We narrowed to 4,001 results for: SPL
-
Plasmid#171899PurposePromoter SPLc19 and RBSc123 in pStA0DepositorInsertPromoter SPLc19 and RBSc123
UseSynthetic BiologyMutationN/AAvailable SinceFeb. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pStA0::SPLc17-RBSc123
Plasmid#171912PurposePromoter SPLc17 and RBSc123 in pStA0DepositorInsertPromoter SPLc17 and RBSc123
UseSynthetic BiologyMutationN/AAvailable SinceFeb. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pStA0::SPLc3-RBSc21
Plasmid#171884PurposePromoter SPLc3 and RBSc21 in pStA0DepositorInsertPromoter SPLc3 and RBSc21
UseSynthetic BiologyMutationN/AAvailable SinceFeb. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
Spike Display_Part 5 Spacer
Plasmid#172733PurposeEncodes Part 5 of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
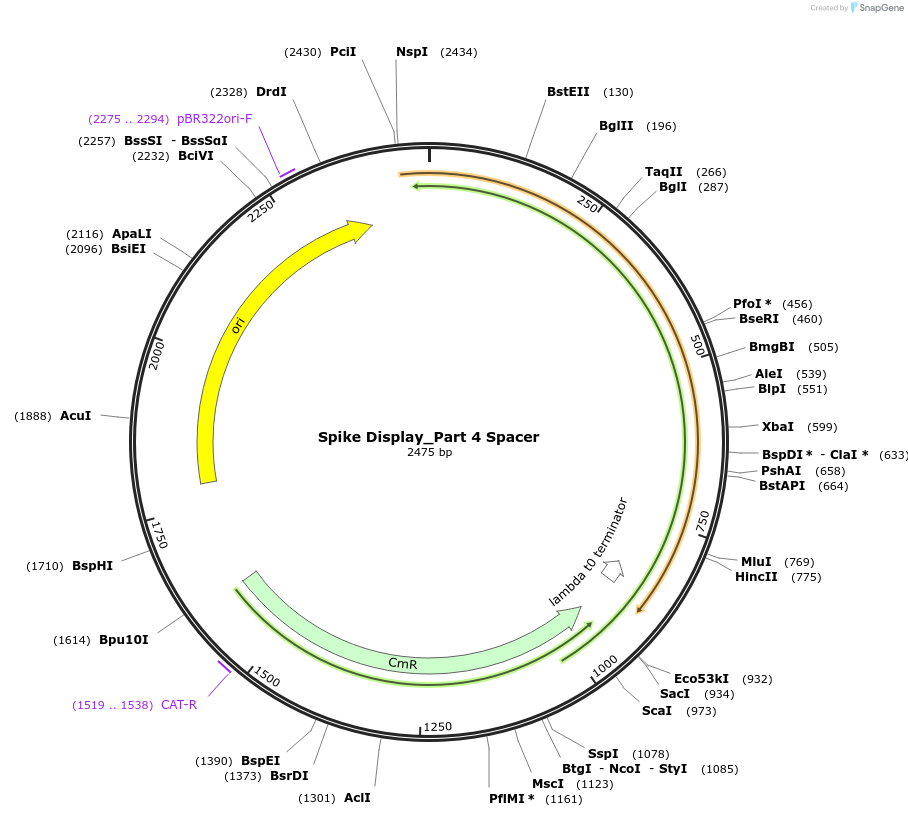
Spike Display_Part 4 Spacer
Plasmid#172732PurposeEncodes Part 4 of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
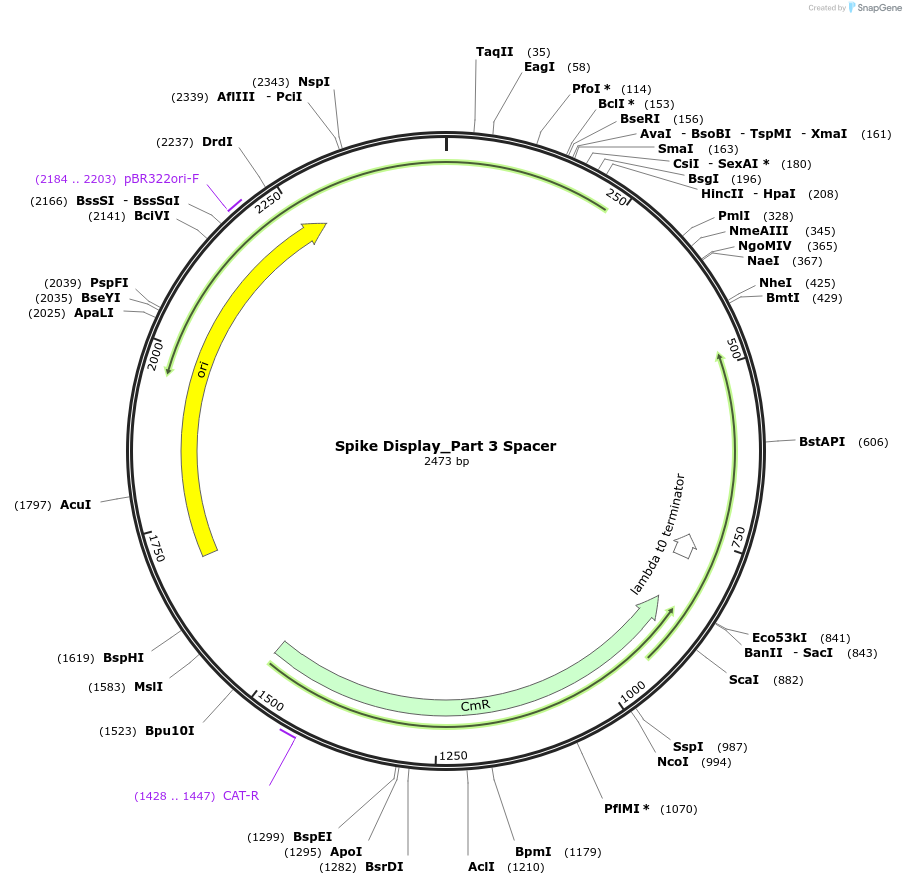
Spike Display_Part 3 Spacer
Plasmid#172731PurposeEncodes Part 3 of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
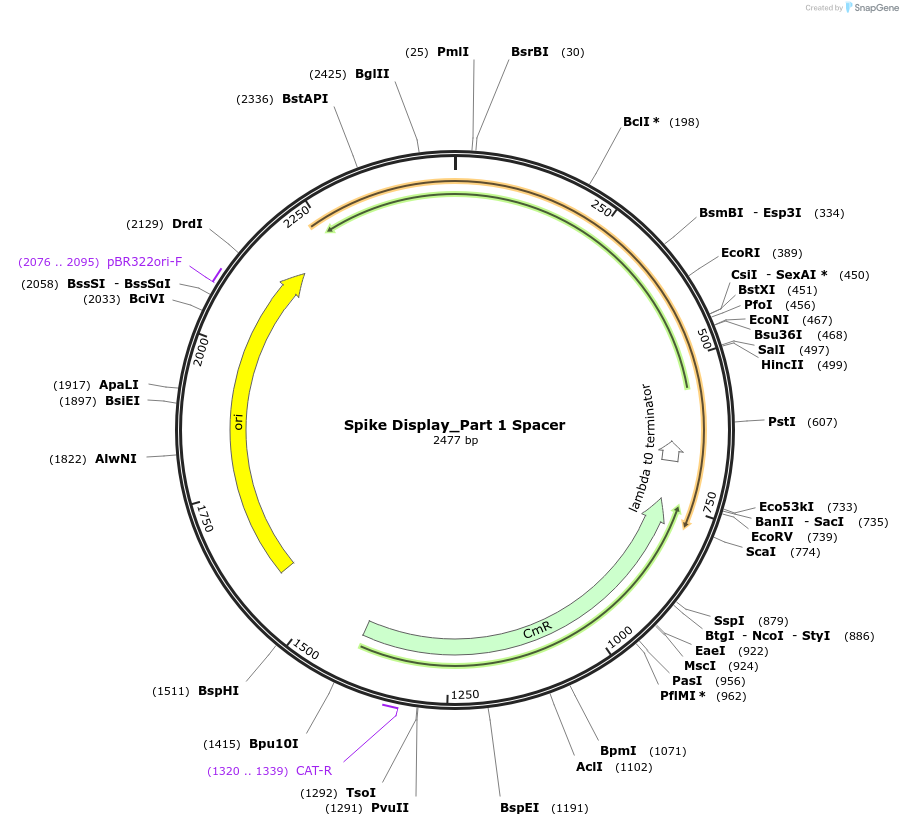
Spike Display_Part 1 Spacer
Plasmid#172729PurposeEncodes Part 1 of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
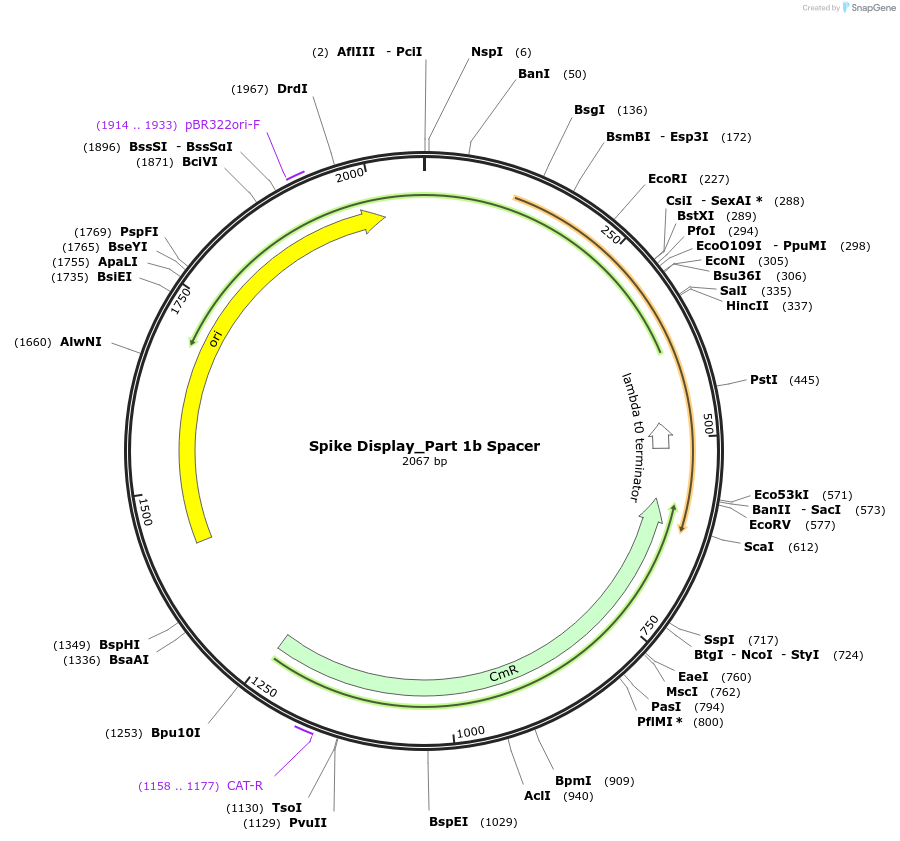
Spike Display_Part 1b Spacer
Plasmid#172728PurposeEncodes Part 1b of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
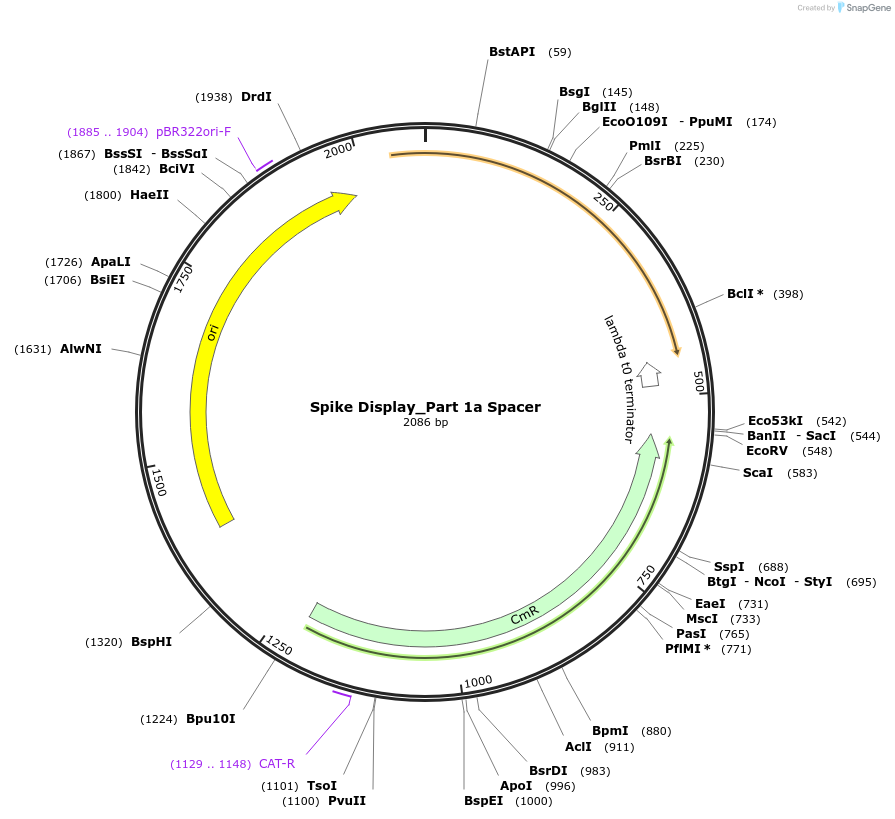
Spike Display_Part 1a Spacer
Plasmid#172727PurposeEncodes Part 1a of HexaPro-D614G spike to be used in the Spike Display systemDepositorInsertSARS-CoV-2 S HexaPro-D614G (S Severe acute respiratory syndrome coronavirus 2)
UseSynthetic BiologyMutationEctodomain only (AAs 1-1208); 682-685 (furin site…Available SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
ESPL1 D7.1 gRNA
Plasmid#90682Purpose3rd generation lentiviral gRNA plasmid targeting human ESPL1DepositorInsertESPL1 (Guide Designation D7.1)
UseCRISPR and LentiviralPromoterU6Available SinceJuly 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
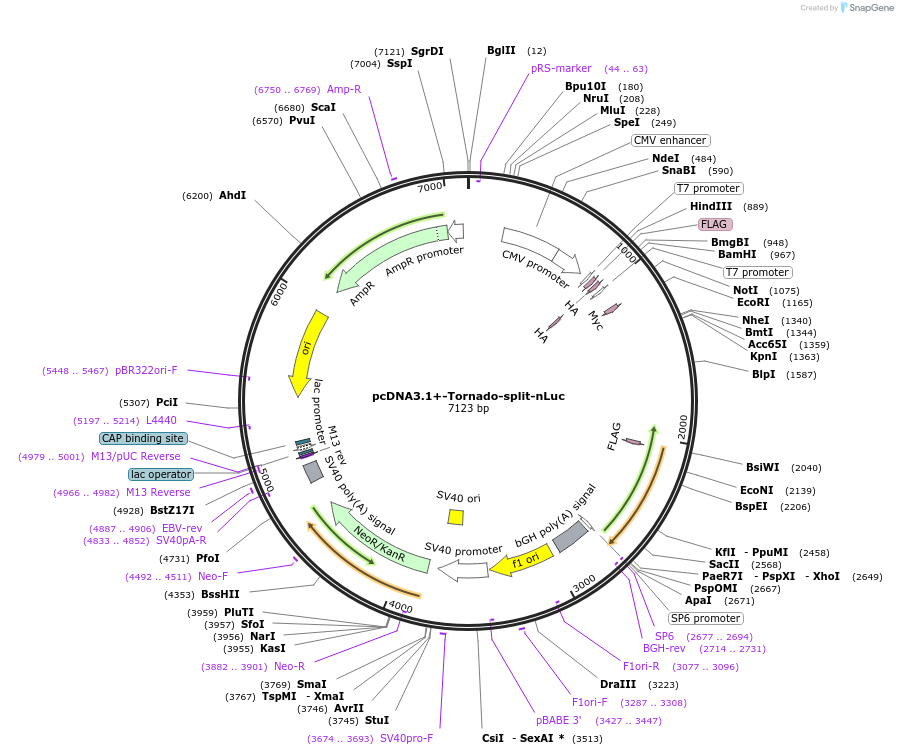
pcDNA3.1+-Tornado-split-nLuc
Plasmid#212612PurposeFor the expression of a split nLuc that produces luminescence only when circularized. To test your IRES of interest, clone into the EcoRI, BsiWI sites.DepositorInsertTornado-CVB3-split-nLuc
UseLuciferaseExpressionMammalianMutationC373T mutation in CVB3PromoterCMVAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
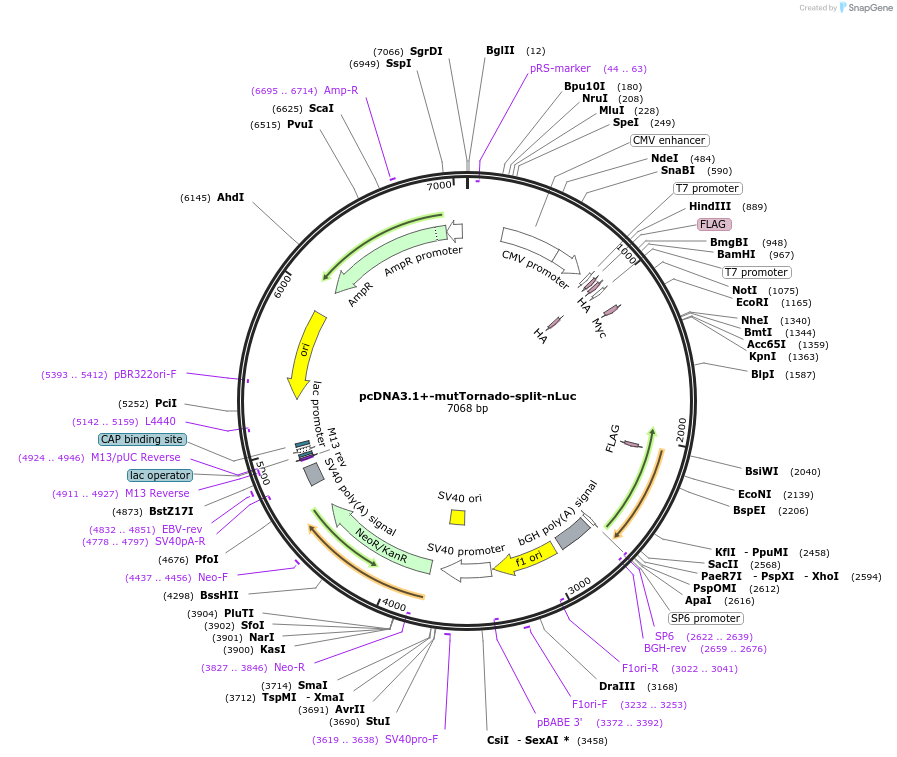
pcDNA3.1+-mutTornado-split-nLuc
Plasmid#212707PurposeTornado split nLuc with a mutated 3' ribozyme so that circularization cannot occur.DepositorInsertmutTornado-CVB3-split-nLuc
MutationDeleted the partial 3'ribozyme sequence in T…Available SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
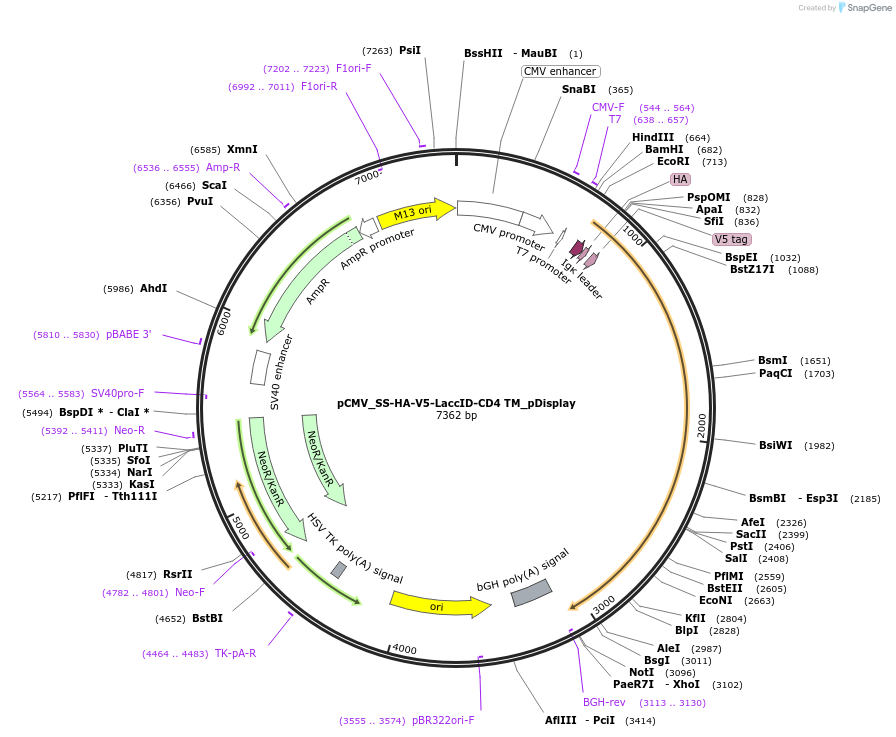
pCMV_SS-HA-V5-LaccID-CD4 TM_pDisplay
Plasmid#234046Purposeexpresses LaccID on the mammalian cell surfaceDepositorInsertLaccID-CD4TM
ExpressionMammalianPromoterCMVAvailable SinceAug. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
Split 'n coexpress CUL3-RBX1
Plasmid#52293Purposebacterial co-expression of N-terminal Cul3, C-terminal Cul3, and Rbx1DepositorTagsGST, IRES, and TEVExpressionBacterialMutationcontains aa 1-309, contains aa 310-768, and conta…PromoterIRES and tacAvailable SinceMarch 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
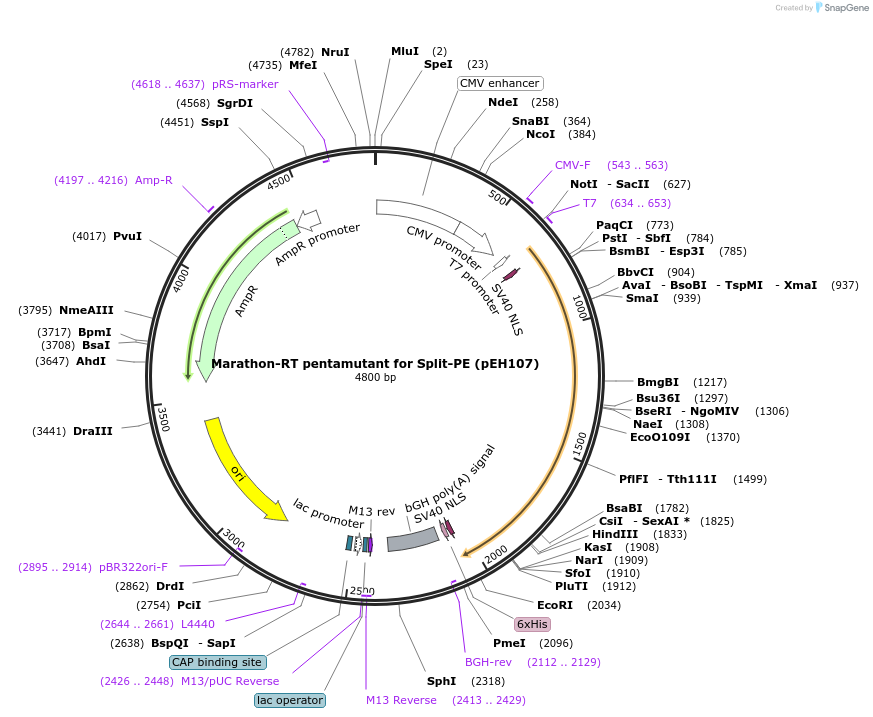
Marathon-RT pentamutant for Split-PE (pEH107)
Plasmid#190110PurposeExpresses Marathon-RT pentamutant (D14R-N26R-D74R-N116K-N197R)DepositorInsertMarathon-RT
TagsbpNLSExpressionMammalianMutationD14R, N26R, D74R, N116K, N197RPromoterCMVAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
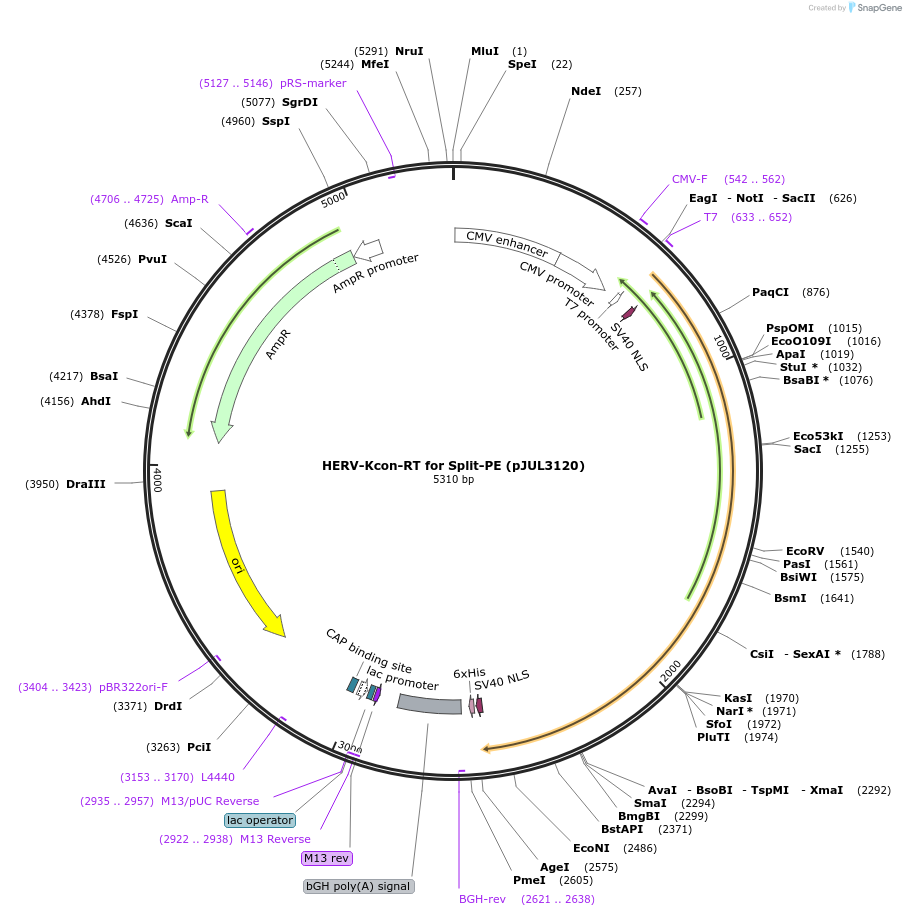
HERV-Kcon-RT for Split-PE (pJUL3120)
Plasmid#190107PurposeExpresses HERV-Kcon RT (human codon optimization)DepositorInsertHERV-Kcon-RT
TagsbpNLSExpressionMammalianPromoterCMVAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
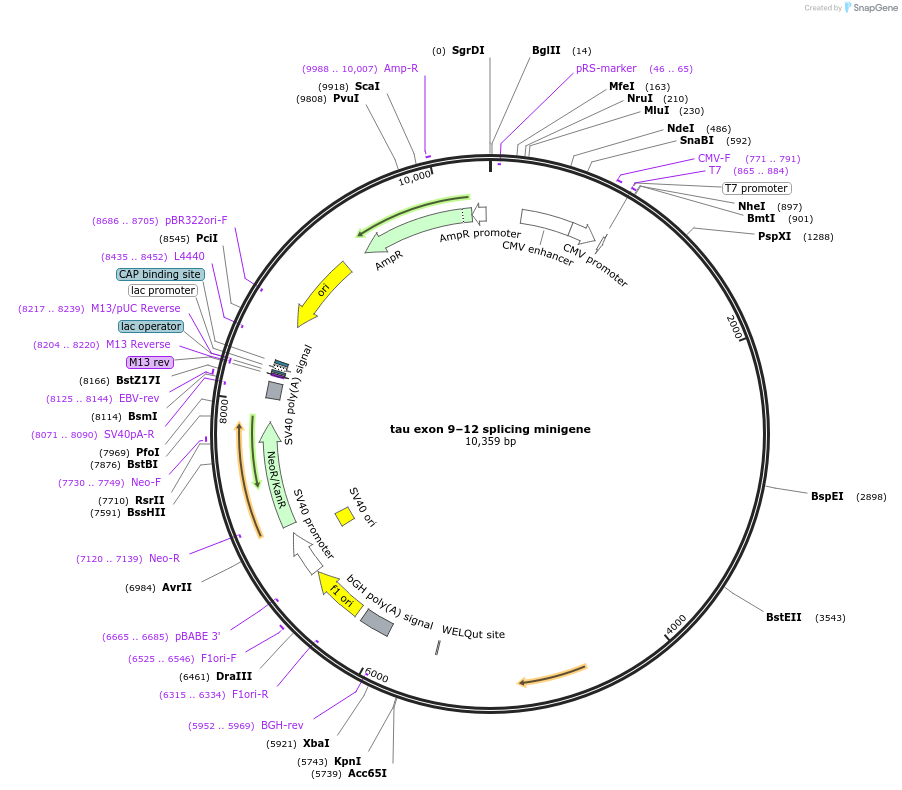
tau exon 9–12 splicing minigene
Plasmid#121165PurposeGenerates circular RNAs from a Tau minigeneDepositorAvailable SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
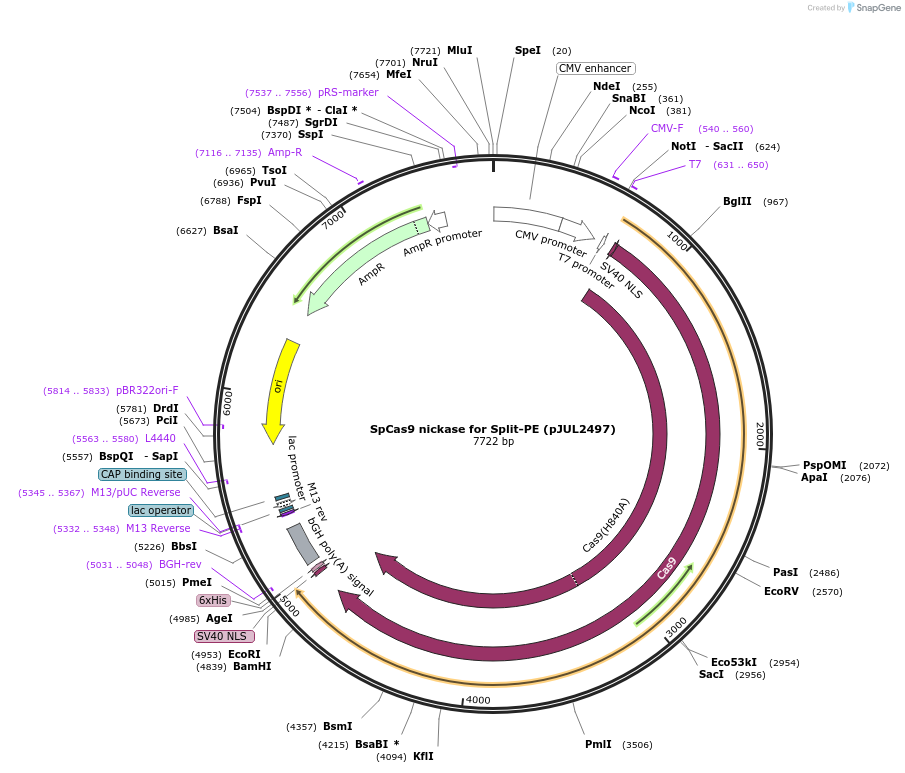
SpCas9 nickase for Split-PE (pJUL2497)
Plasmid#190104PurposeExpresses nickase SpCas9(H840A) for Split-PE useDepositorInsertnSpCas9
TagsXTEN-bpNLS and bpNLSExpressionMammalianMutationH840APromoterCMVAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
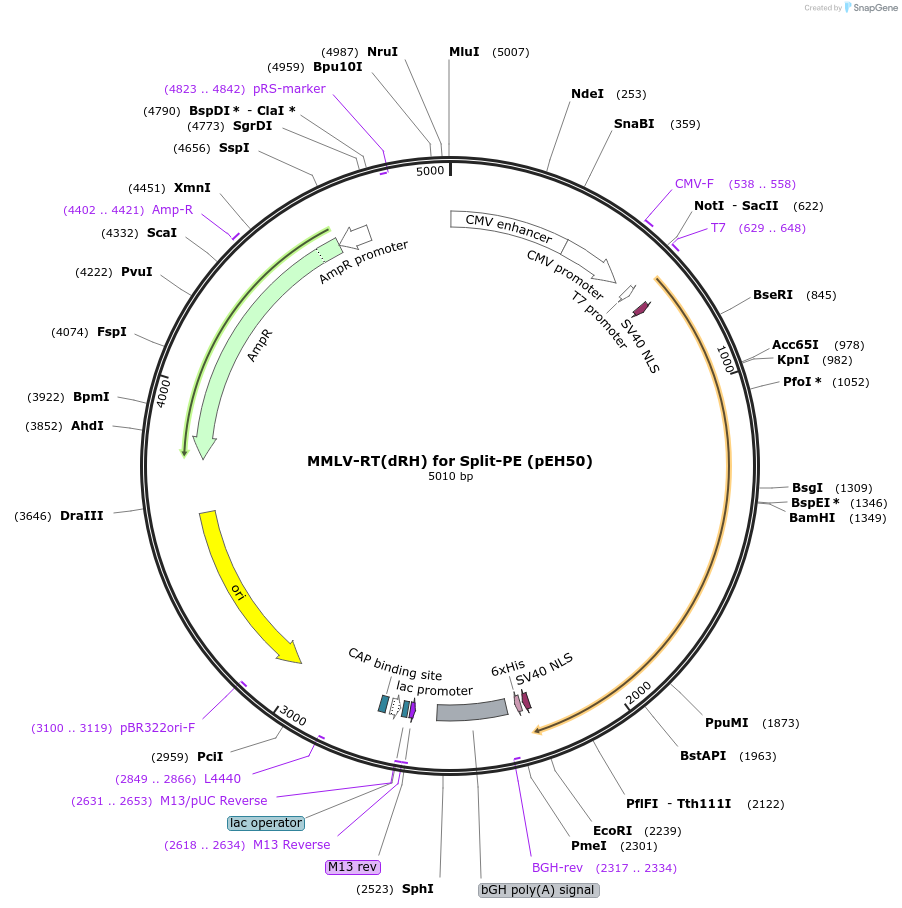
MMLV-RT(dRH) for Split-PE (pEH50)
Plasmid#190106PurposeExpresses MMLV-RT (from PE2) with truncated RNAse H domain (dRH) for Split-PE useDepositorInsertMMLV-RT(delta RNAseH)
TagsbpNLSExpressionMammalianMutationmutations from RT in PE2 and truncation of RNAse …PromoterCMVAvailable SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
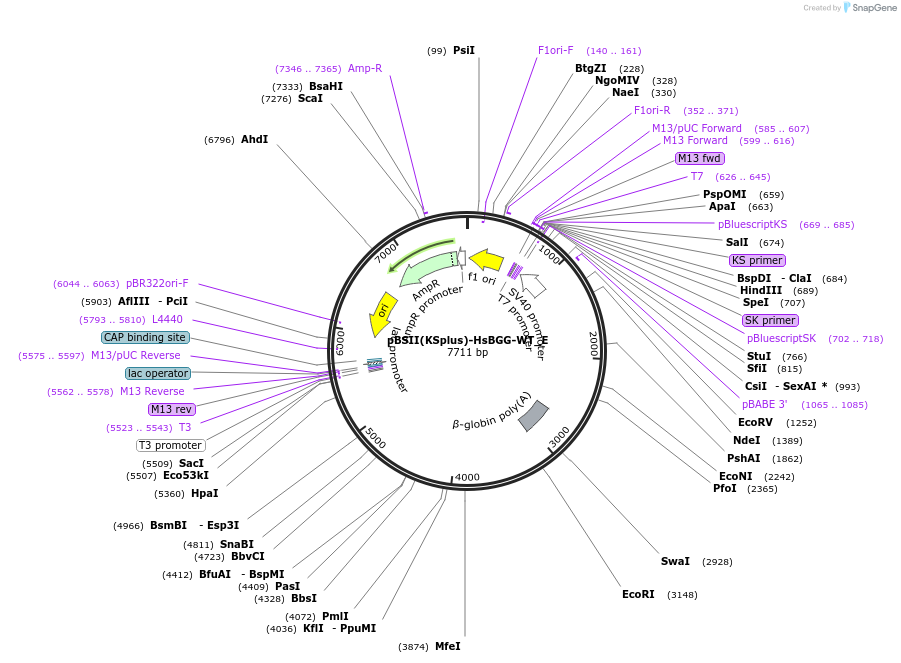
pBSII(KSplus)-HsBGG-WT_E
Plasmid#146187PurposeBacterial Expression of HsBGG-WTDepositorInsertHsBGG-WT
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only