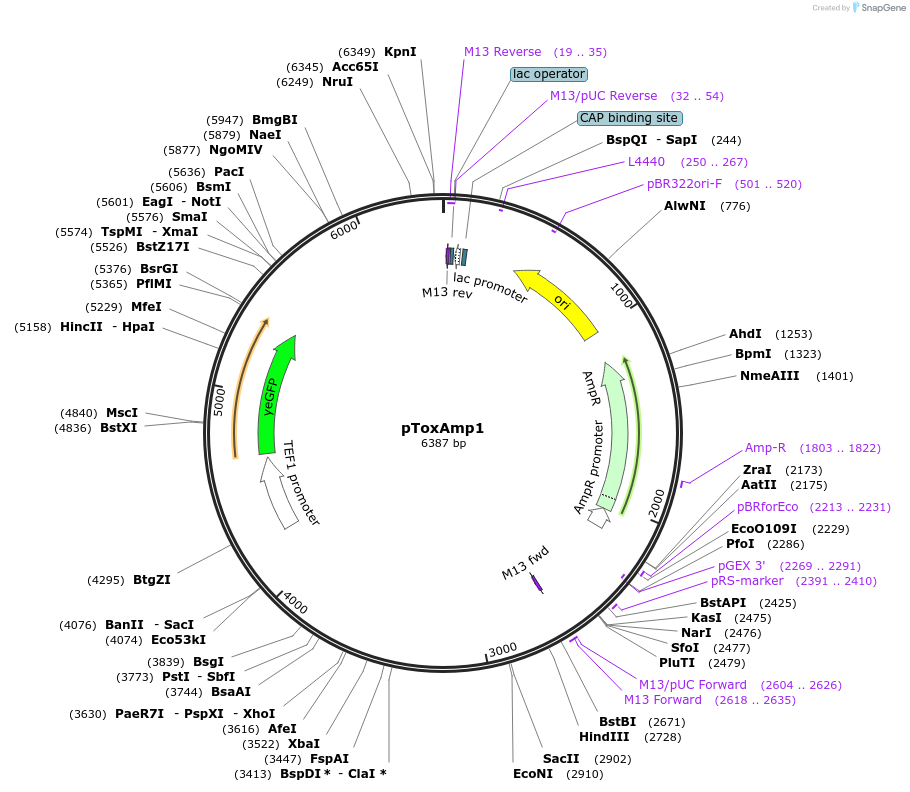
pToxAmp1
Plasmid#218285PurposeAmplifying the gene copy number of heterologous genes (yEGFP and RelB) through ToxAmp (toxin-antitoxin-driven gene amplification) mechanismDepositorInsertHO(-253, -1)-pRPL8B>RelB>tPDC1-pTEF1>yEGFP>tURA3-ARS712-HO(-731, -264)
ExpressionYeastAvailable SinceMay 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
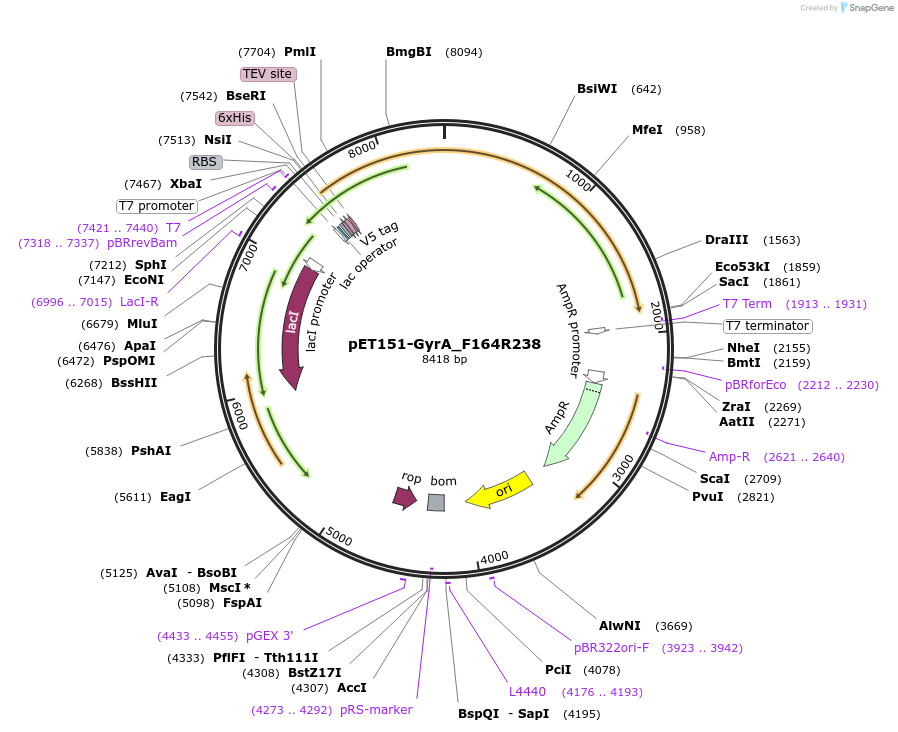
pET151-GyrA_F164R238
Plasmid#216230PurposeS. aureus USA300 GyrA mutant F164 R238 residues codon-optimized for E. coliDepositorInsertDNA gyrase
Tags6xHisExpressionBacterialMutationChanged arginine 238 to serine, phenylalanine 168…PromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
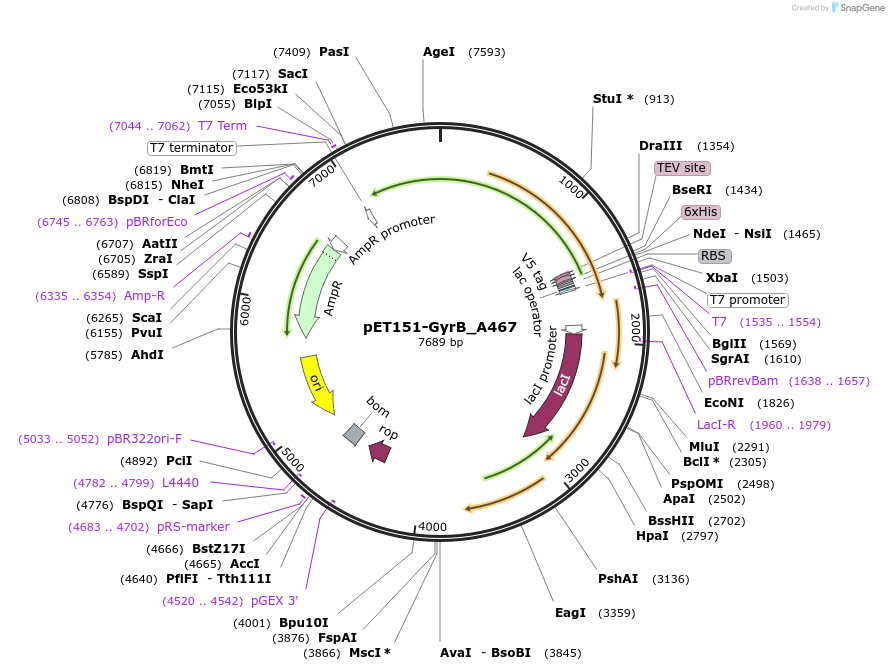
pET151-GyrB_A467
Plasmid#216234PurposeS. aureus USA300 GyrB mutant A467 residue codon-optimized for E. coliDepositorInsertDNA gyrase
Tags6xHisExpressionBacterialMutationChanged alanine 467 to valinePromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
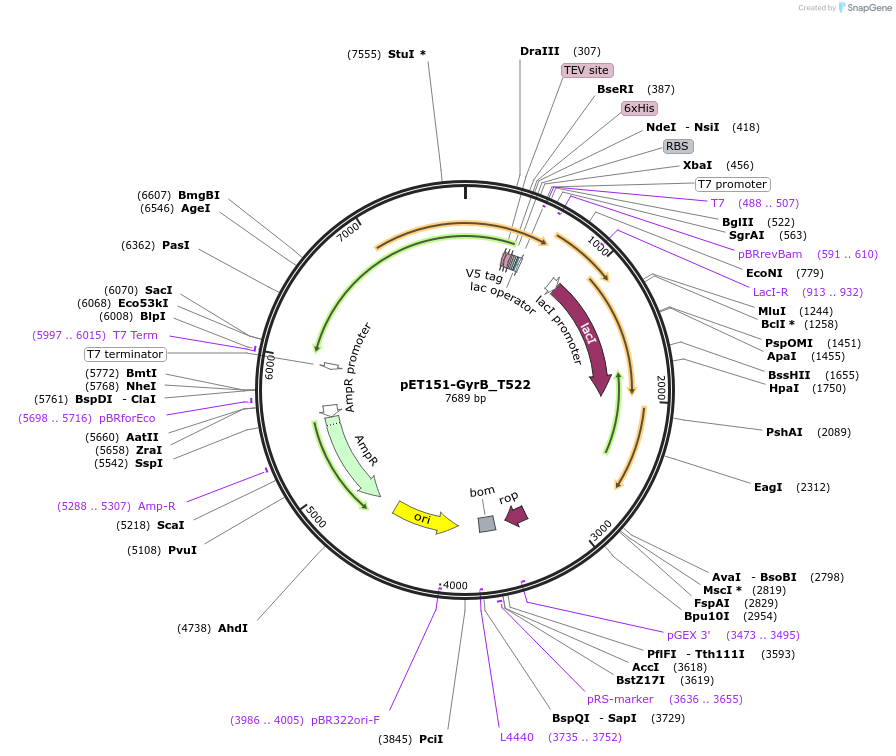
pET151-GyrB_T522
Plasmid#216235PurposeS. aureus USA300 GyrB mutant T522 residue codon-optimized for E. coliDepositorInsertDNA gyrase
Tags6xHisExpressionBacterialMutationChanged threonine 522 to isoleucinePromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
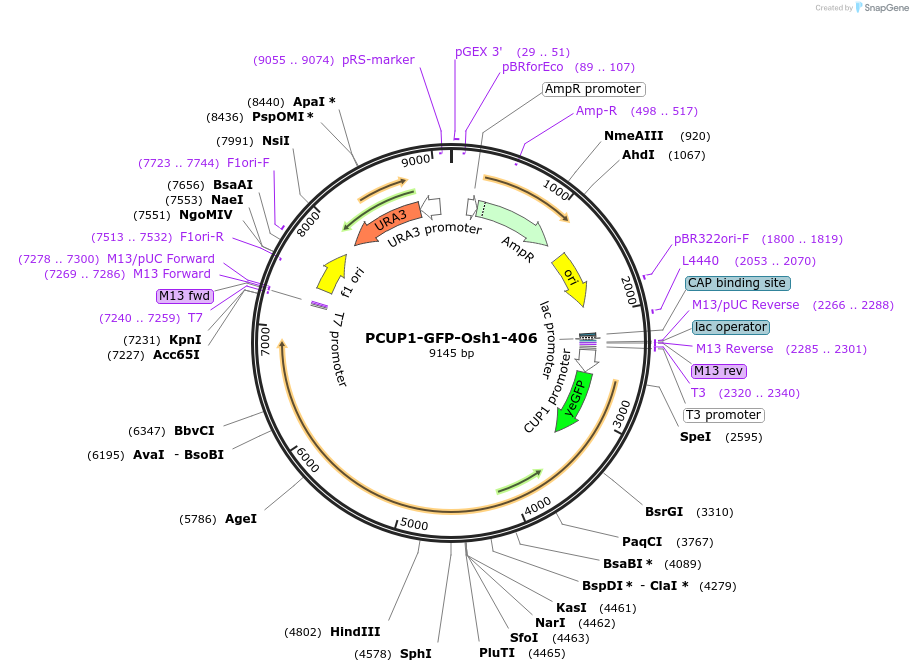
PCUP1-GFP-Osh1-406
Plasmid#207007PurposeIntegration of Osh1-GFP as an additional copy in genome, expressed under CUP1 promoter. Uses auxotrophic marker URA3 (Saccharomyces cerevisiae) for selection. Yeast PMN pathway substrate.DepositorInsertOSH1
TagsGFPExpressionYeastAvailable SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
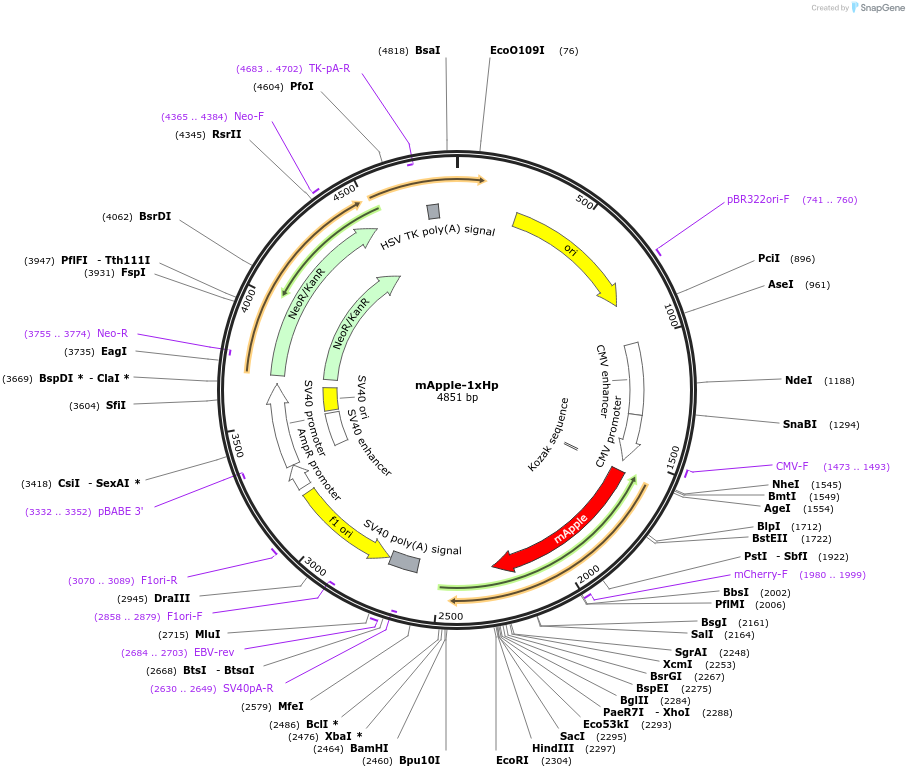
mApple-1xHp
Plasmid#214409PurposeLipid droplet and ER localization of 1xHp of Spastin fused to mAppleDepositorAvailable SinceMarch 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
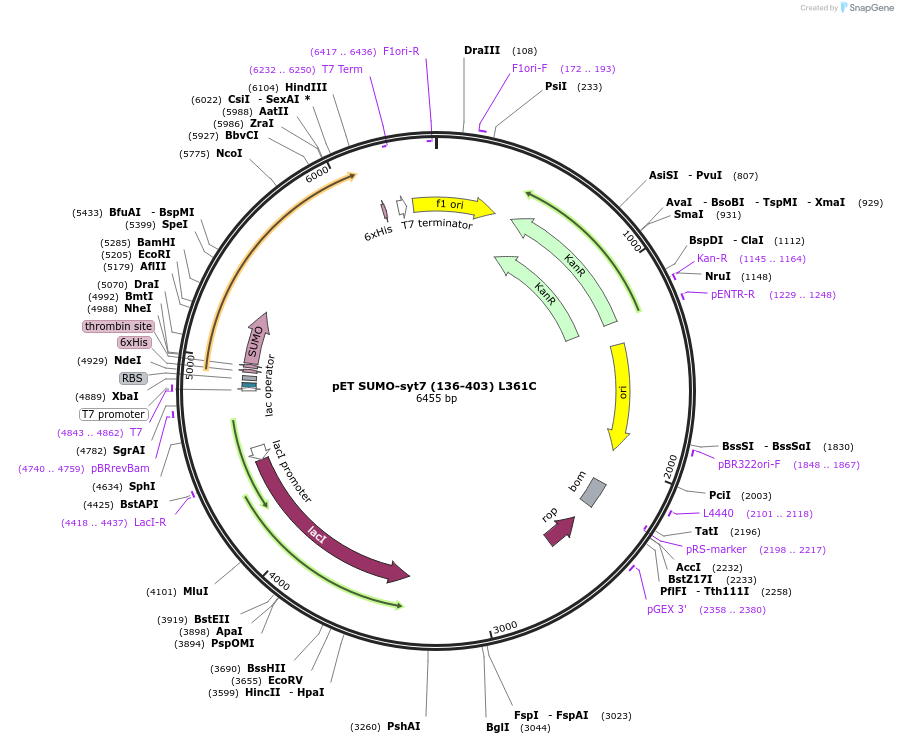
pET SUMO-syt7 (136-403) L361C
Plasmid#213622PurposeBacterial expression of SUMO-tagged mouse syt7 (136-403) L361CDepositorAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
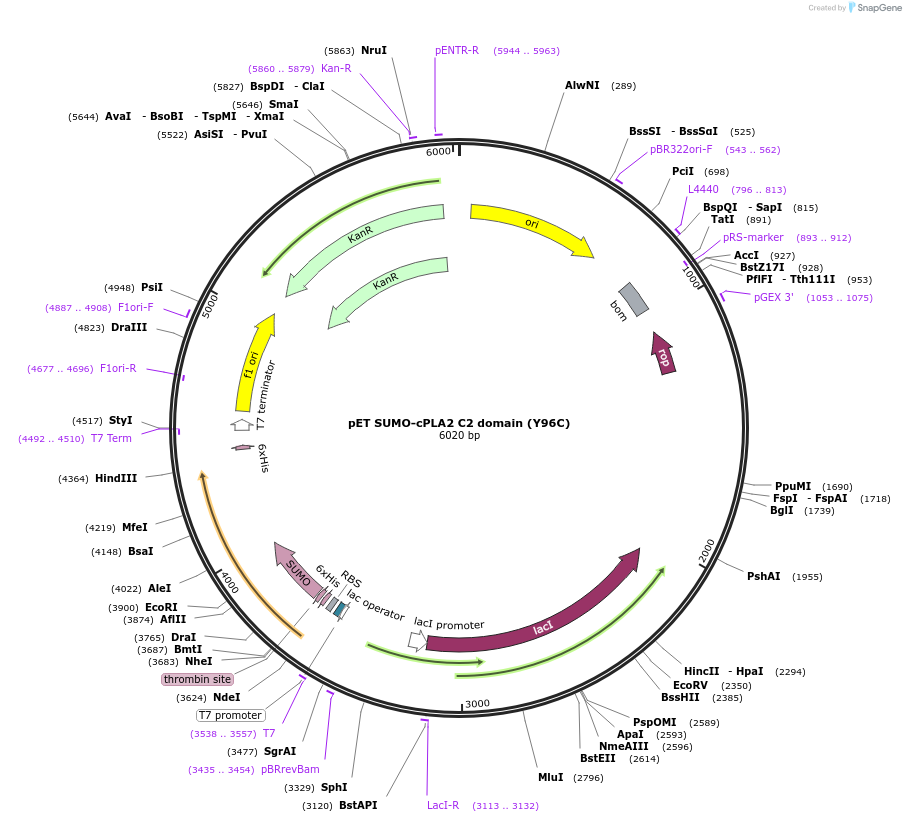
pET SUMO-cPLA2 C2 domain (Y96C)
Plasmid#213633PurposeBacterial expression of SUMO-tagged human cPLA2 C2 domain (Y96C)DepositorInsertcPLA2 C2 domain (Y96C) (PLA2G4A Human)
TagsSUMOExpressionBacterialMutationY96C, C139A, C141SAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
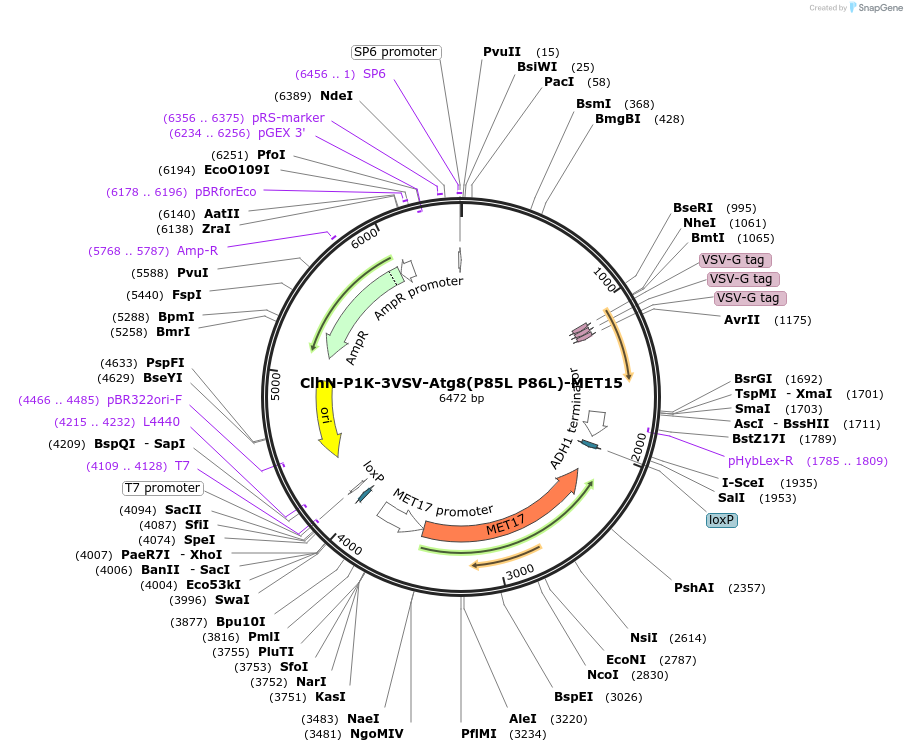
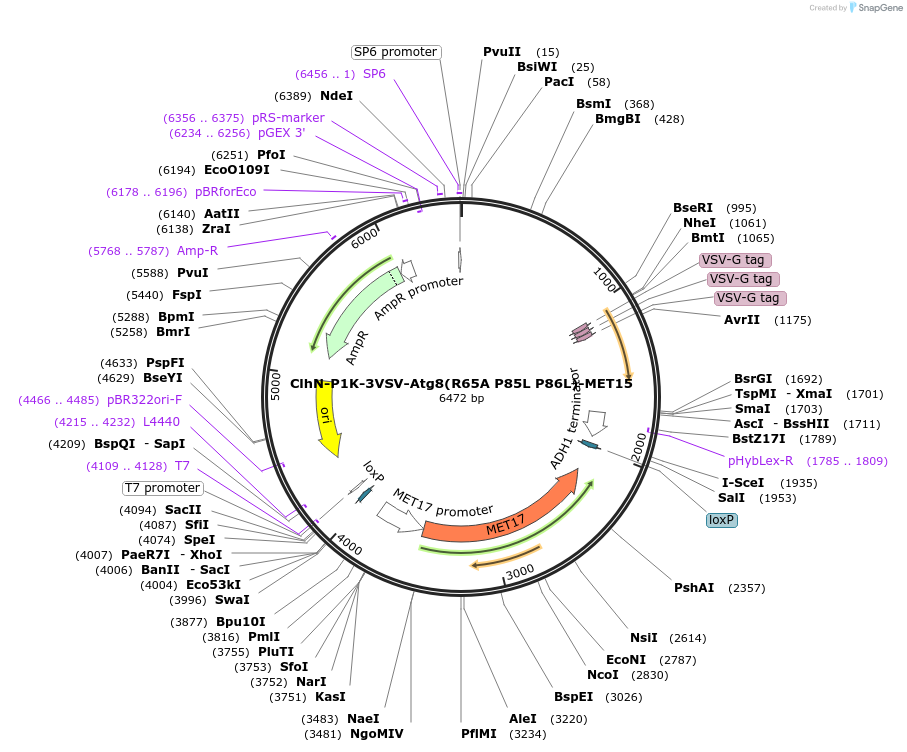
ClhN-P1K-3VSV-Atg8(P85L P86L)-MET15
Plasmid#207050PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationP85L P86LAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
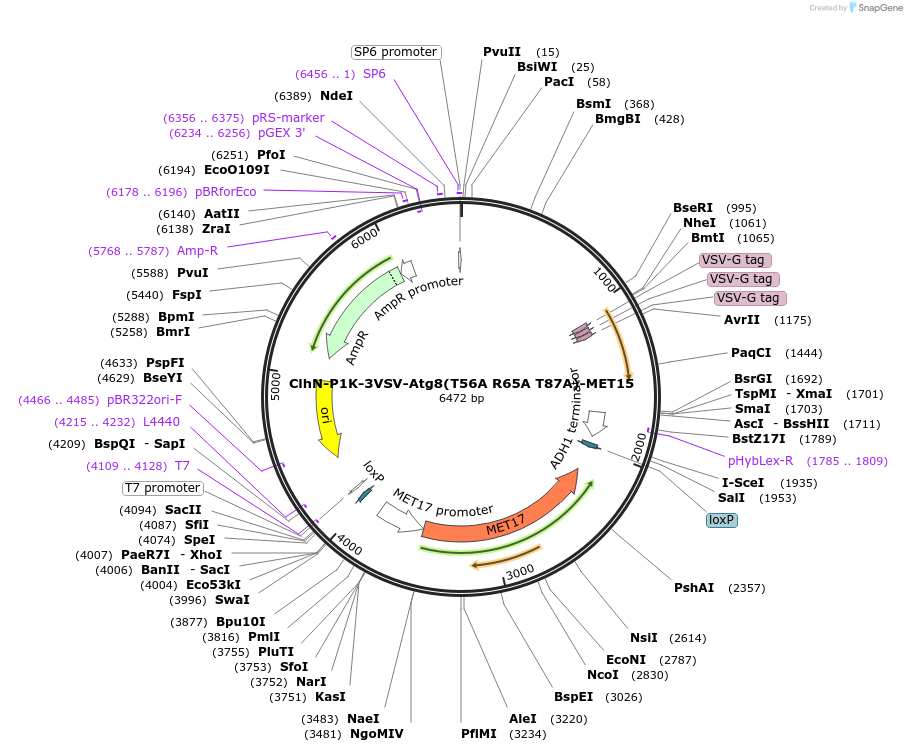
ClhN-P1K-3VSV-Atg8(T56A R65A T87A)-MET15
Plasmid#207051PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationT56A R65A T87AAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
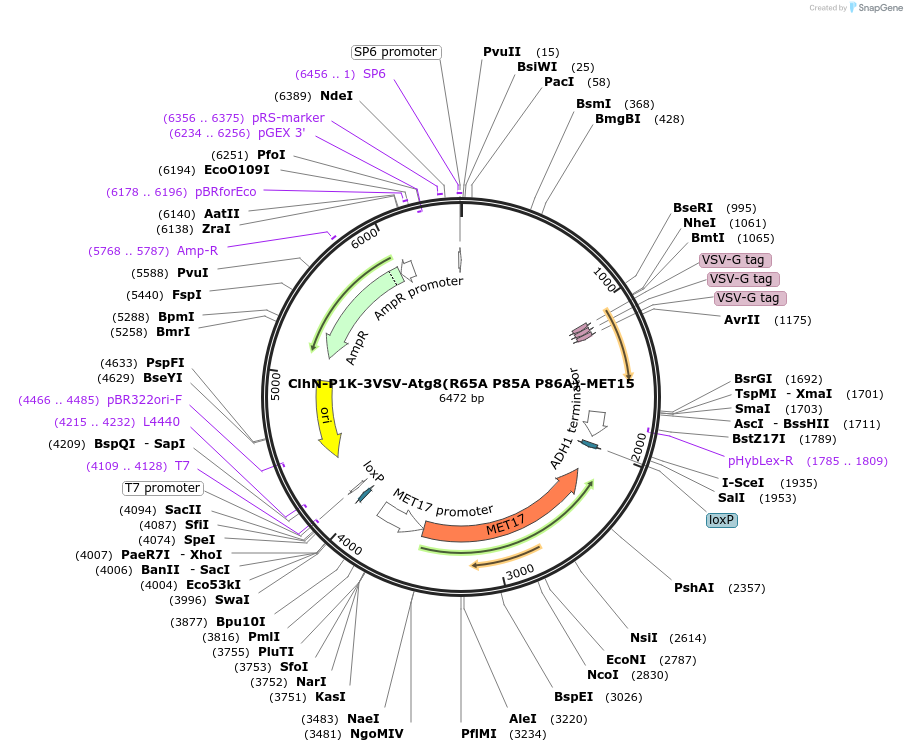
ClhN-P1K-3VSV-Atg8(R65A P85A P86A)-MET15
Plasmid#207052PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationR65A P85A P86AAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
ClhN-P1K-3VSV-Atg8(R65A P85L P86L)-MET15
Plasmid#207053PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationR65A P85L P86LAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
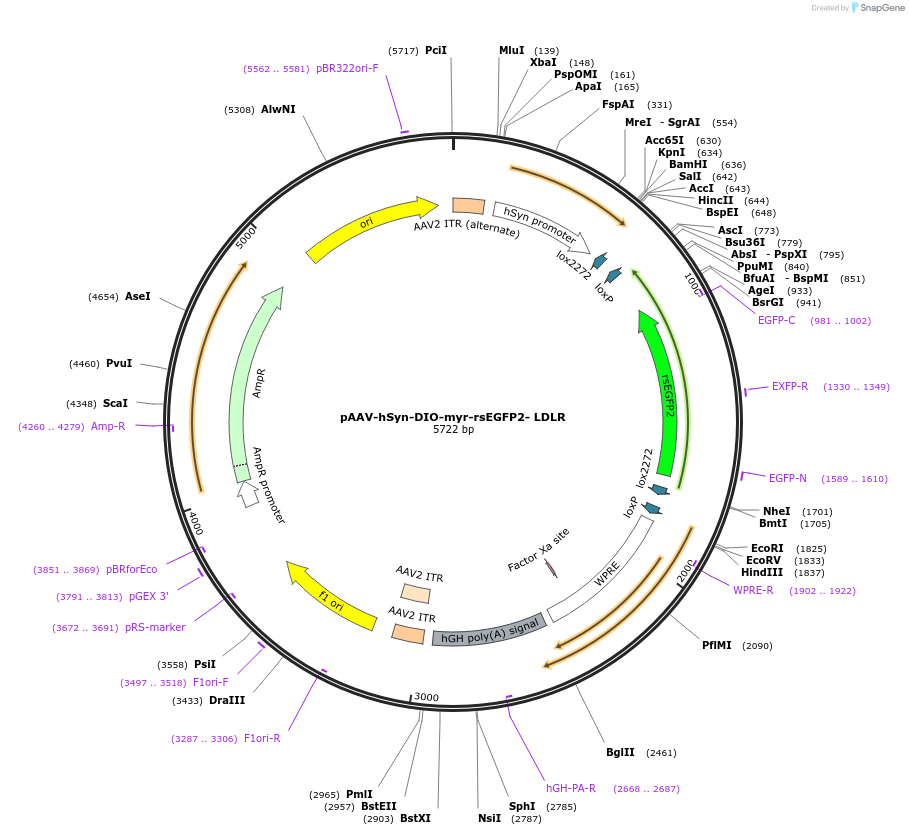
pAAV-hSyn-DIO-myr-rsEGFP2- LDLR
Plasmid#213399Purposedouble floxed, myristoylation site (myr) and LDLR, fused to reversibly switchable EGFP rsEGFP2DepositorInsertrsEGFP2
UseAAVTagsC-terminal (Ct) cytoplasmic domains of low densit…ExpressionMammalianAvailable SinceFeb. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
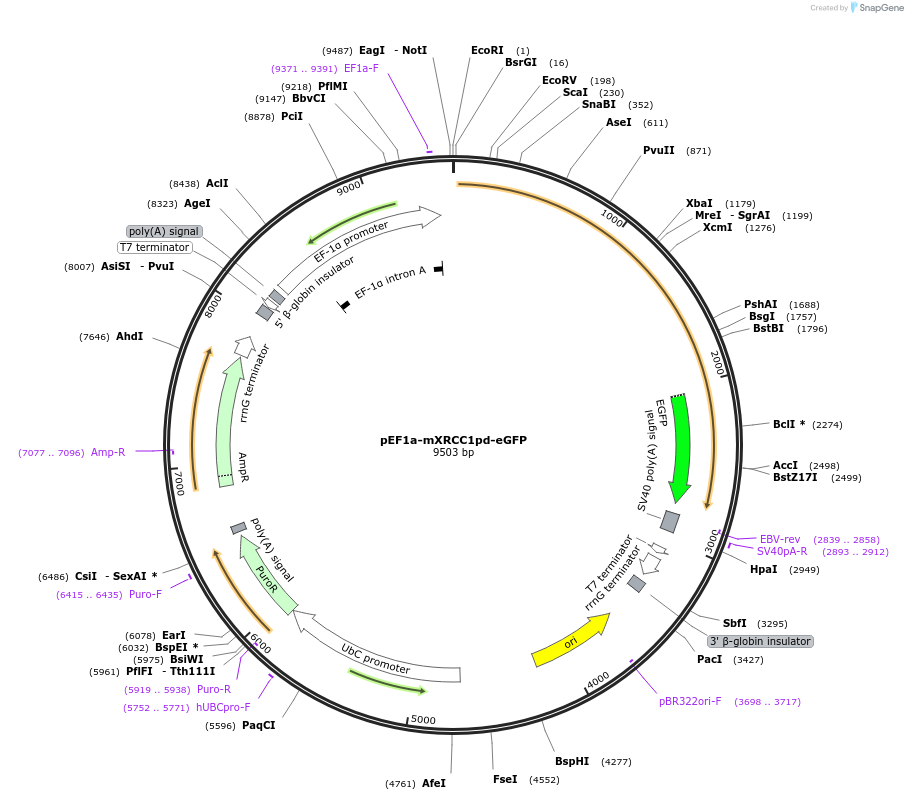
pEF1a-mXRCC1pd-eGFP
Plasmid#206035PurposeMammalian expression of PAR-deficient mXRCC1pd coupled to eGFP under the control of a EF1a promoterDepositorInsertmXRCC1pd
TagseGFPExpressionMammalianMutationS105A, S186A, K188A, S195A, S221A, S222A,S238A, S…PromoterEf1aAvailable SinceJan. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
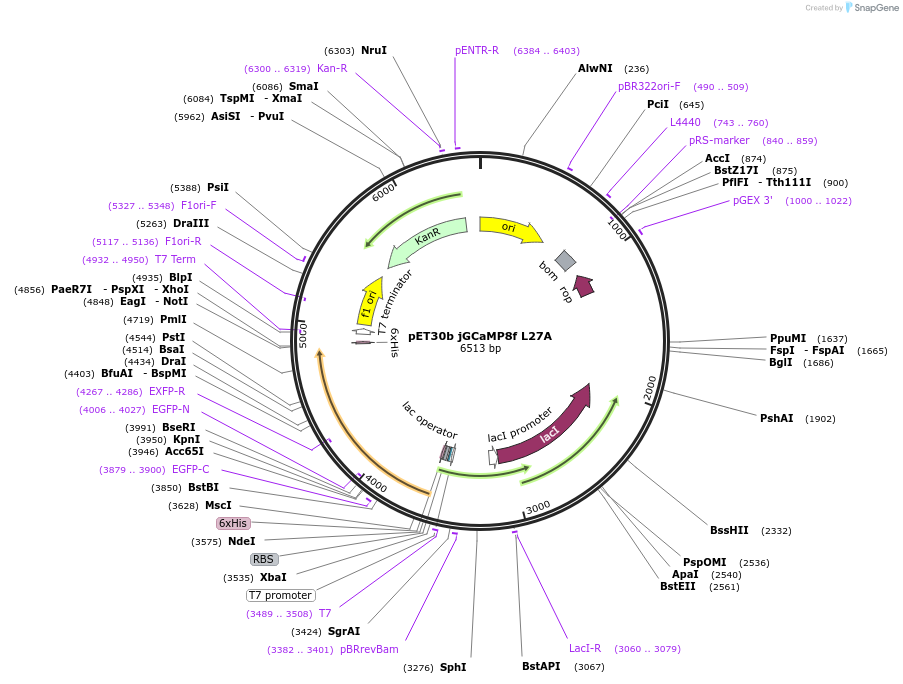
pET30b jGCaMP8f L27A
Plasmid#204138PurposeProduction of recombinant calcium sensor jGCaMP8f L27A fast decay variant in E.coliDepositorInsertjGCaMP8f L27A
TagsN-terminal His tagExpressionBacterialMutationLeucine 27 changed to AlaninePromoterT7Available SinceDec. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
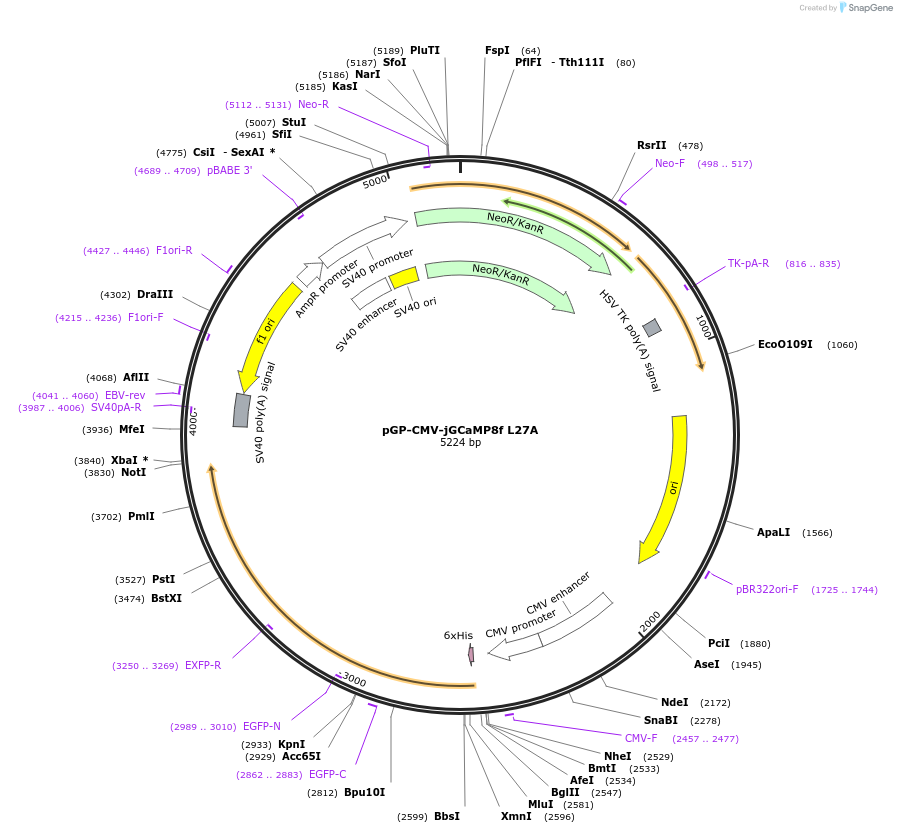
pGP-CMV-jGCaMP8f L27A
Plasmid#204140PurposeFluorescent reporter for calcium imaging in mammalian cells - jGCaMP8f with L27A mutationDepositorInsertjGCaMP8f L27A
TagsN-terminal His tagExpressionMammalianMutationLeucine 27 changed to AlaninePromoterCMVAvailable SinceDec. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
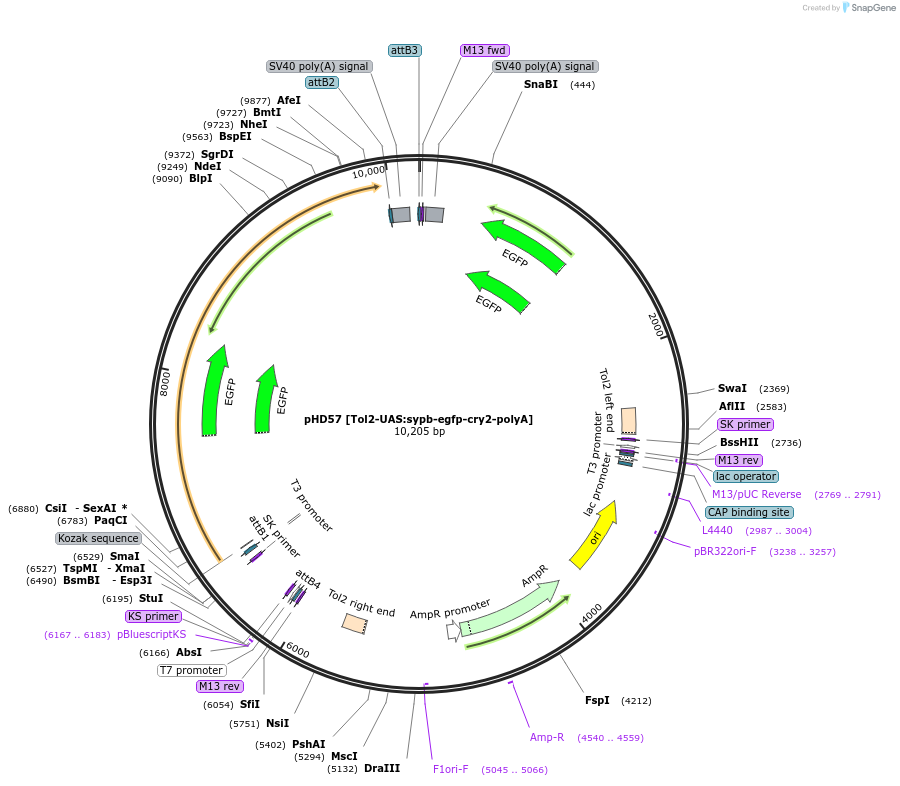
pHD57 [Tol2-UAS:sypb-egfp-cry2-polyA]
Plasmid#198381PurposeExpression of SYPB::CRY2olig(535) in neurons of Danio rerioDepositorInsertUAS:sypb-egfp-cry2
UseTol2TagsEGFPMutationD387APromoterUASAvailable SinceNov. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
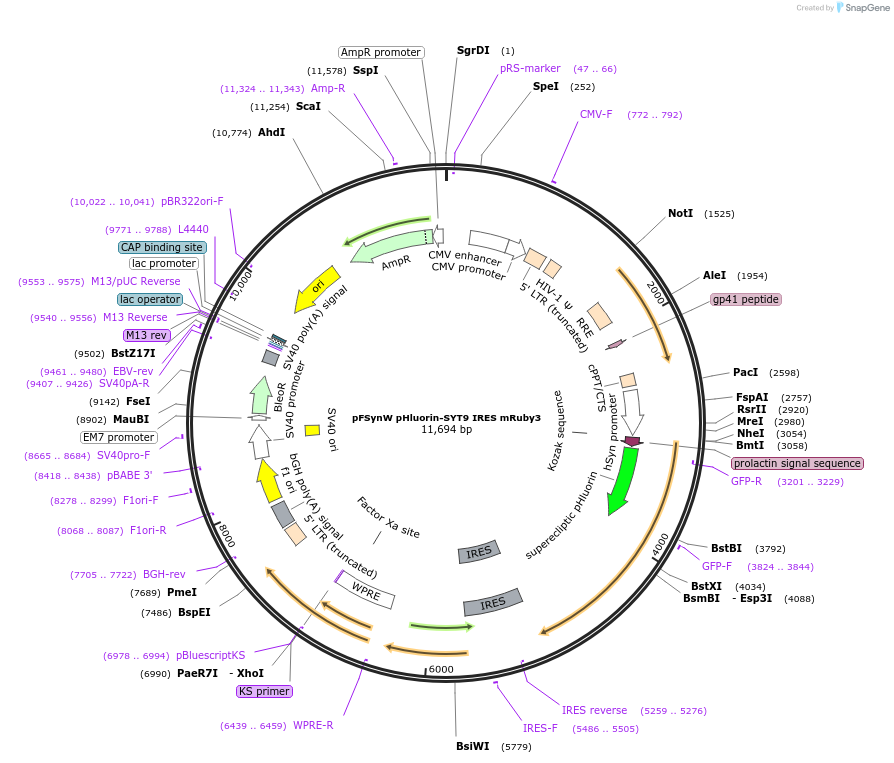
pFSynW pHluorin-SYT9 IRES mRuby3
Plasmid#195698PurposeLentiviral plasmid encoding SYT9 with an N-terminal pHluorin tag followed by an internal ribosomal entry site followed by mRuby3 under the human synapsin promoterDepositorAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
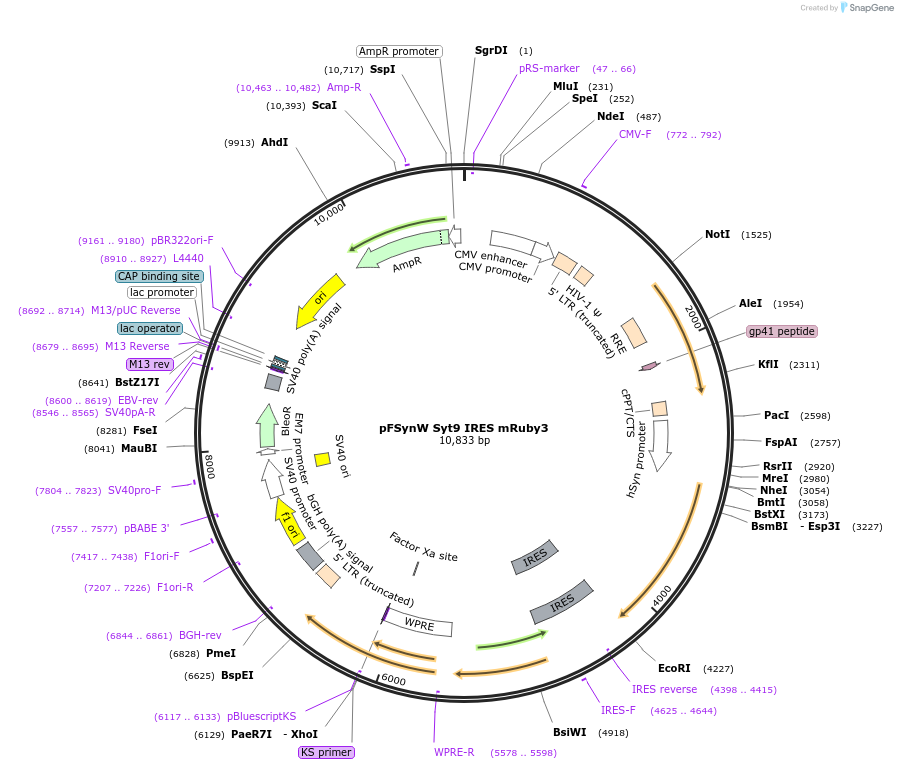
pFSynW Syt9 IRES mRuby3
Plasmid#195700PurposeLentiviral plasmid encoding full-length mouse SYT9 followed by an internal ribosomal entry site followed by mRuby under the human synapsin promoterDepositorAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only