We narrowed to 11,193 results for: ENA
-
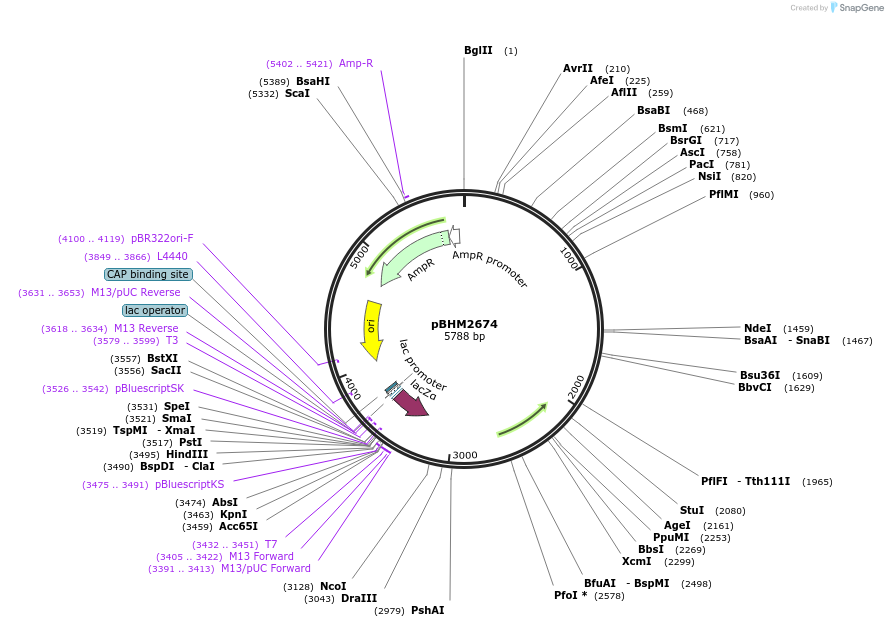
Plasmid#241727PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains BSR marker for selection on blasticidin SDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only
-
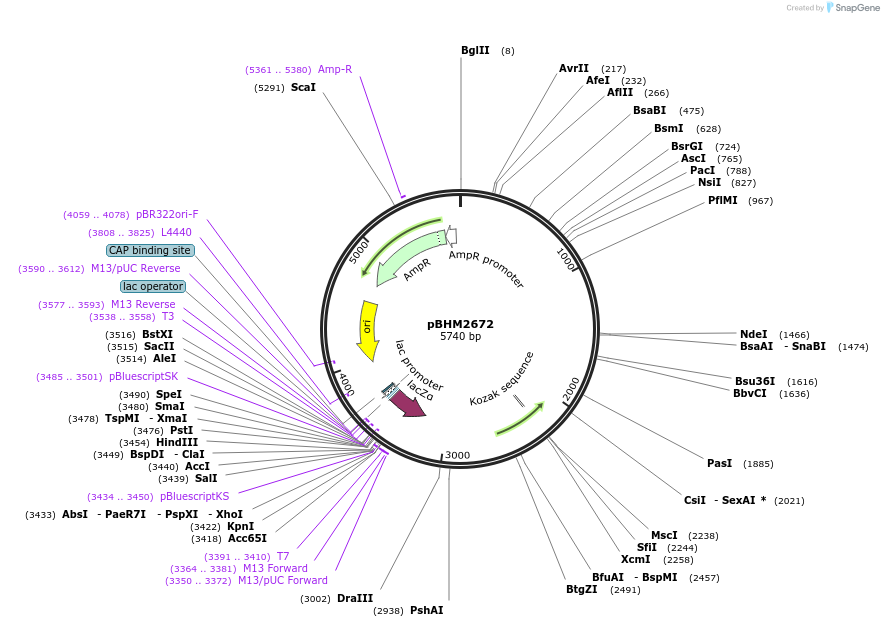
pBHM2672
Plasmid#241719PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains BLE marker for selection on phleomycinDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
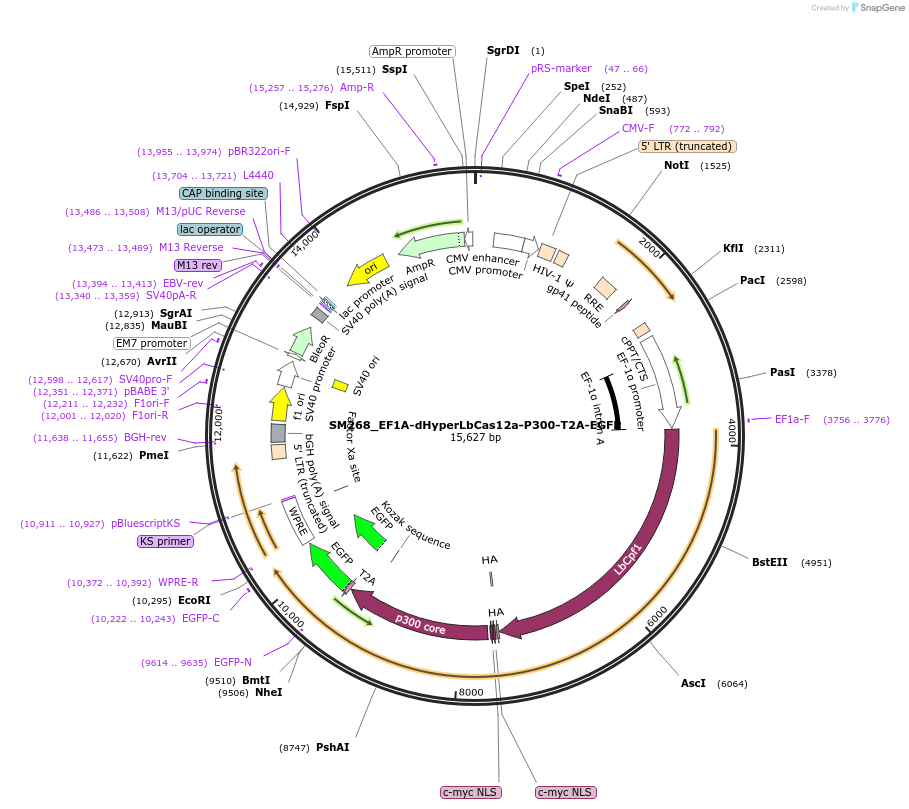
SM268_EF1A-dHyperLbCas12a-P300-T2A-EGFP
Plasmid#244206PurposeMammalian expression of dHyperLbCas12a-P300 with EGFP selectable markerDepositorInsertdHyperLbCas12a-P300
UseCRISPR and LentiviralExpressionMammalianMutationdHyperLbCas12a: D156R, D235R, E292R, E350R, and D…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
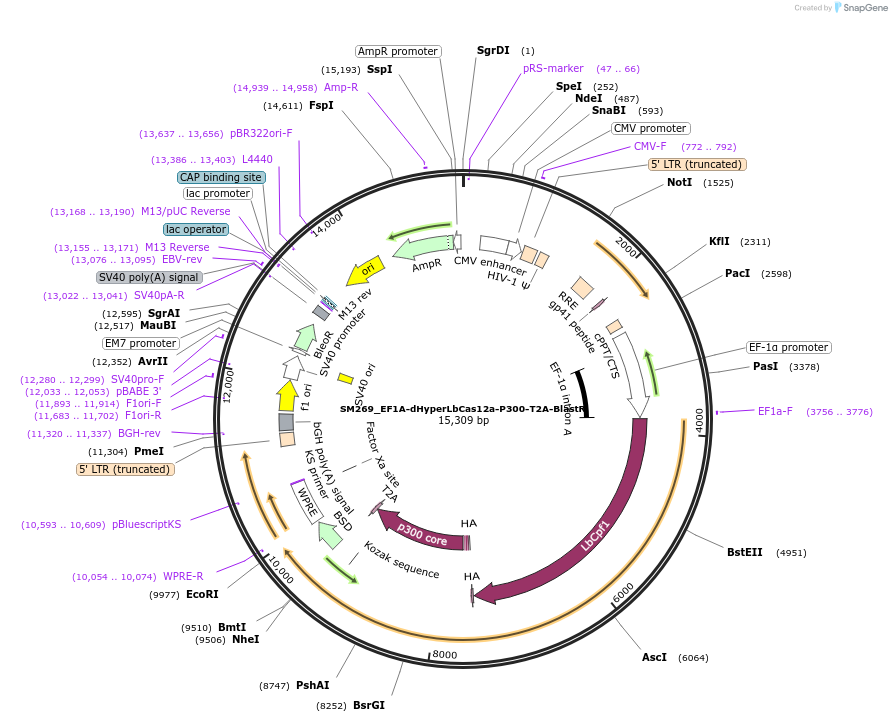
SM269_EF1A-dHyperLbCas12a-P300-T2A-BlastR
Plasmid#244207PurposeMammalian expression of dHyperLbCas12a-P300 with Blasticidin resistance selectable markerDepositorInsertdHyperLbCas12a-P300
UseCRISPR and LentiviralExpressionMammalianMutationdHyperLbCas12a: D156R, D235R, E292R, E350R, and D…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
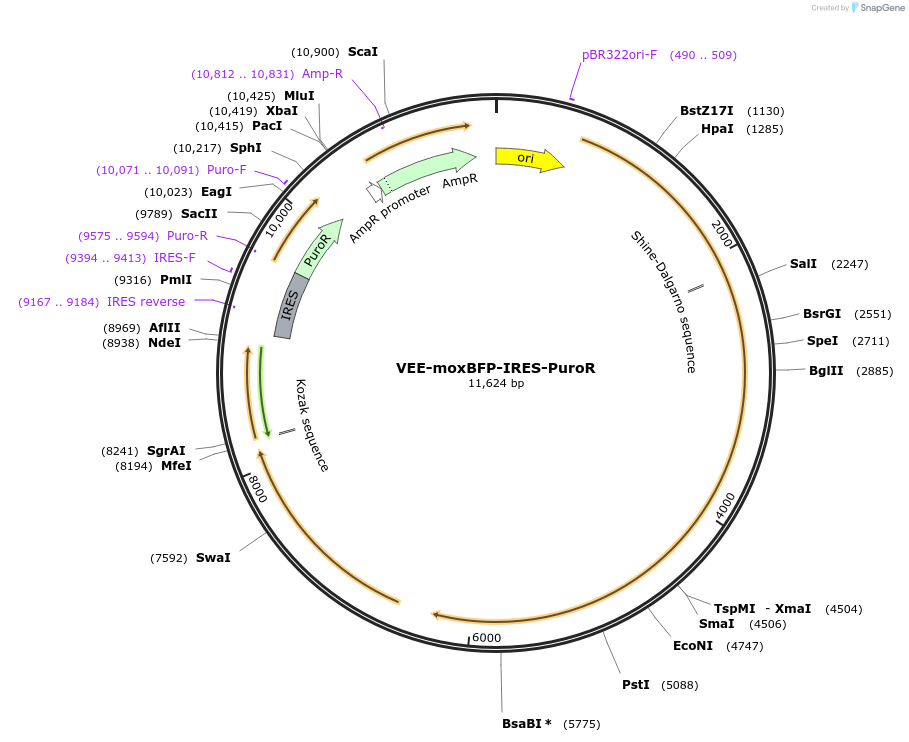
VEE-moxBFP-IRES-PuroR
Plasmid#242413PurposeSpectral unmixing: moxBFP onlyDepositorInsertsmoxBFP
IRES-PuroR
UseSelf-amplifying rnaExpressionMammalianAvailable SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
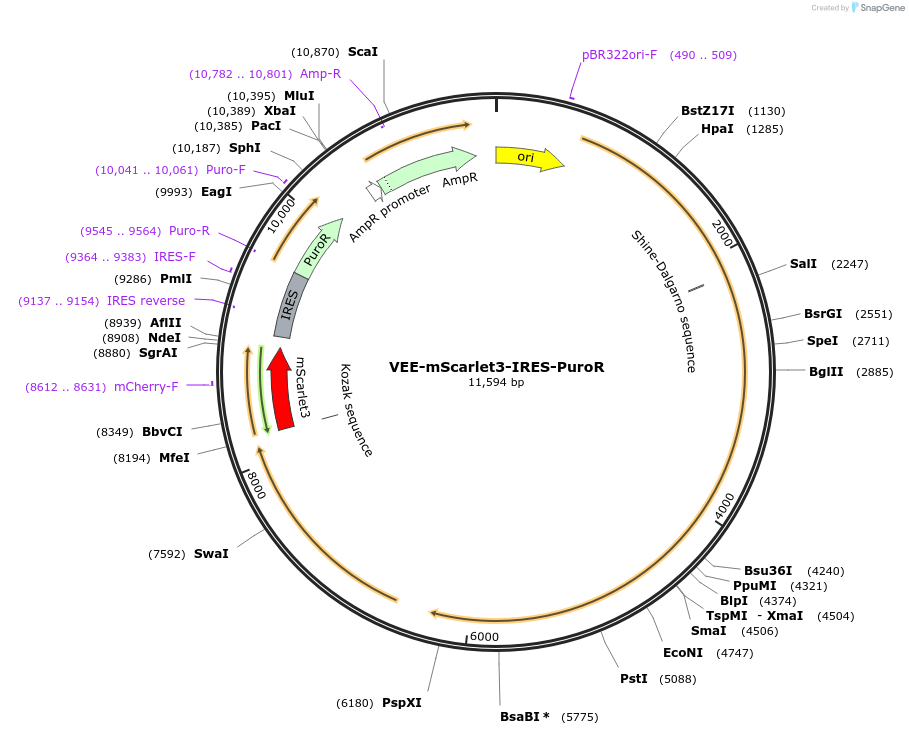
VEE-mScarlet3-IRES-PuroR
Plasmid#242415PurposeSpectral unmixing: mScarlet3 onlyDepositorInsertsmScarlet3
IRES-PuroR
UseSelf-amplifying rnaExpressionMammalianAvailable SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
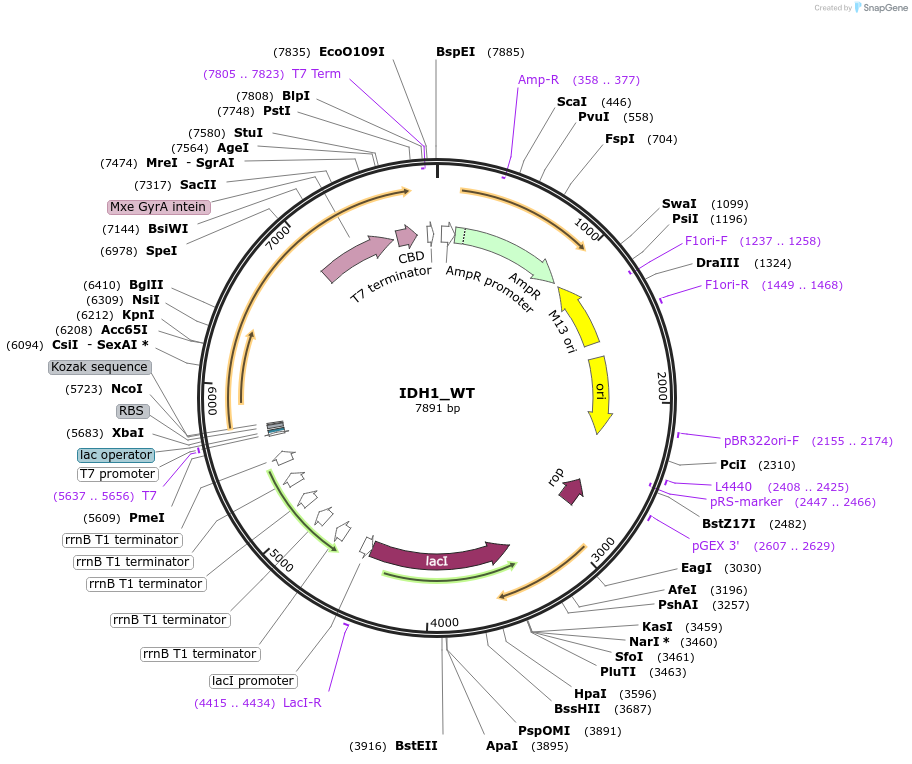
IDH1_WT
Plasmid#224142PurposeIPTG-inducible expression of C. elegans protein IDH-1 with a chitin tagDepositorAvailable SinceAug. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
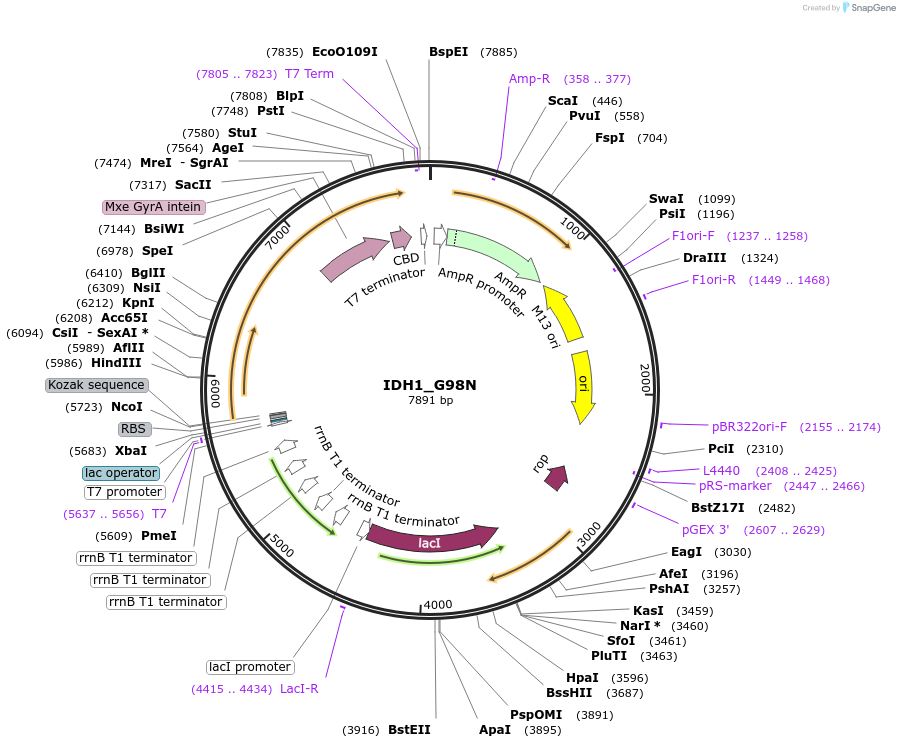
IDH1_G98N
Plasmid#224143PurposeIPTG-inducible expression of C. elegans protein IDH-1(G98N) with a chitin tagDepositorInsertIDH-1(G98N)
Tagsintein-CBDExpressionBacterialMutationG98NAvailable SinceAug. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
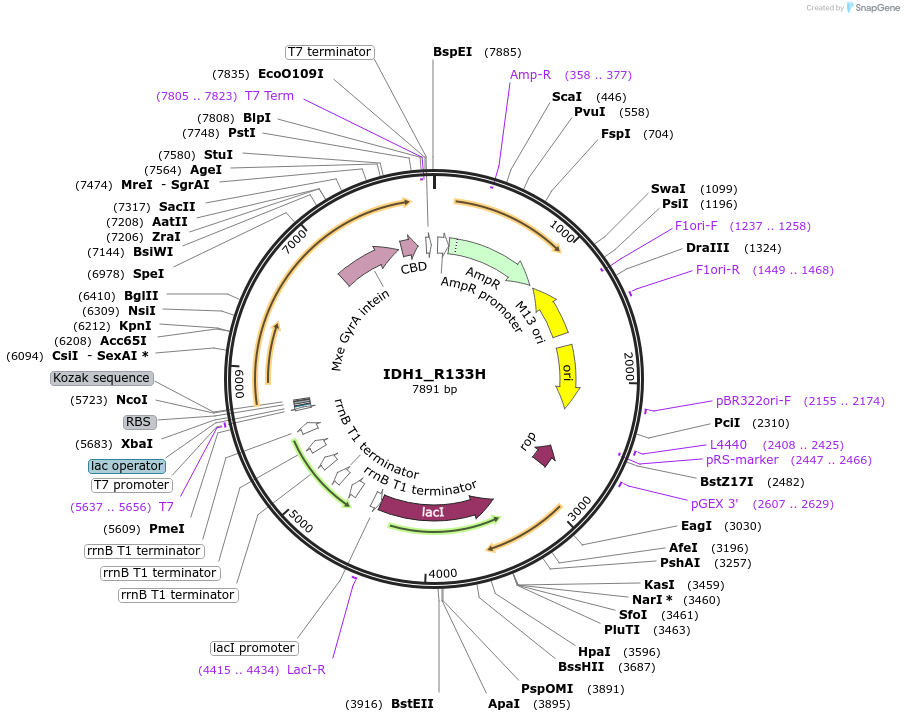
IDH1_R133H
Plasmid#224144PurposeIPTG-inducible expression of C. elegans protein IDH-1(R133H) with a chitin tagDepositorInsertIDH-1(R133H)
Tagsintein-CBDExpressionBacterialMutationR133HAvailable SinceAug. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
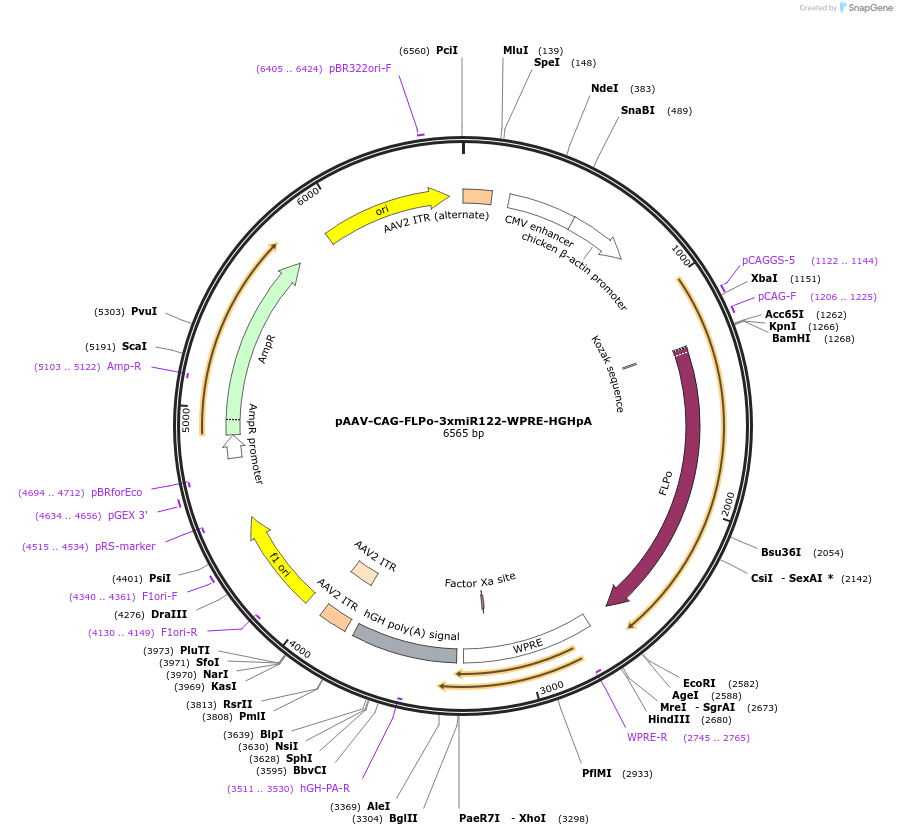
pAAV-CAG-FLPo-3xmiR122-WPRE-HGHpA
Plasmid#220938PurposeFLPo recombinaseDepositorInsertCodon-optimized FLPe recombinase
UseAAVAvailable SinceJuly 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
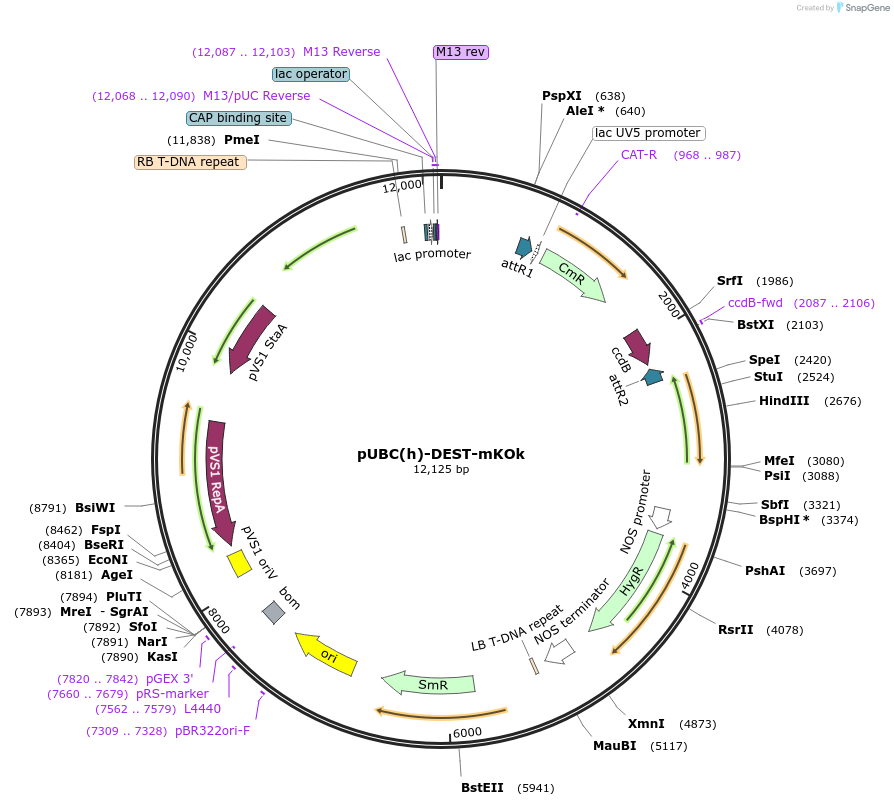
pUBC(h)-DEST-mKOk
Plasmid#224830PurposeEmpty Gateway DEST vector for expressing C-terminal tagged proteins with mKusabiraOrangekand hygromycin selection in plantsDepositorTypeEmpty backboneTagsmKusabiraOrangekExpressionPlantAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
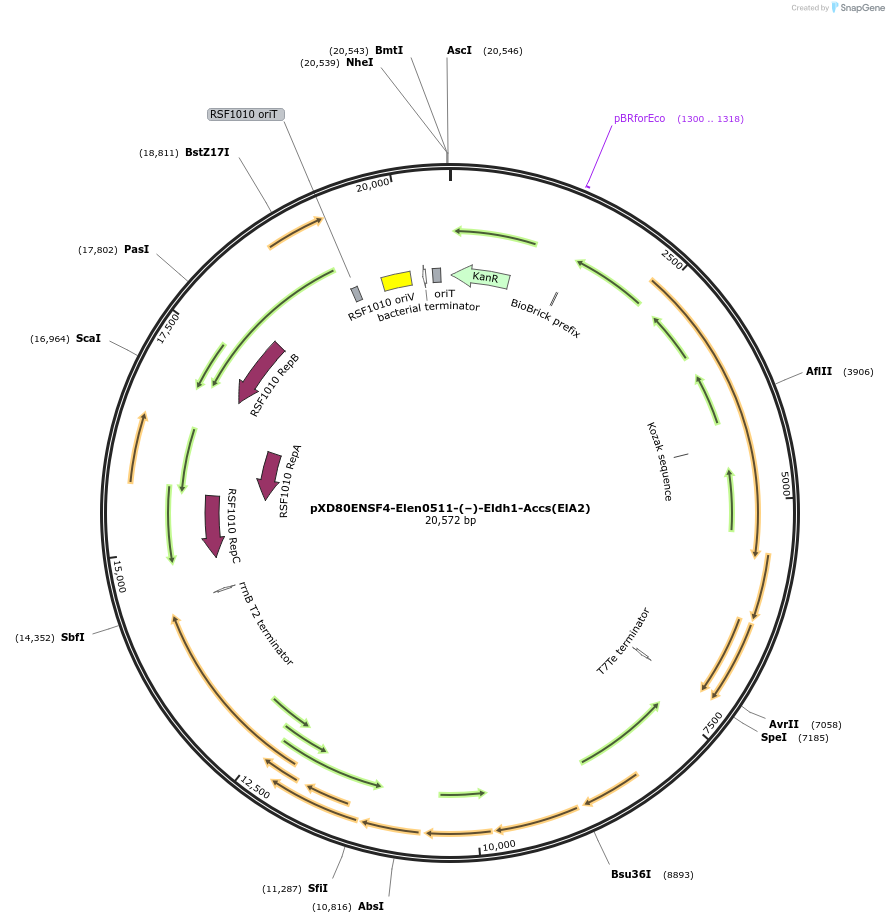
pXD80ENSF4-Elen0511-(–)-Eldh1-Accs(ElA2)
Plasmid#225299PurposeExpresses Eggerthella lenta A2 (–)-Eldh1 in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh1
ExpressionBacterialAvailable SinceMay 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
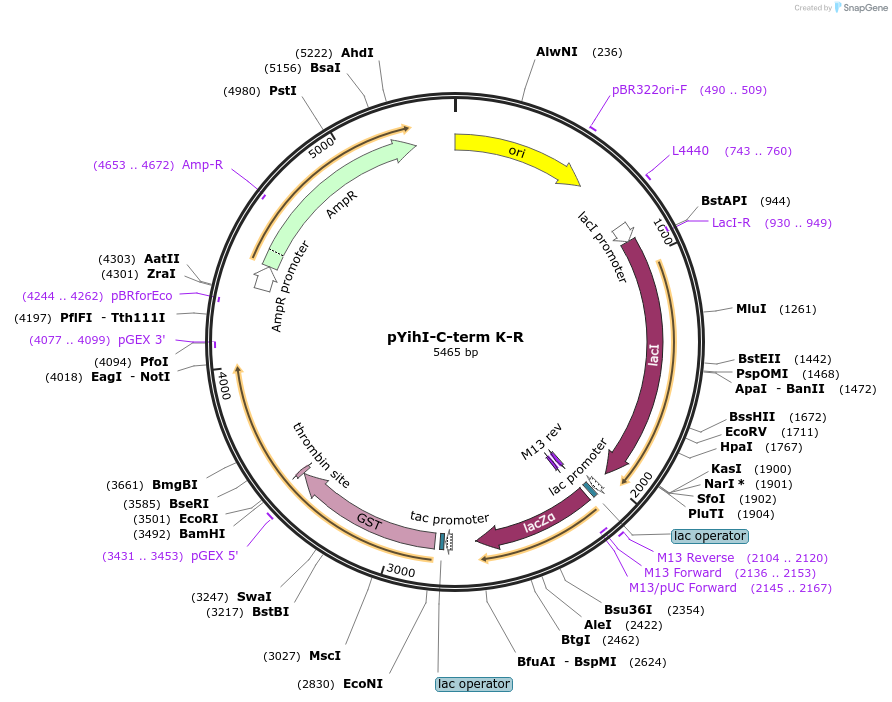
pYihI-C-term K-R
Plasmid#233092PurposeExpression of GST-YihI with C-term lysines (K) mutated to arginine (R)DepositorInsertGST-YihI with C-term lysines (K) mutated to arginine (R)
TagsGSTExpressionBacterialMutationC-term lysines (K) mutated to arginine (R)Available SinceMay 12, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
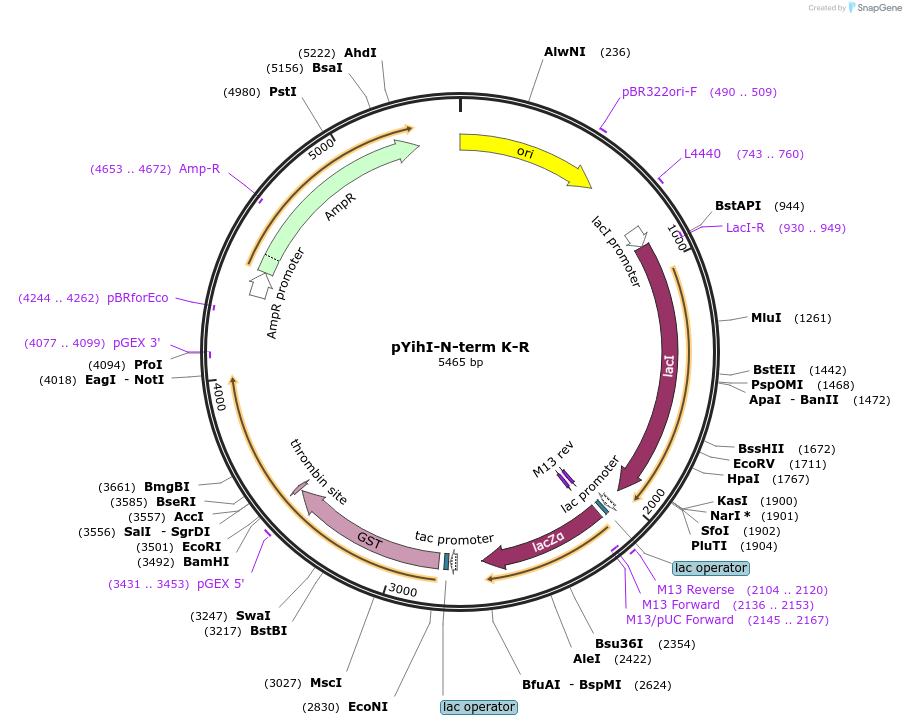
pYihI-N-term K-R
Plasmid#233091PurposeExpression of GST-YihI with N-term lysines (K) mutated to arginine (R)DepositorInsertYihI with N-term lysines (K) mutated to arginine (R)
TagsGSTExpressionBacterialMutationN-terminal lysine residues mutated to arginineAvailable SinceMay 12, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
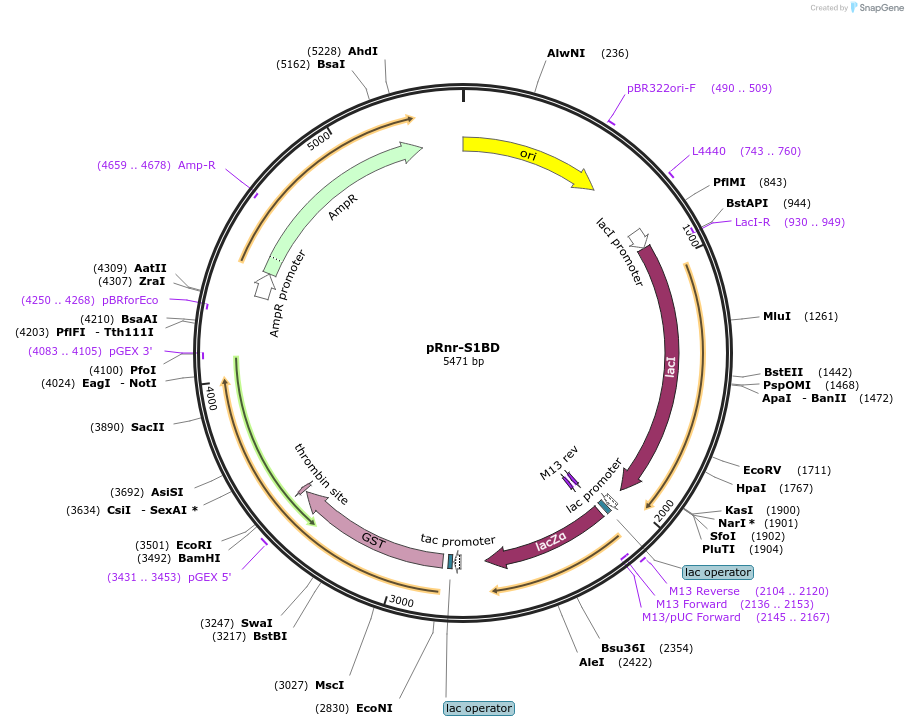
pRnr-S1BD
Plasmid#232580PurposeExpression of the Rnr S1 and basic domains (residues 1930 to 2442) as a GST-fusion.DepositorInsertRnr S1 + basic domain (residues 1930 to 2442)
TagsGSTExpressionBacterialAvailable SinceMay 12, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
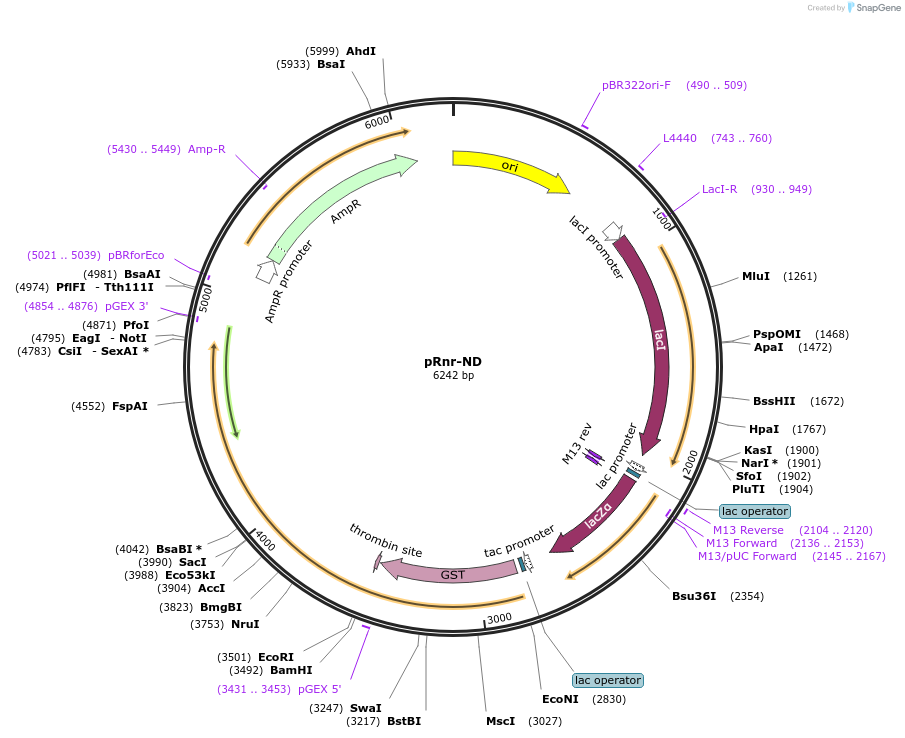
pRnr-ND
Plasmid#232579PurposeExpression of the Rnr nuclease domain (residues 649 to 1929) as a GST-fusion.DepositorInsertRnr nuclease domain (residues 649 to 1929)
TagsGSTExpressionBacterialAvailable SinceMay 12, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
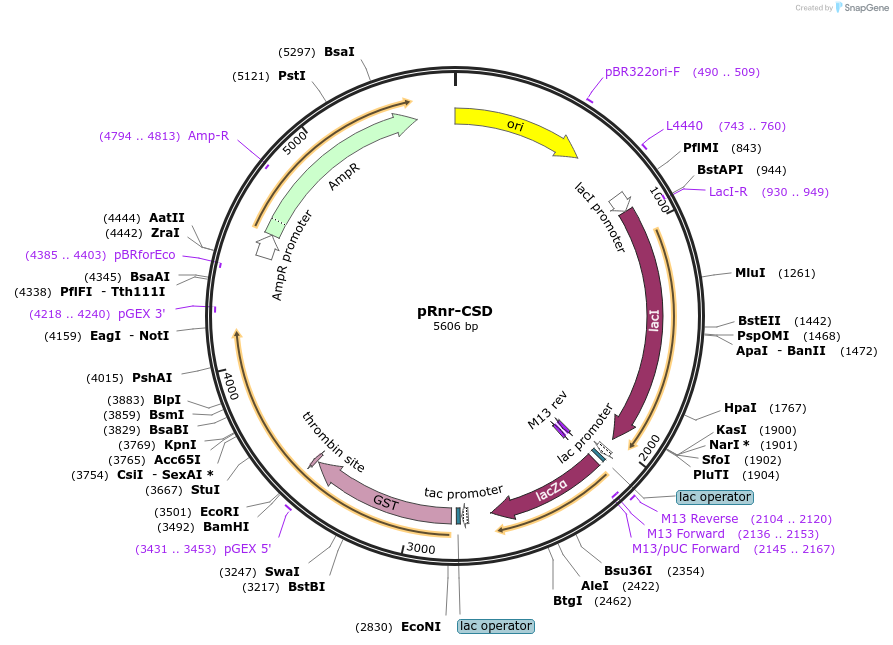
pRnr-CSD
Plasmid#232578PurposeExpression of the Rnr cold shock domains I and II (residues 1 to 648) as a GST-fusion.DepositorInsertRnr cold shock domains I and II (residues 1 to 648)
TagsGSTExpressionBacterialAvailable SinceMay 12, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
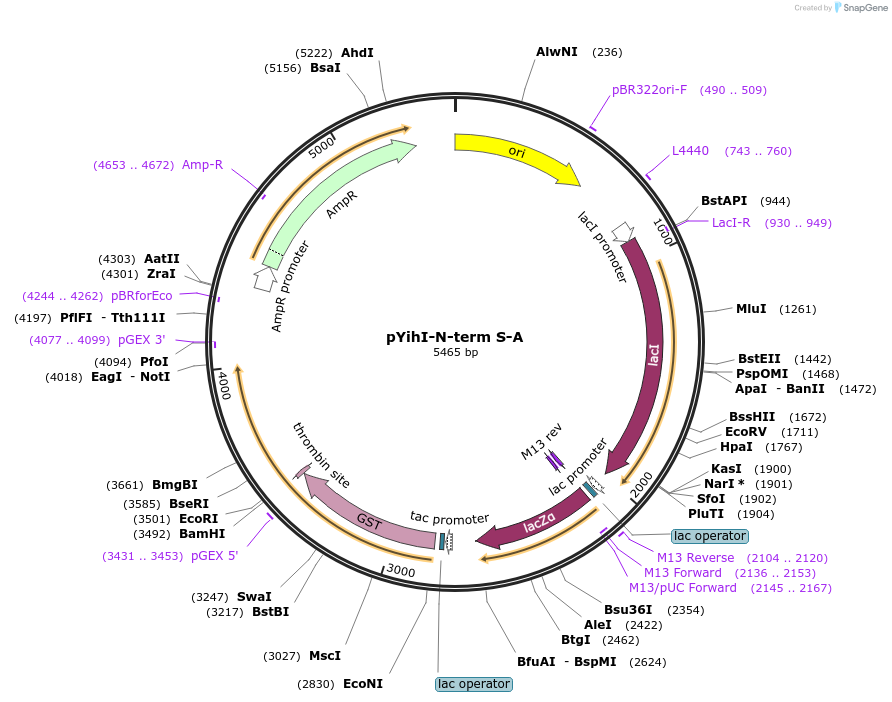
pYihI-N-term S-A
Plasmid#233096PurposeExpression of GST-YihI with N-term serines (S) mutated to alanine (A)DepositorInsertYihI with N-term serines (S) mutated to alanine (A)
TagsGSTExpressionBacterialMutationN-term serines (S) mutated to alanine (A)Available SinceMay 7, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
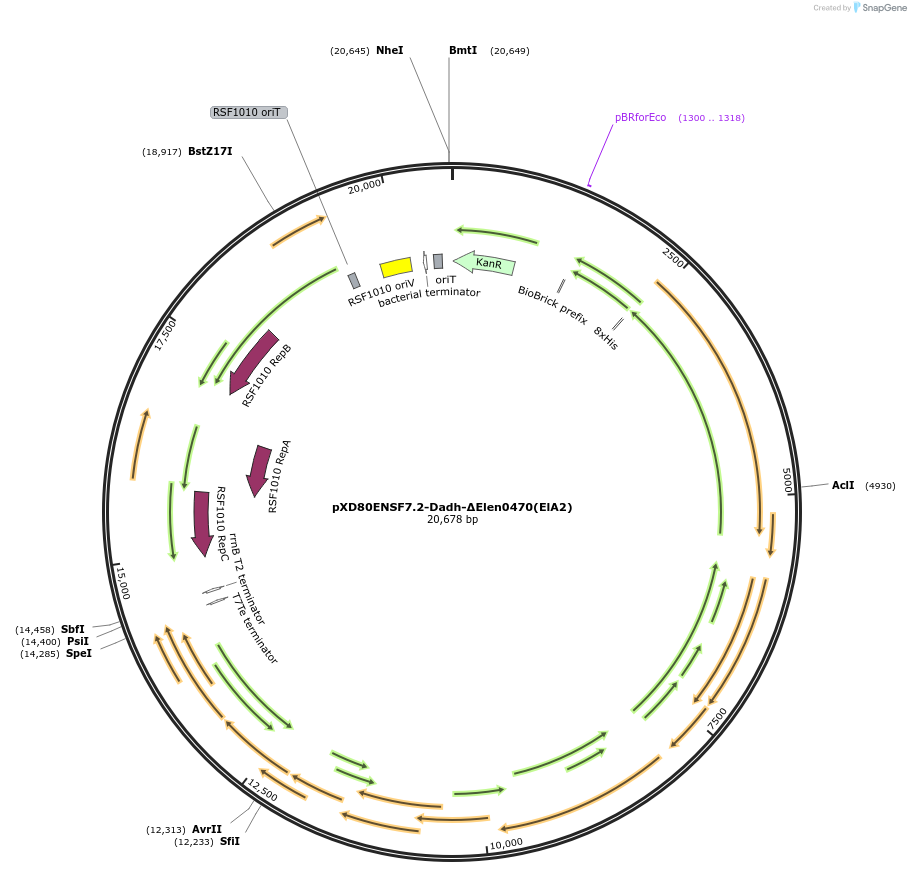
pXD80ENSF7.2-Dadh-ΔElen0470(ElA2)
Plasmid#225317PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0470) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0470)
TagsHis8ExpressionBacterialAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
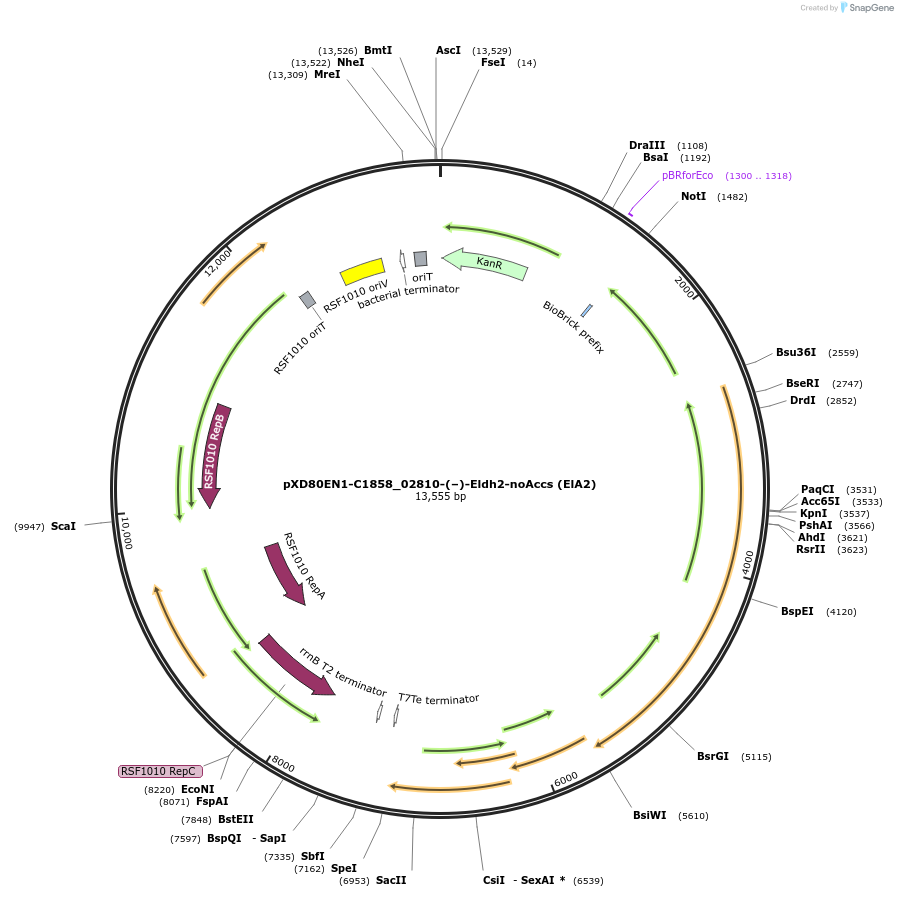
pXD80EN1-C1858_02810-(–)-Eldh2-noAccs (ElA2)
Plasmid#225309PurposeExpresses Eggerthella lenta A2 (–)-Eldh2 without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh2
ExpressionBacterialAvailable SinceApril 2, 2025AvailabilityAcademic Institutions and Nonprofits only