We narrowed to 19,943 results for: INO
-
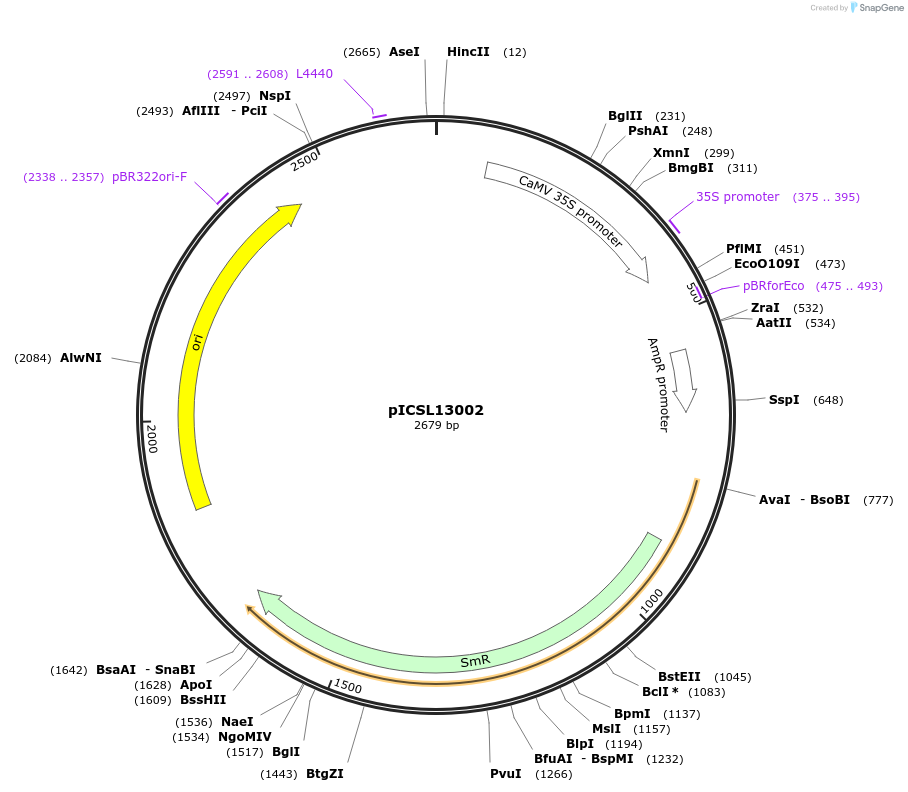
Plasmid#50266DepositorInsertpromoter (0.4 kb ) + 5'UTR, 35s (Cauliflower Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
pBABE Rad N88
Plasmid#11218DepositorInsertRad (RRAD Human)
UseRetroviralExpressionMammalianMutationN88: deletion of amino acids 1-88Available SinceMarch 1, 2006AvailabilityAcademic Institutions and Nonprofits only -
RCAS(A) cTbx5 EnR (CT#635)
Plasmid#13967DepositorAvailable SinceFeb. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
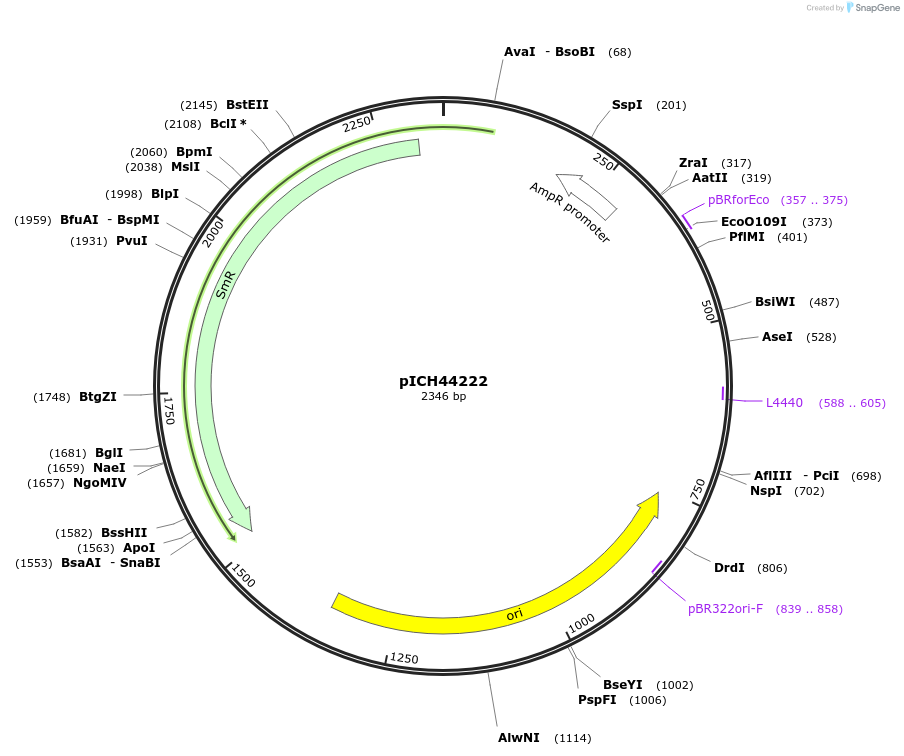
pICH44222
Plasmid#50287DepositorInsert5'UTR CMV1 (Cucumber Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
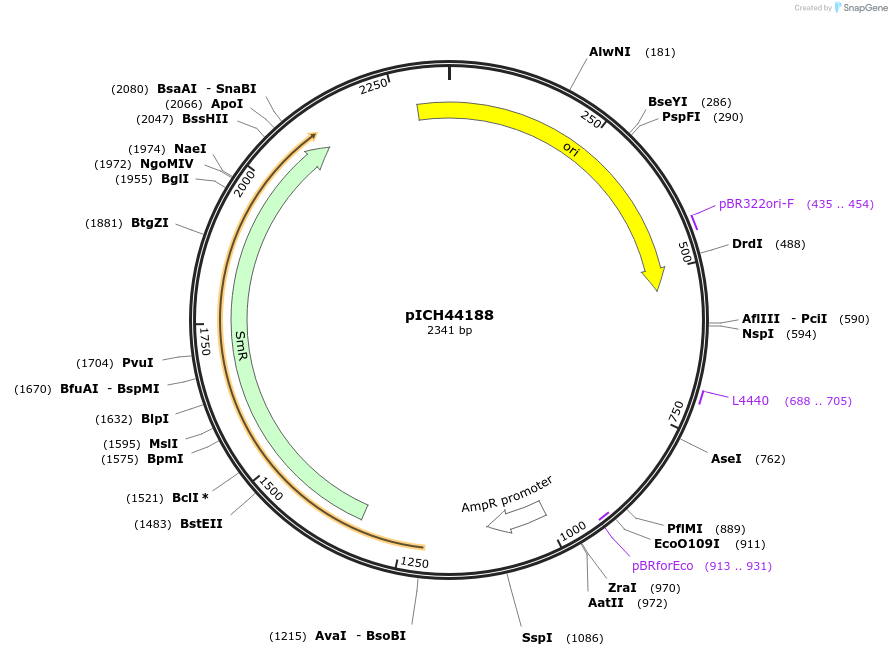
pICH44188
Plasmid#50289DepositorInsert5'UTR BSMV (Barley Stripe Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
KIF3B
Plasmid#25254PurposeBacterial expression for structure determination; may not be full ORFDepositorInsertKIF3B:S6-Q359 (KIF3B Human)
TagsHisExpressionBacterialMutationContains amino acids S6-Q359Available SinceAug. 13, 2010AvailabilityIndustry, Academic Institutions, and Nonprofits -
IQGAP3
Plasmid#25173PurposeBacterial expression for structure determination; may not be full ORFDepositorInsertIQGAP3:G1529-K1631 (IQGAP3 Human)
TagsHisExpressionBacterialMutationContains amino acids G1529-K1631Available SinceAug. 9, 2010AvailabilityIndustry, Academic Institutions, and Nonprofits -
FKBP03
Plasmid#25209PurposeBacterial expression for structure determination; may not be full ORFDepositorInsertfkbp03.001.073 (FKBP3 Human)
TagsHisExpressionBacterialMutationContains amino acids 1-73Available SinceAug. 12, 2010AvailabilityIndustry, Academic Institutions, and Nonprofits -
pWZ414-F14
Plasmid#23063DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 deleted amino acids 3-29Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
CBX3 (3KUP)
Plasmid#25332PurposeBacterial expression for structure determination; may not be full ORFDepositorAvailable SinceAug. 17, 2010AvailabilityIndustry, Academic Institutions, and Nonprofits -
RCAS(A) cTbx5 VP16 (CT#630)
Plasmid#13969DepositorAvailable SinceFeb. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
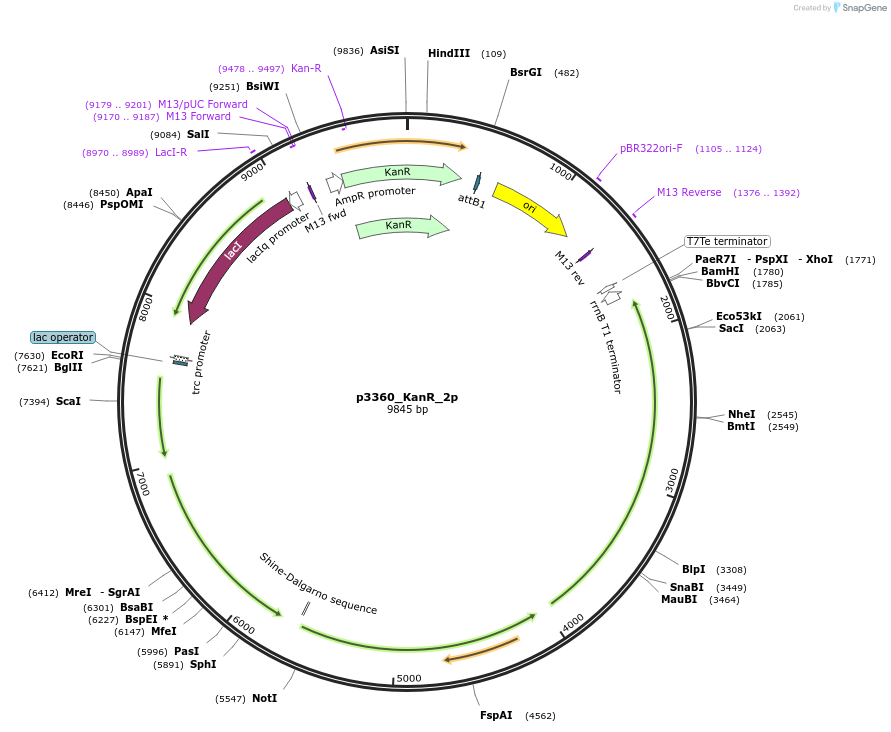
p3360_KanR_2p
Plasmid#247302PurposeProduction of pinocembrin in Escherichia coli, cloned by BASIC assembly, based on 3360 in Carbonell et al. 2018DepositorInsertsAtCHI
AtCHS
Sc4CL
AtPAL
ExpressionBacterialPromoterPtrc with lac operonAvailable SinceNov. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
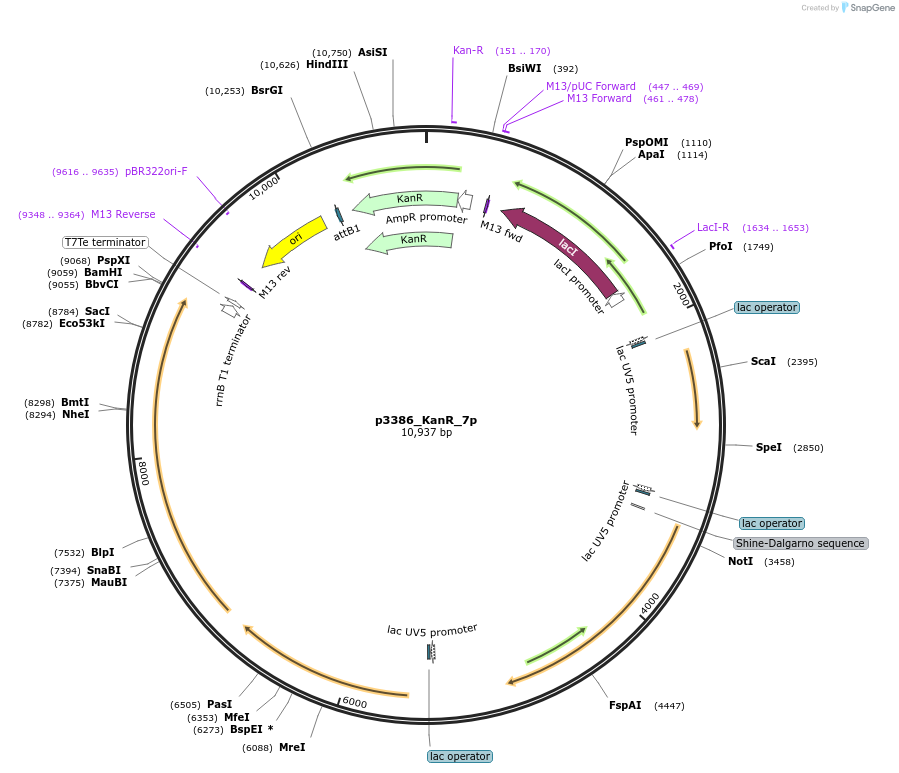
p3386_KanR_7p
Plasmid#247309PurposeProduction of pinocembrin in Escherichia coli, cloned by BASIC assembly, based on 3386 in Carbonell et al. 2018DepositorInsertsAtCHI
Sc4CL
AtCHS
AtPAL
ExpressionBacterialPromoterPlacUV5 with lac operonAvailable SinceNov. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
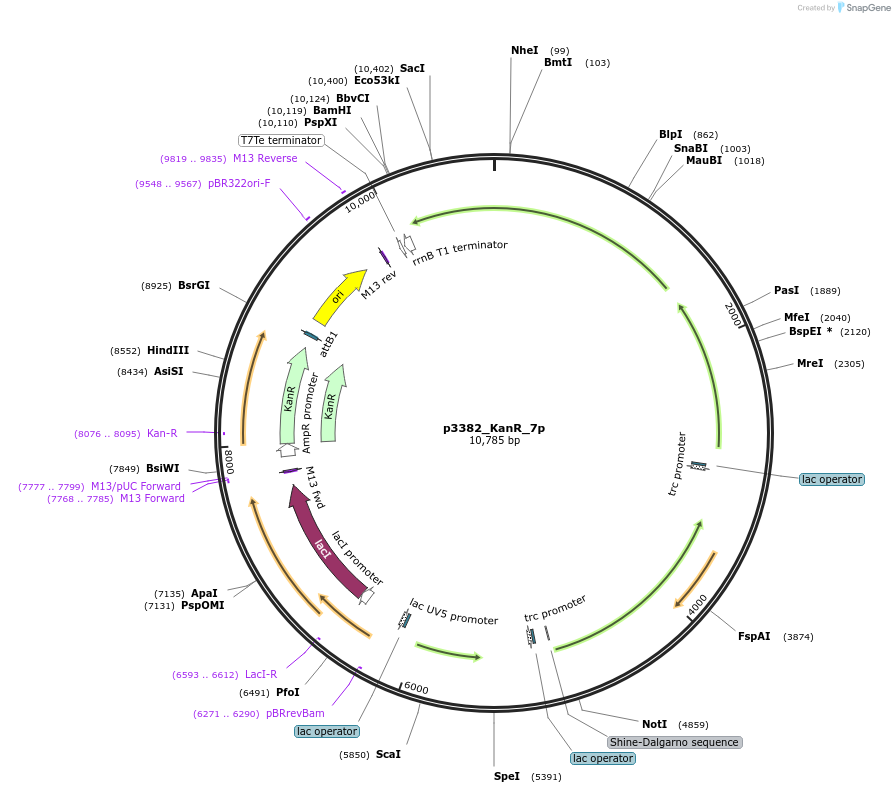
p3382_KanR_7p
Plasmid#247308PurposeProduction of Pinocembrin in Escherichia coli, cloned by BASIC assembly, based on 3382 in Carbonell et al. 2018DepositorInsertsAtCHI
Sc4CL
AtCHS
AtPAL
ExpressionBacterialPromoterPLacUV5 with lac operator and Ptrc with lac opera…Available SinceNov. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
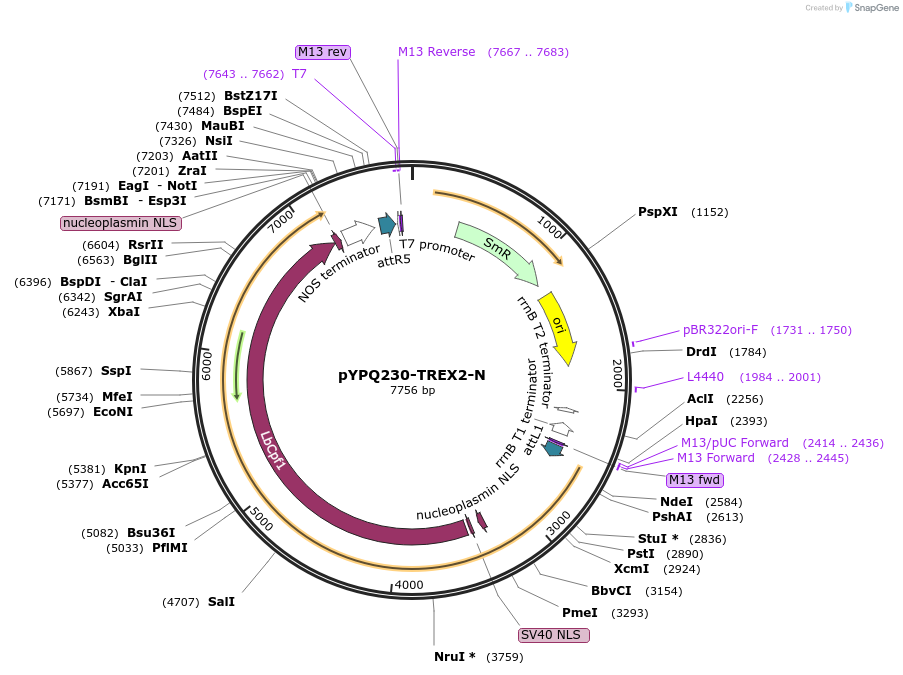
pYPQ230-TREX2-N
Plasmid#244025PurposeGateway entry plasmid (attL1& attR5) expressing TREX2 exonuclease fused to the N-terminus of LbCas12a, connected by a flexible XTEN linker, without promoterDepositorInsertCas12a
UseCRISPRTagsTREX2Available SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
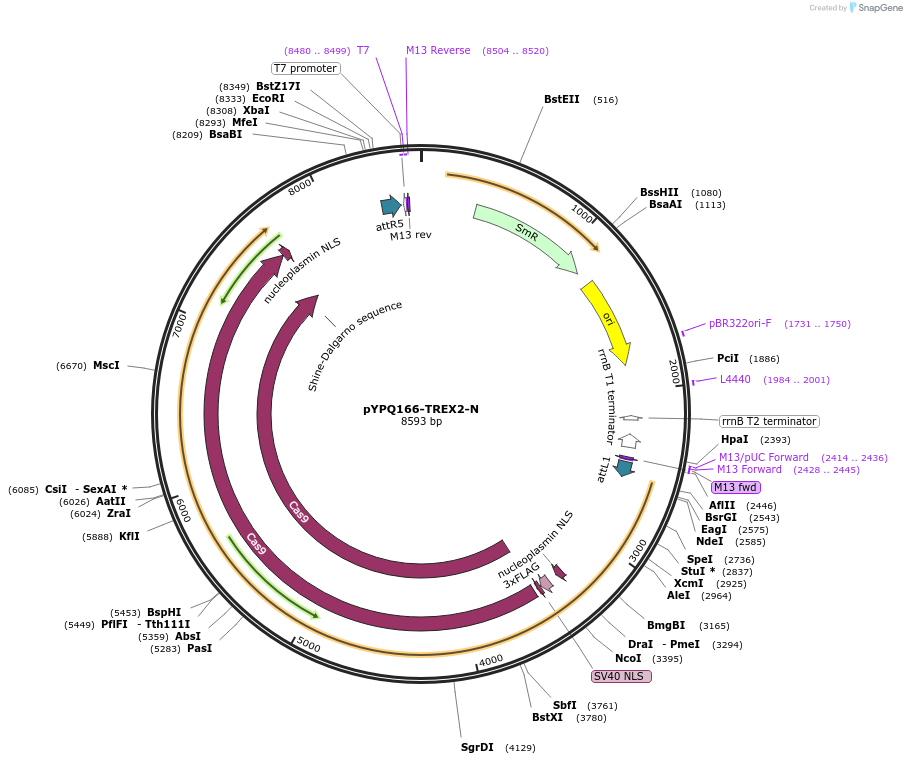
pYPQ166-TREX2-N
Plasmid#244022PurposeGateway entry plasmid (attL1& attR5) expressing TREX2 exonuclease fused to the N-terminus of zSpCas9, connected by a flexible XTEN linker, without promoterDepositorInsertCas9
UseCRISPRTagsTREX2Available SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
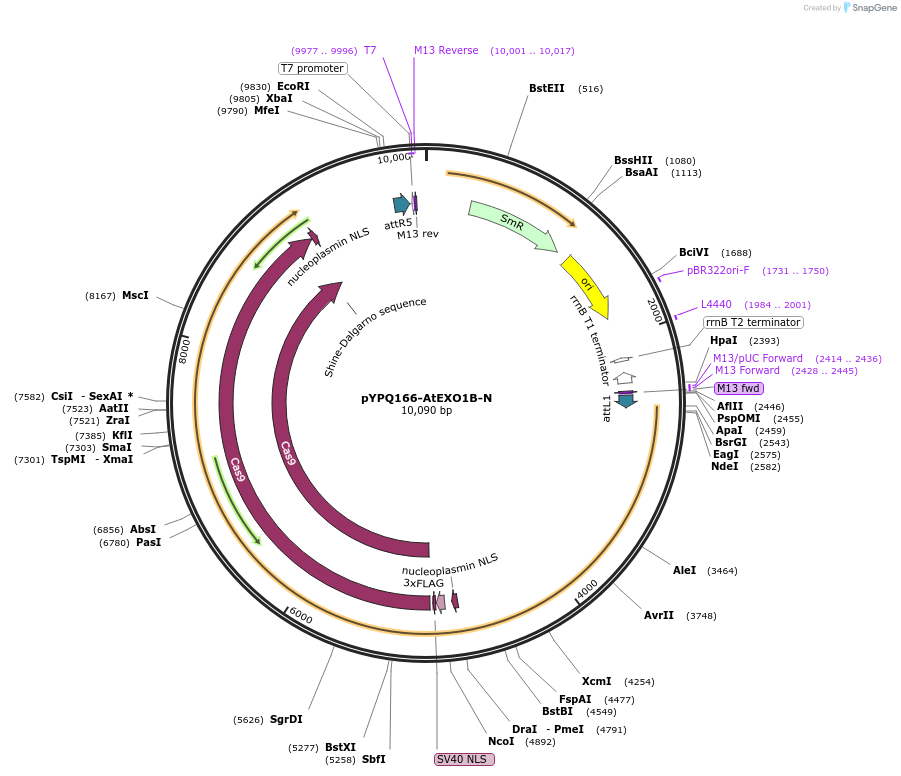
pYPQ166-AtEXO1B-N
Plasmid#244023PurposeGateway entry plasmid (attL1& attR5) expressing AtEXO1B exonuclease fused to the N-terminus of zSpCas9, connected by a flexible XTEN linker, without promoterDepositorInsertCas9
UseCRISPRTagsAtEXO1BAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
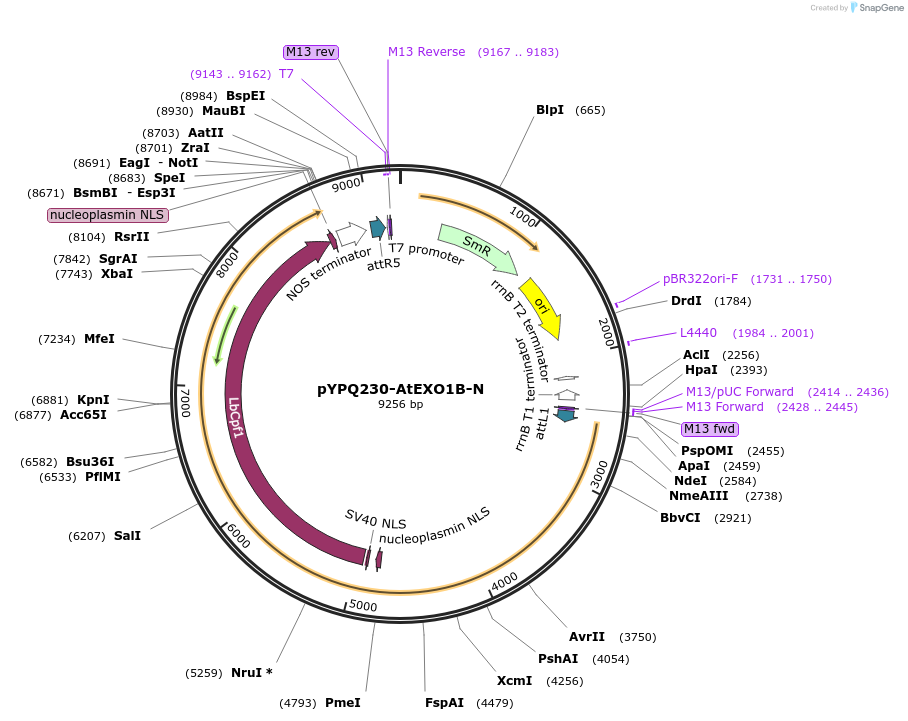
pYPQ230-AtEXO1B-N
Plasmid#244024PurposeGateway entry plasmid (attL1& attR5) expressing AtEXO1B exonuclease fused to the N-terminus of LbCas12a, connected by a flexible XTEN linker, without promoterDepositorInsertCas12a
UseCRISPRTagsAtEXO1BAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
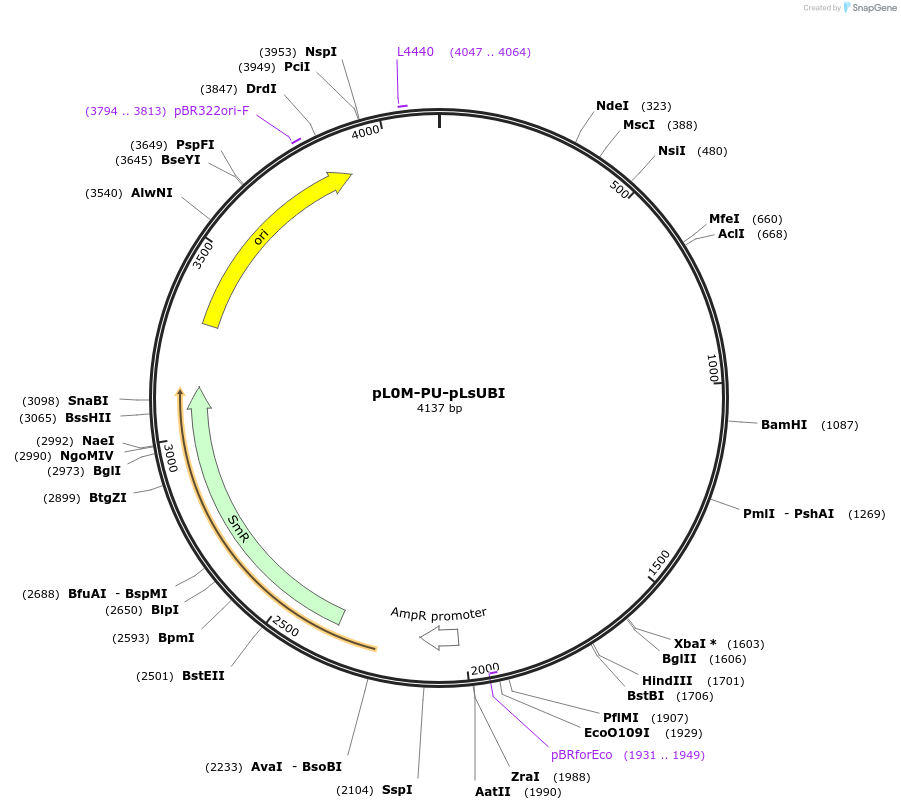
pL0M-PU-pLsUBI
Plasmid#244448PurposeLevel 0 Golden Gate "PU" module containing the domesticated sequence of the promoter of the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)DepositorInsertDomesticated sequence of the promoter for the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)
ExpressionPlantMutationThe promoter sequence 96336283-96337681 from NC_0…PromoterPolyubiquitin 4 gene (LOC111919935) from lettuceAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
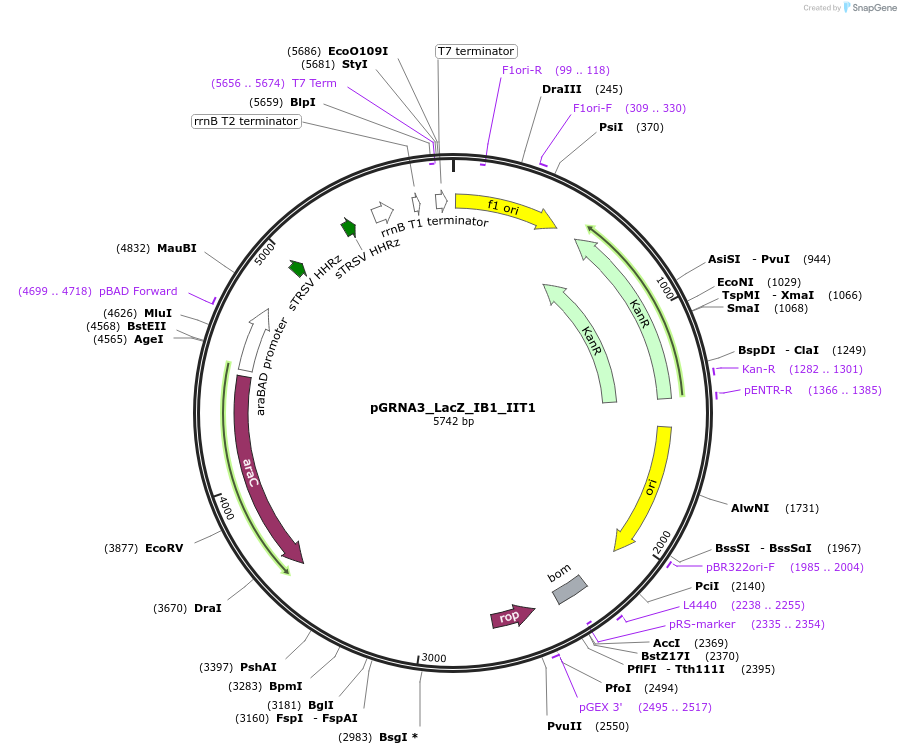
pGRNA3_LacZ_IB1_IIT1
Plasmid#247080PurposeL-arabinose inducible crRNA of type I-E CRISPR cas (Escherichia coli K-12). encoding 105 nt spacer which targets both Ecoli LacZ B1 and T1 site. riboJ/RNaseIII site are encoded between two crRNADepositorInserttype I-E CRISPR cas crRNA
UseCRISPRExpressionBacterialAvailable SinceNov. 4, 2025AvailabilityAcademic Institutions and Nonprofits only