We narrowed to 20,061 results for: INO
-
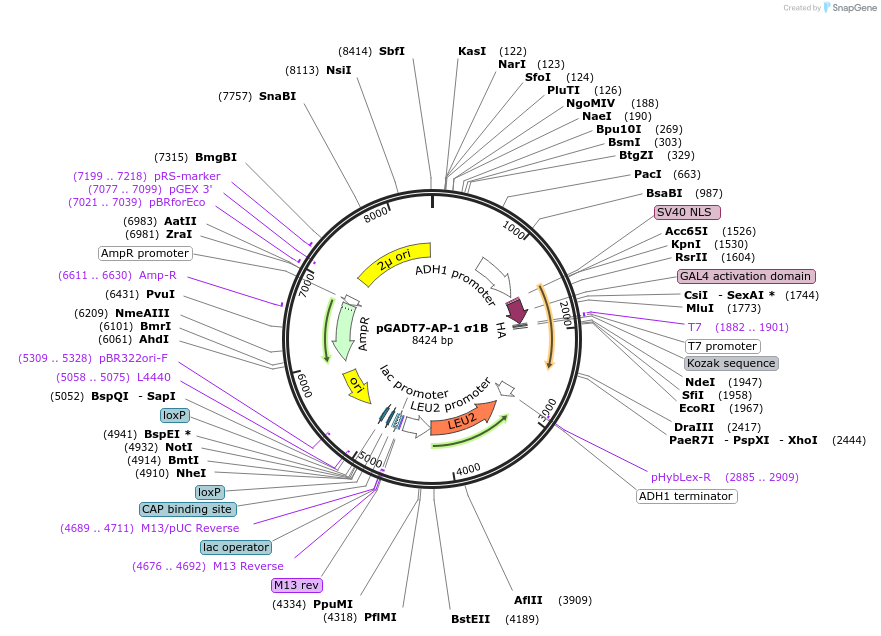
Plasmid#198335PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-1 σ1B fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 σ1B
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastMutationExpresses a LELQMNRRY nonapeptide after the last …PromoterADH1Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
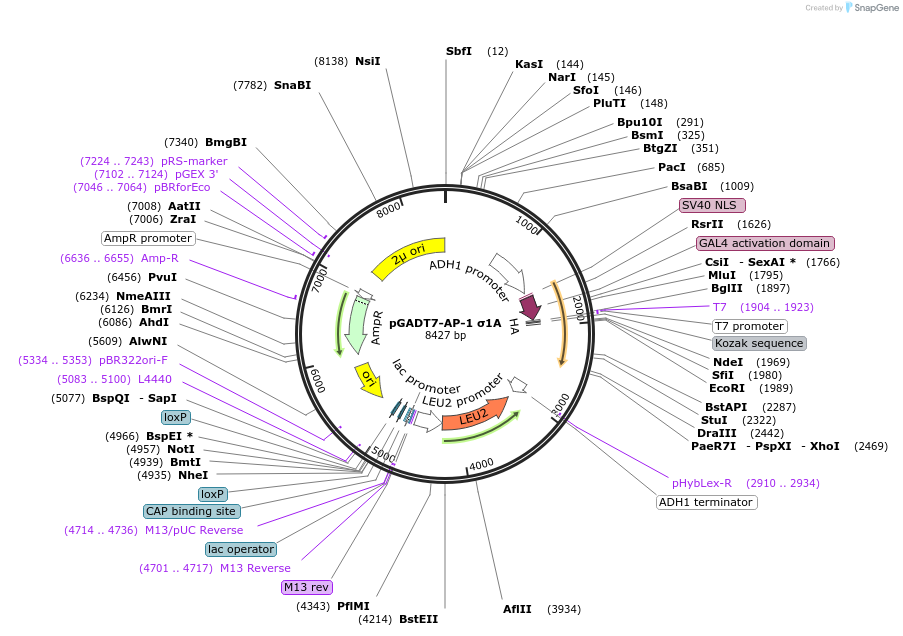
pGADT7-AP-1 σ1A
Plasmid#198334PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-1 σ1A fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 σ1A
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastMutationExpresses a LELQMNRRY nonapeptide after the last …PromoterADH1Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
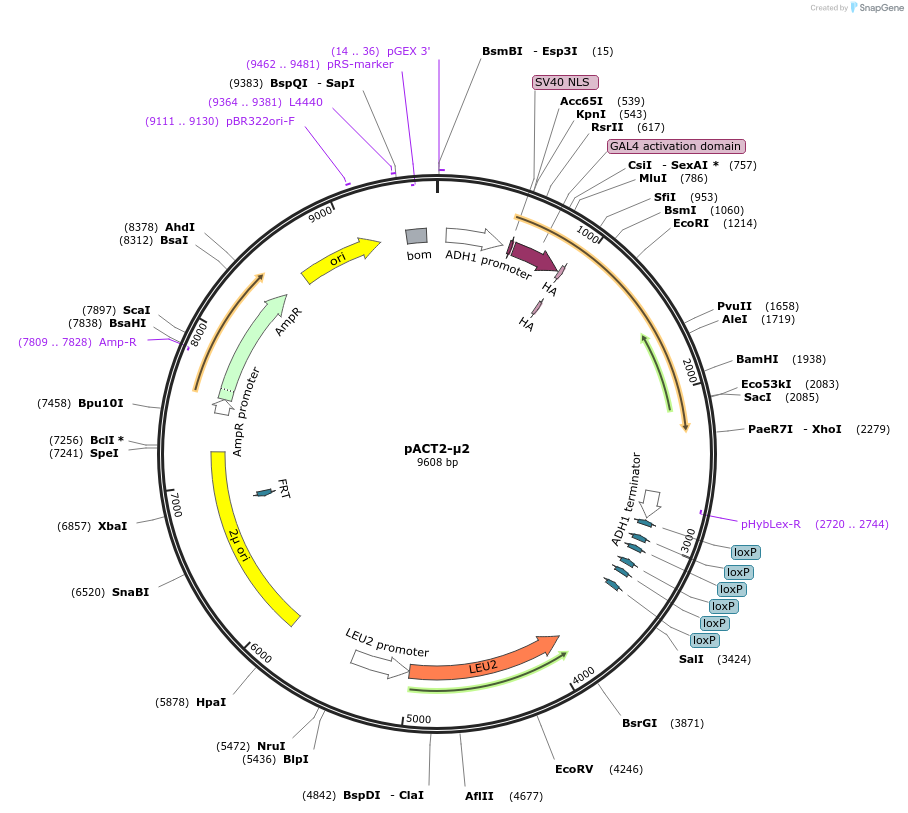
pACT2-μ2
Plasmid#198173PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-2 μ2 fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-2 μ2
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastPromoterADH1Available SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
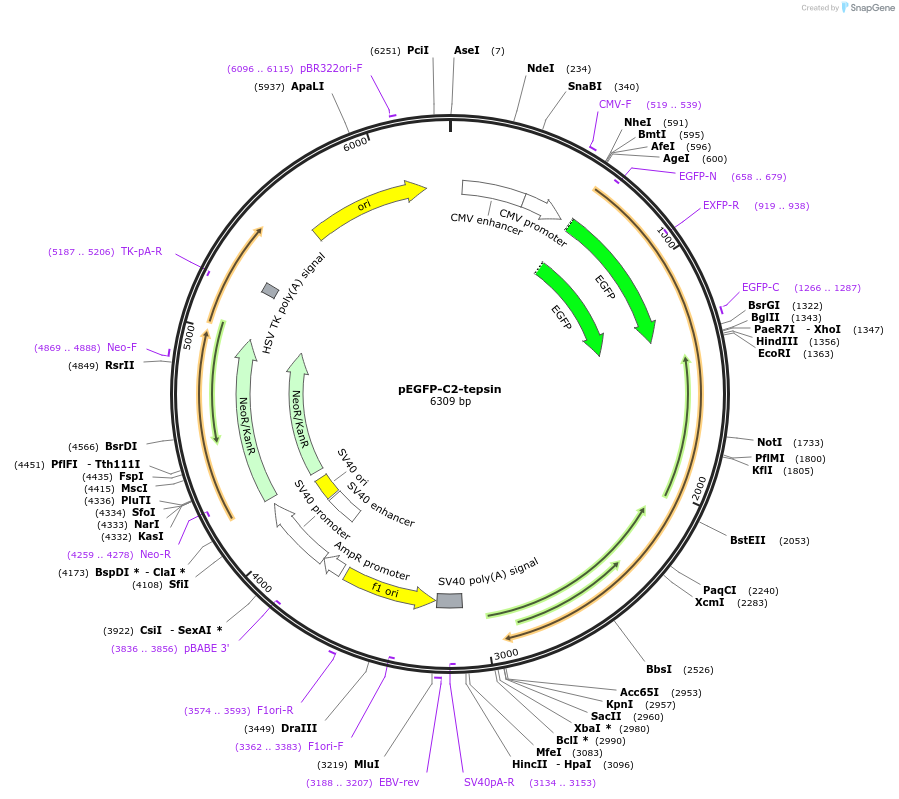
pEGFP-C2-tepsin
Plasmid#196707PurposeExpresses GFP-tepsin in mammalian cellsDepositorInserttepsin (TEPSIN Human)
TagsEGFPExpressionMammalianMutationsilent substitution in codon 199 to eliminate int…PromoterCMVAvailable SinceMarch 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
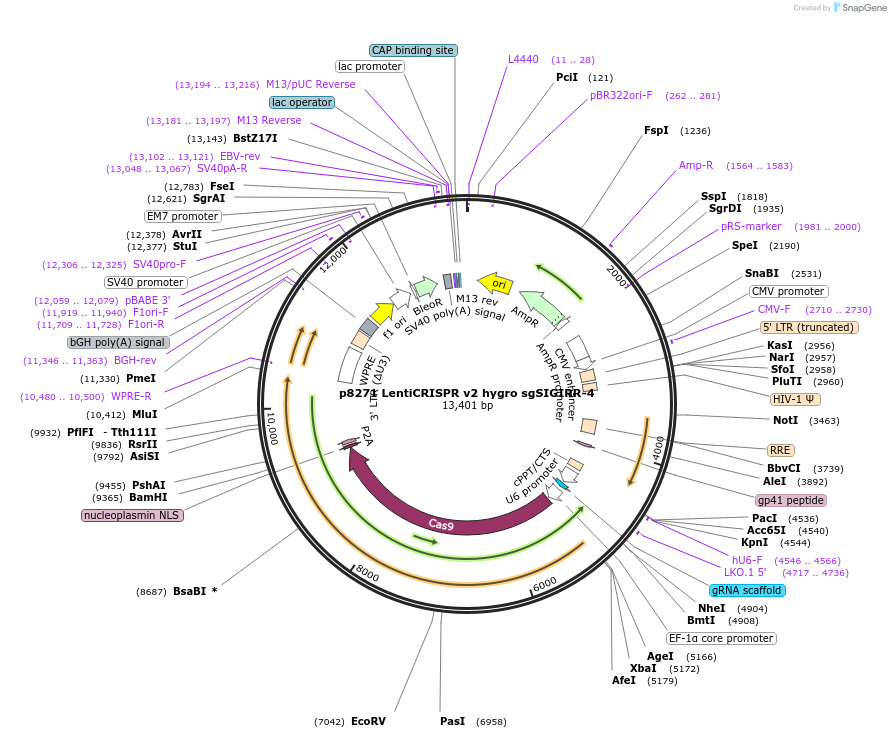
p8271 LentiCRISPR v2 hygro sgSIGIRR-4
Plasmid#193980PurposeExpression of spCas9 and sgRNA targeting SIGIRRDepositorInsertspCas9 and sgRNA targeting SIGIRR (SIGIRR Human)
UseLentiviralAvailable SinceJan. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
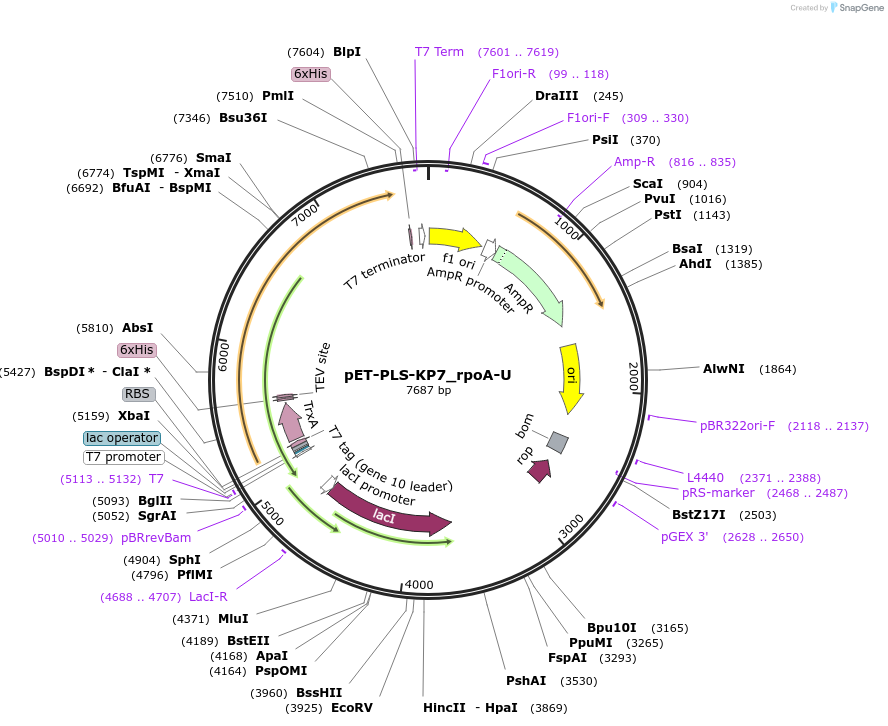
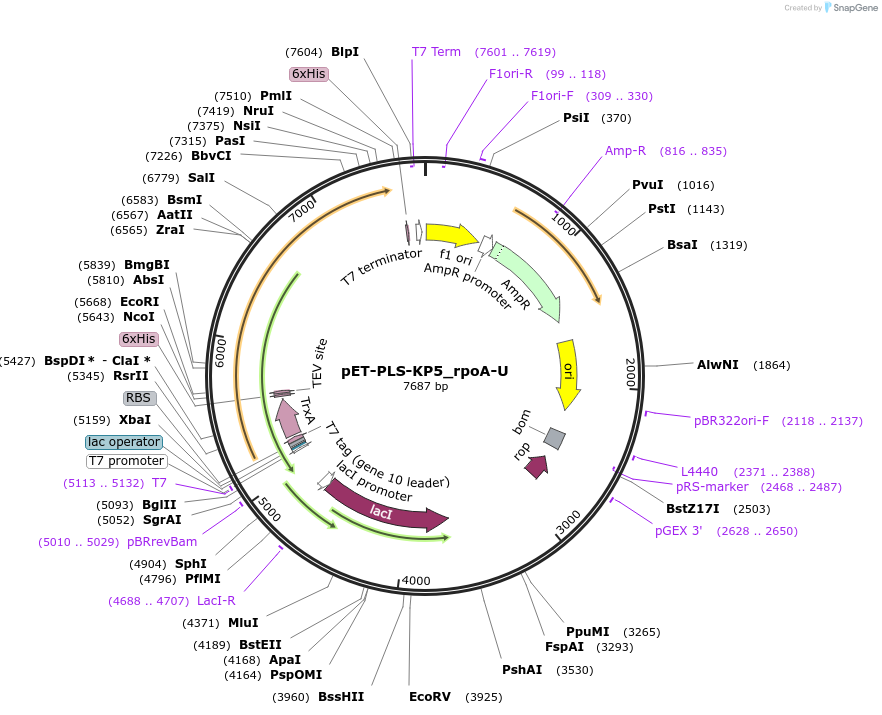
pET-PLS-KP7_rpoA-U
Plasmid#190974PurposeTest the U-to-C editing activity of KP7 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP7
TagsHisExpressionBacterialPromoterT7Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
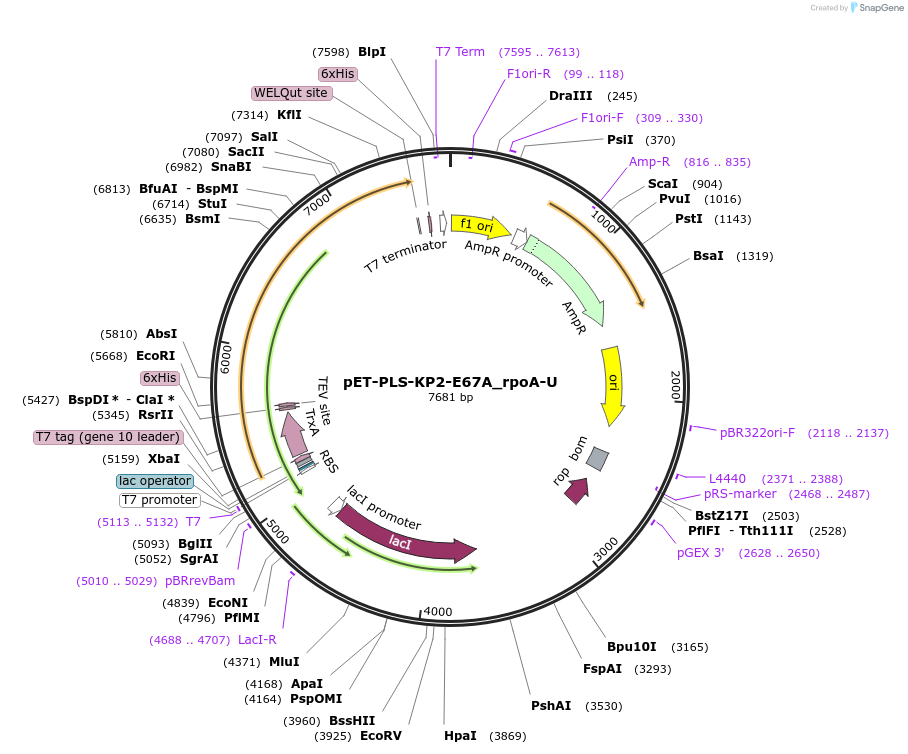
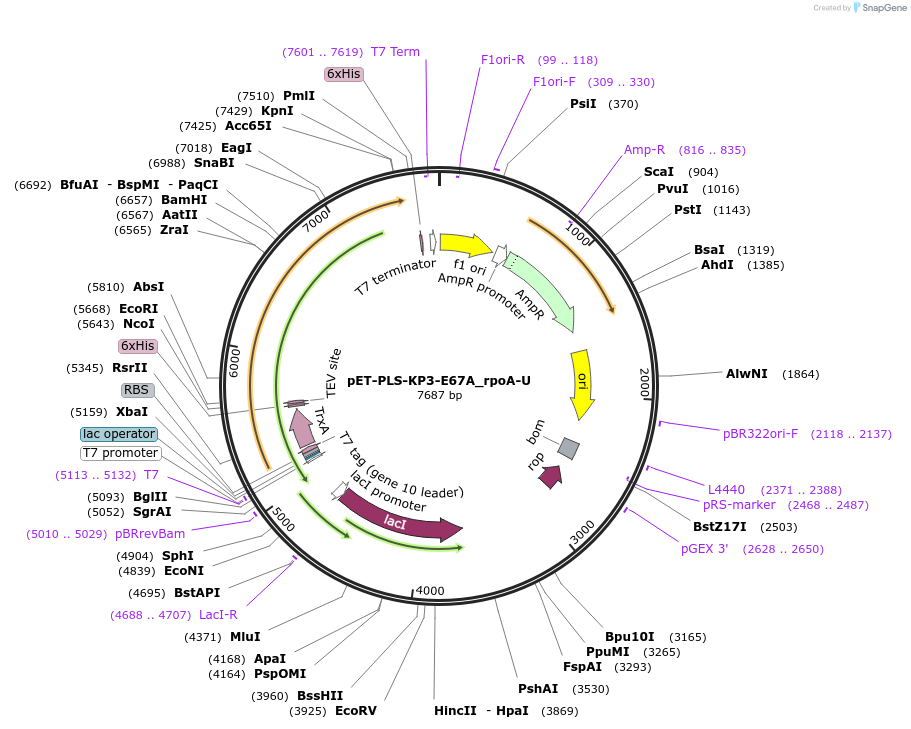
pET-PLS-KP2-E67A_rpoA-U
Plasmid#190959PurposeTest the U-to-C editing activity of KP2:E67A on AtrpoA editing site, in E. coliDepositorInsertPLS-KP2
TagsHisExpressionBacterialMutationDYW-KP:E67APromoterT7Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
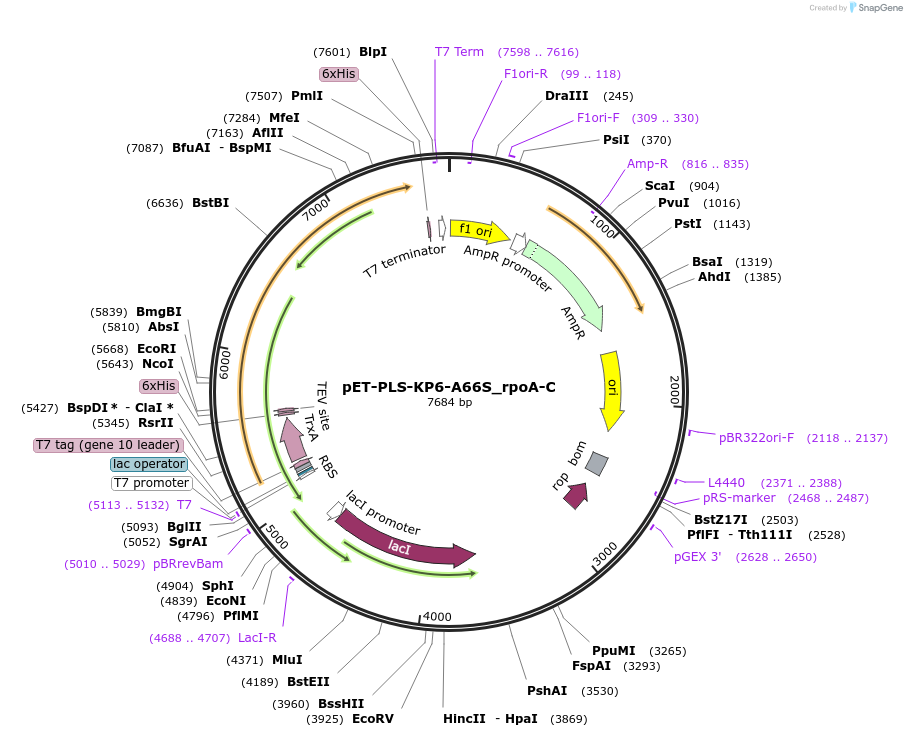
pET-PLS-KP6-A66S_rpoA-C
Plasmid#190968PurposeTest the C-to-U editing activity of KP6:A66S on AtrpoA editing site, in E. coliDepositorInsertPLS-KP6
TagsHisExpressionBacterialMutationDYW-KP:A66SPromoterT7Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
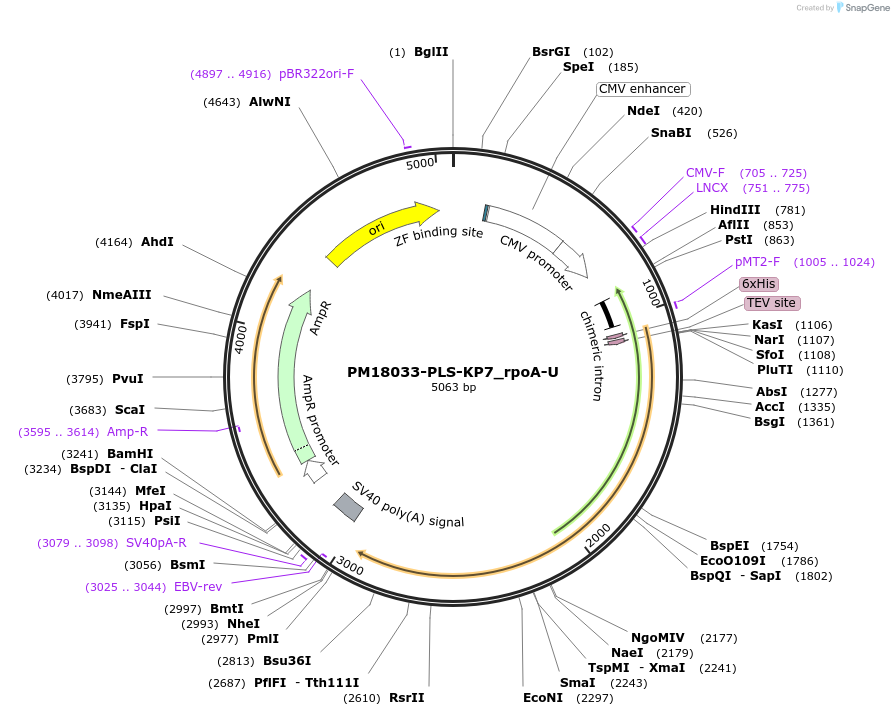
PM18033-PLS-KP5_rpoA-U
Plasmid#190985PurposeTest the U-to-C editing activity of KP5 on AtrpoA editing site, in human cellsDepositorInsertPLS-KP5
TagsHisExpressionMammalianPromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
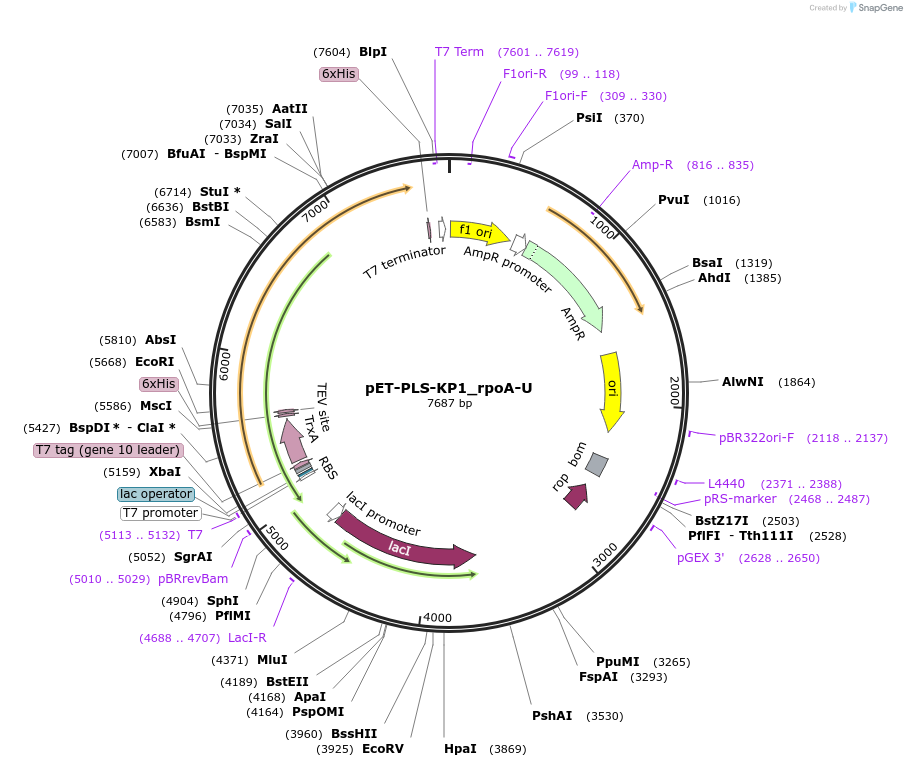
pET-PLS-KP1_rpoA-U
Plasmid#190955PurposeTest the U-to-C editing activity of KP1 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP1
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
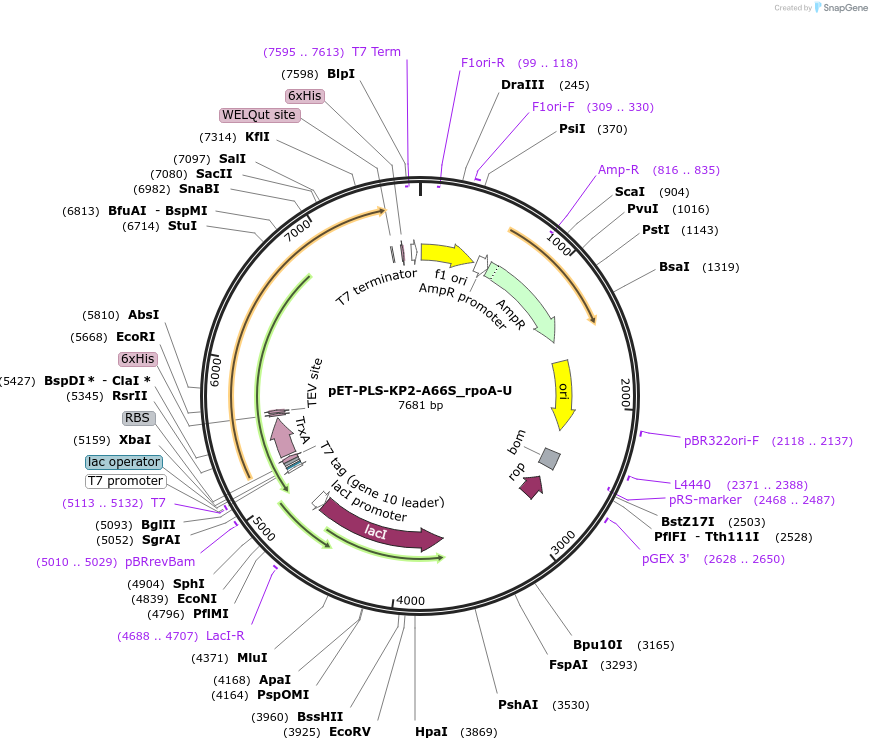
pET-PLS-KP2-A66S_rpoA-U
Plasmid#190957PurposeTest the U-to-C editing activity of KP2:A66S on AtrpoA editing site, in E. coliDepositorInsertPLS-KP2
TagsHisExpressionBacterialMutationDYW-KP:A66SPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
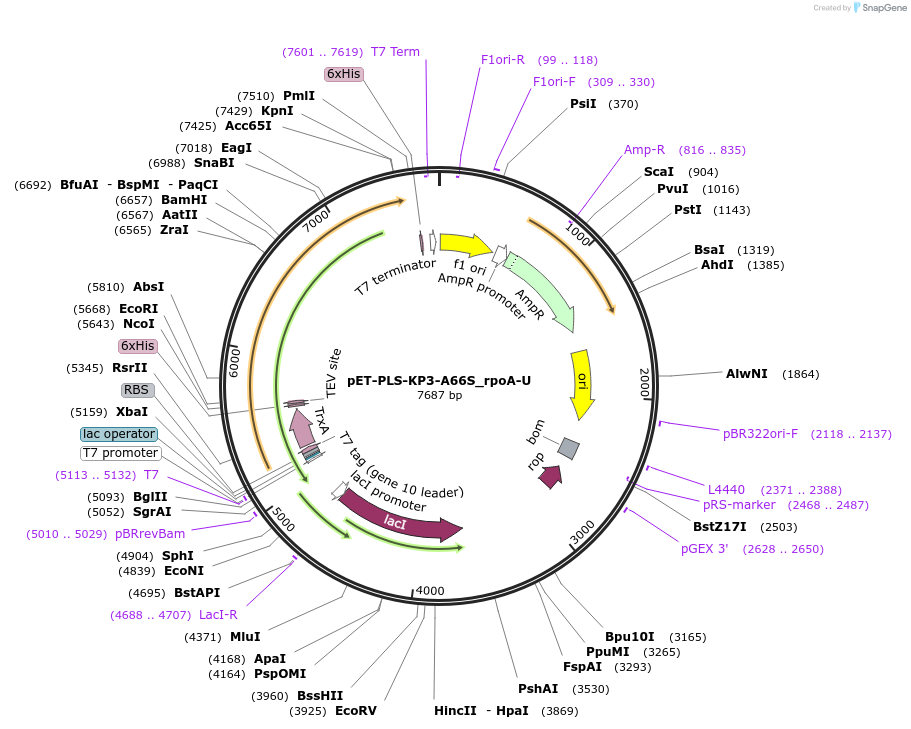
pET-PLS-KP3-A66S_rpoA-U
Plasmid#190961PurposeTest the U-to-C editing activity of KP3:A66S on AtrpoA editing site, in E. coliDepositorInsertPLS-KP3
TagsHisExpressionBacterialMutationDYW-KP:A66SPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
PM18033-PLS-KP7_rpoA-U
Plasmid#190994PurposeTest the U-to-C editing activity of KP7 on AtrpoA editing site, in human cellsDepositorInsertPLS-KP7
TagsHisExpressionMammalianPromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-PLS-KP3-E67A_rpoA-U
Plasmid#190963PurposeTest the U-to-C editing activity of KP3:E67A on AtrpoA editing site, in E. coliDepositorInsertPLS-KP3
TagsHisExpressionBacterialMutationDYW-KP:E67APromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
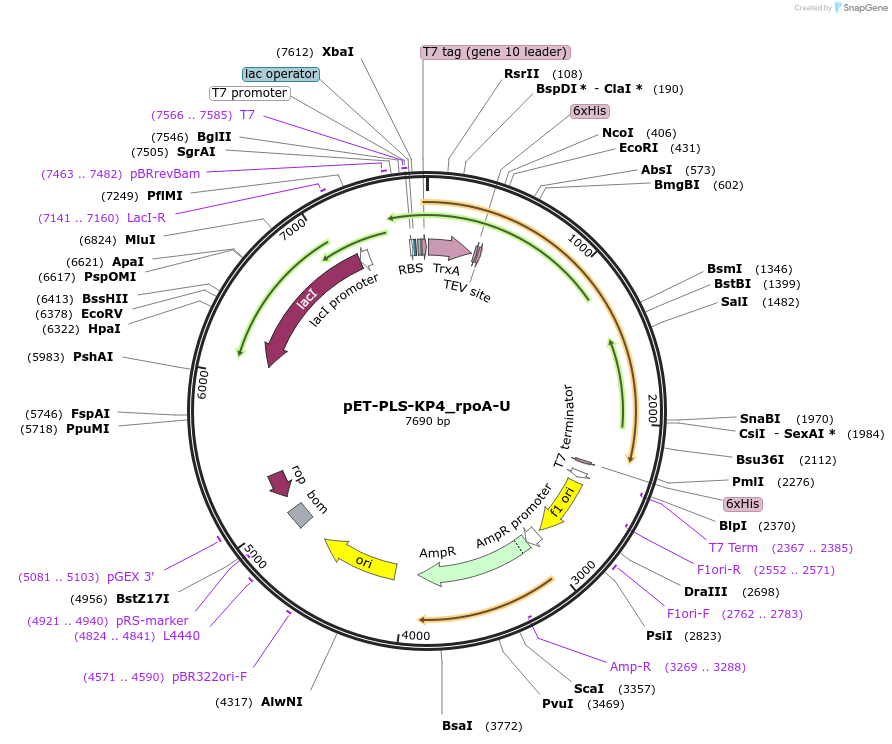
pET-PLS-KP4_rpoA-U
Plasmid#190964PurposeTest the U-to-C editing activity of KP4 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP4
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
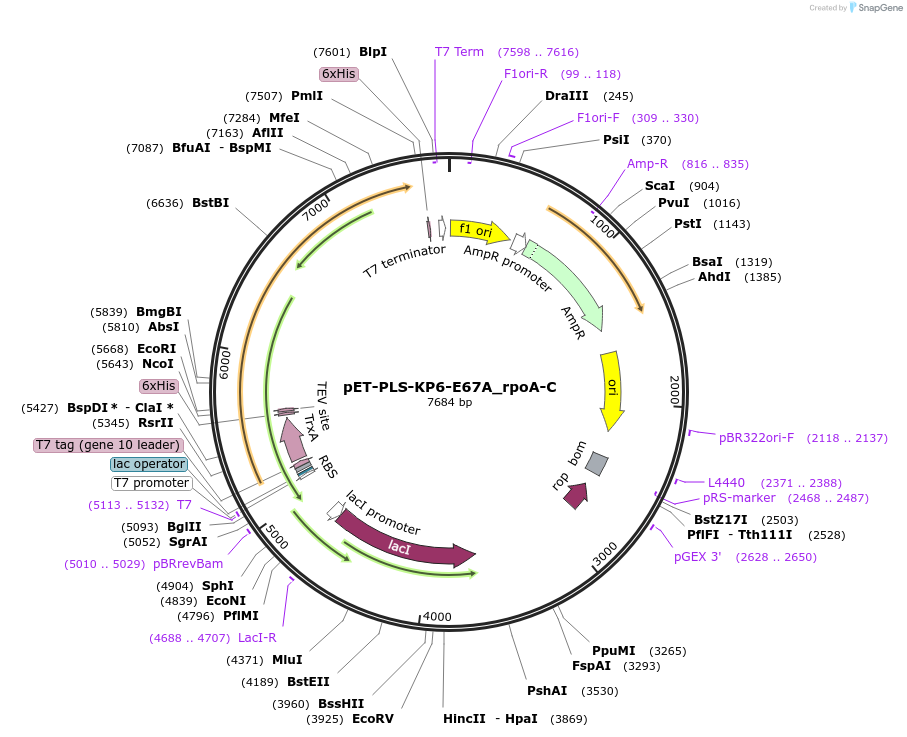
pET-PLS-KP6-E67A_rpoA-C
Plasmid#190972PurposeTest the C-to-U editing activity of KP6:E67A on AtrpoA editing site, in E. coliDepositorInsertPLS-KP6
TagsHisExpressionBacterialMutationDYW-KP:E67APromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
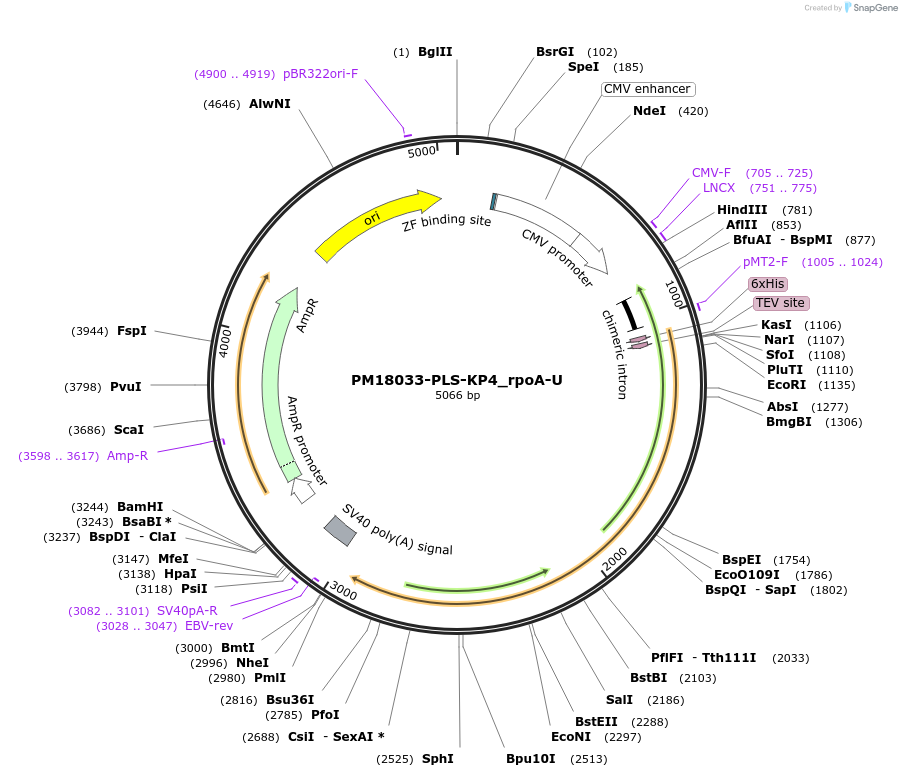
PM18033-PLS-KP4_rpoA-U
Plasmid#190984PurposeTest the U-to-C editing activity of KP4 on AtrpoA editing site, in human cellsDepositorInsertPLS-KP4
TagsHisExpressionMammalianPromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-PLS-KP5_rpoA-U
Plasmid#190965PurposeTest the U-to-C editing activity of KP5 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP5
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
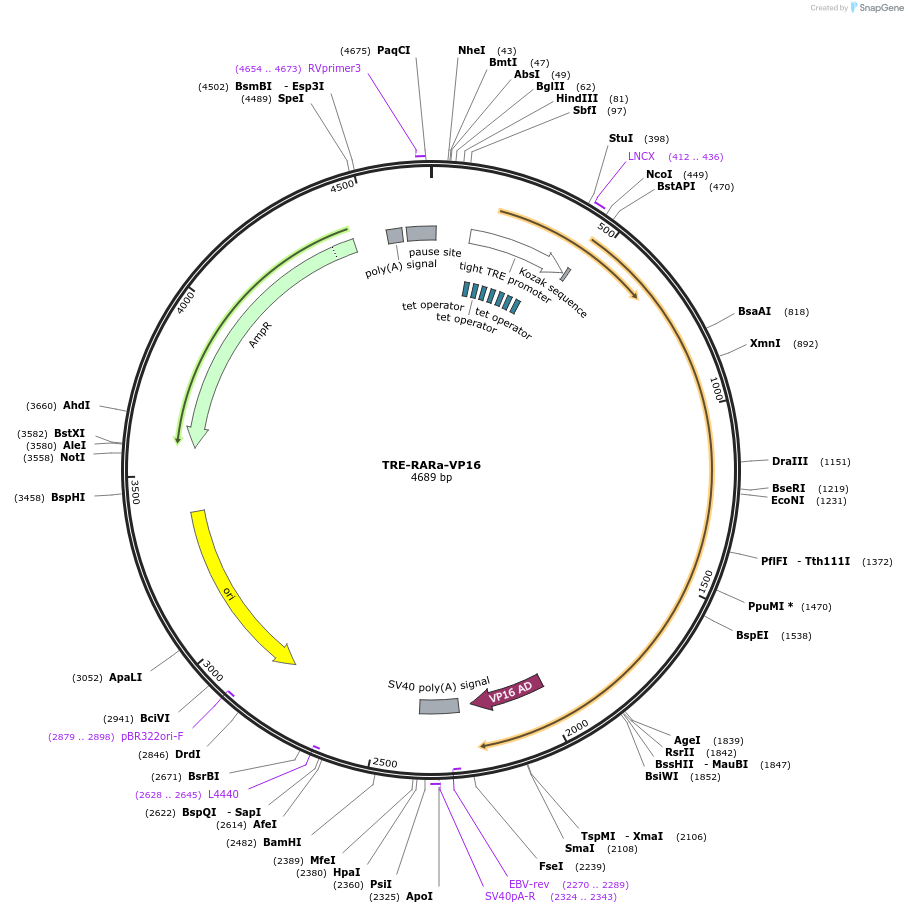
TRE-RARa-VP16
Plasmid#185561PurposeDoxycycline inducible expression of constitutively active Retinoic Acid Receptor alphaDepositorAvailable SinceJune 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
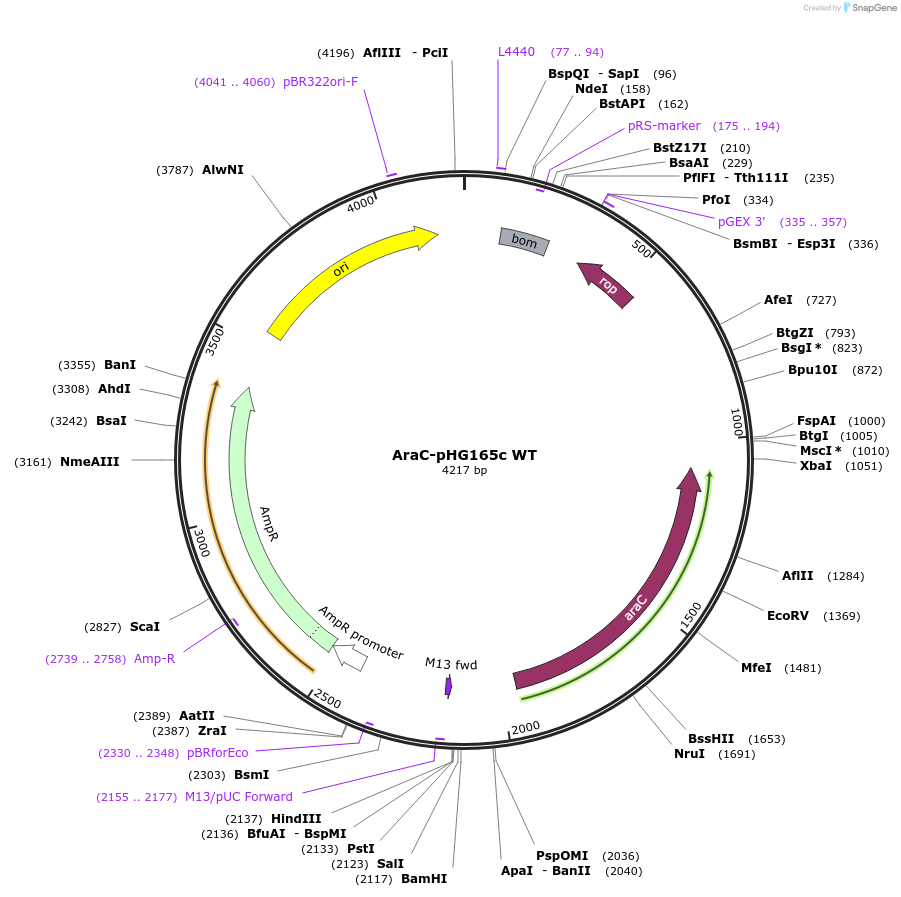
AraC-pHG165c WT
Plasmid#170114PurposeConstitutively expressed, wild-type E coli AraC transcription activator; activated by L-Arabinose.DepositorInsertAraC
ExpressionBacterialAvailable SinceAug. 17, 2021AvailabilityAcademic Institutions and Nonprofits only