We narrowed to 12,964 results for: BASE
-
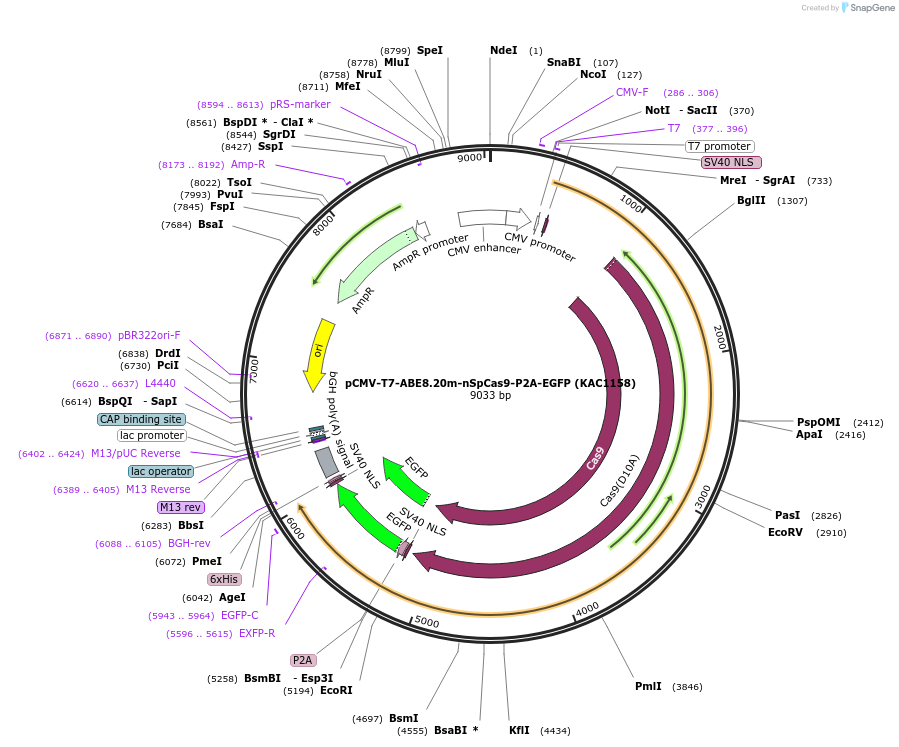
Plasmid#185915PurposepCMV and pT7 plasmid encoding human codon optimized ABE8.20m A-to-G base editor with nickase SpCas9(D10A) and P2A-EGFPDepositorInserthuman codon optimized ABE8.20m-nSpCas9-P2A-EGFP
UseIn vitro transcription; t7 promoterTagsBPNLS and BPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationnSpCas9=D10APromoterCMV and T7Available SinceJuly 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
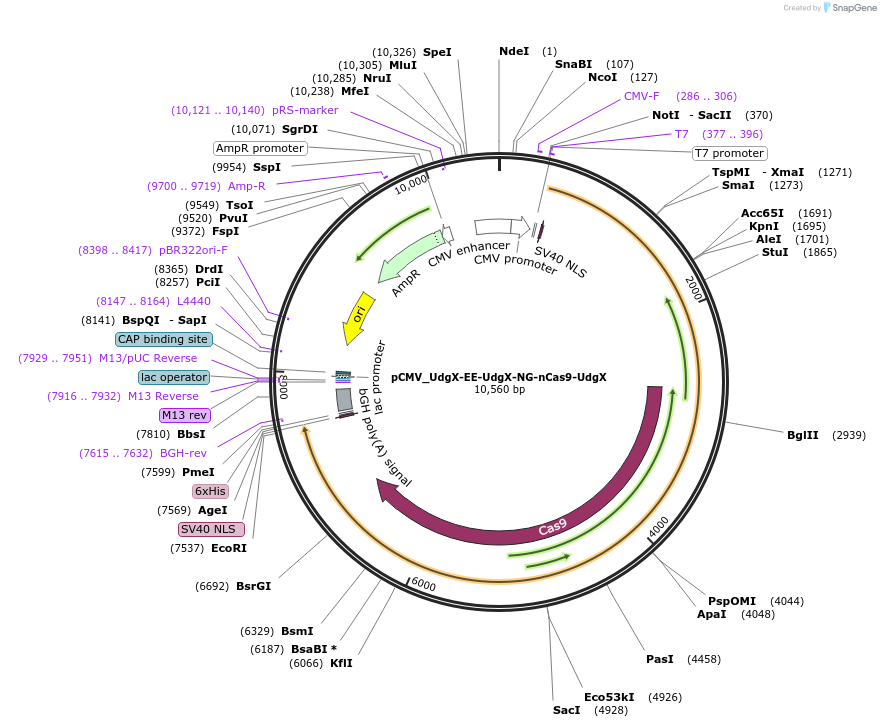
pCMV_UdgX-EE-UdgX-NG-nCas9-UdgX
Plasmid#163572PurposeMammalian CG-to-GC base editingDepositorInsertUdgX-EE-UdgX-NG-nCas9-UdgX
UseCRISPRExpressionMammalianMutationnCas9 (D10A)EE (R126E, R132E)NG (L111R, D1135V, G…Available SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
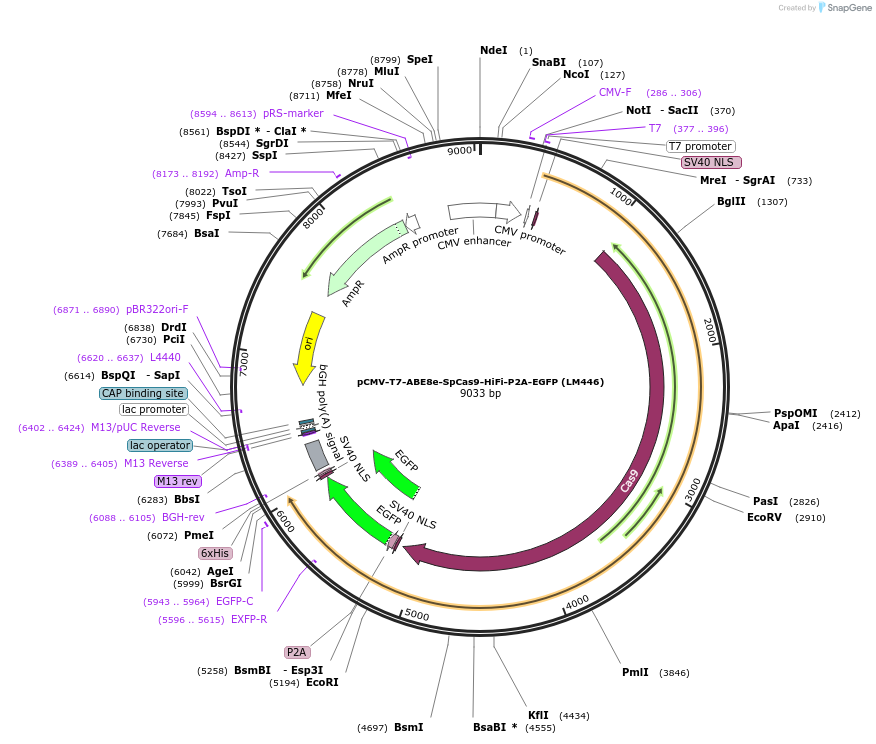
pCMV-T7-ABE8e-SpCas9-HiFi-P2A-EGFP (LM446)
Plasmid#197506PurposeCMV and T7 promoter expression plasmid for human codon optimized ABE8e A-to-G base editor with nSpCas9-HiFi(D10A) and P2A-EGFPDepositorInsertpCMV-T7-BPNLS-ABE8e-nSpCas9-HiFi-BPNLS-P2A-EGFP
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLS and BPNLS-P2A-EGFPExpressionMammalianMutationABE8e mutations in TadA and nSpCas9-HiFi(D10A/R69…PromoterCMV and T7Available SinceMarch 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
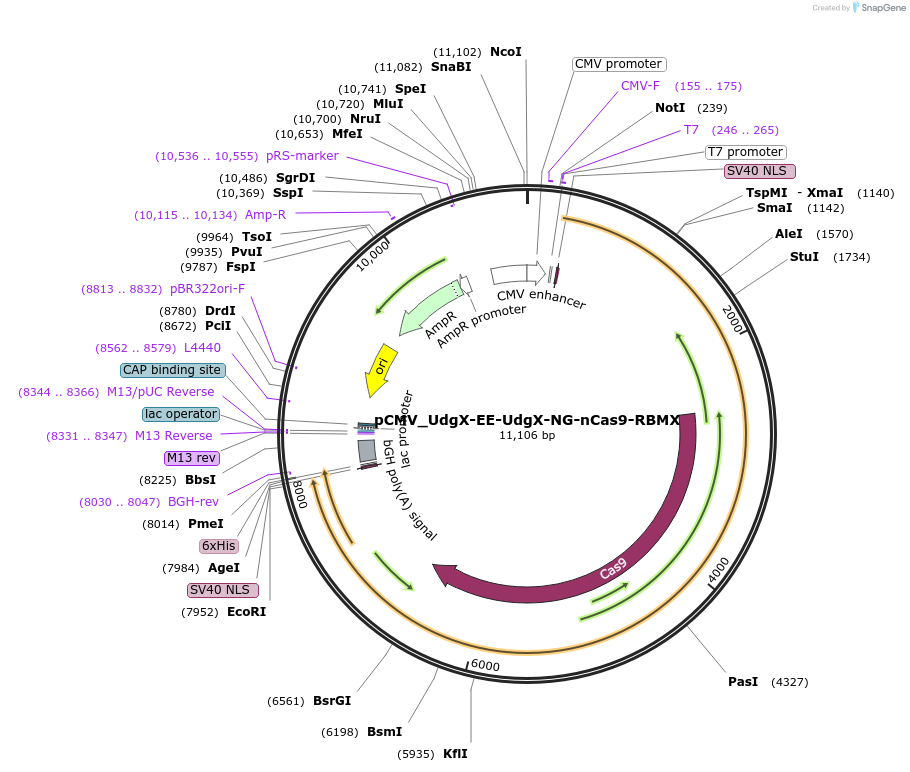
pCMV_UdgX-EE-UdgX-NG-nCas9-RBMX
Plasmid#163571PurposeMammalian CG-to-GC base editingDepositorInsertUdgX-EE-UdgX-NG-nCas9-RBMX
UseCRISPRExpressionMammalianMutationnCas9 (D10A)EE (R126E, R132E)NG (L111R, D1135V, G…Available SinceDec. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
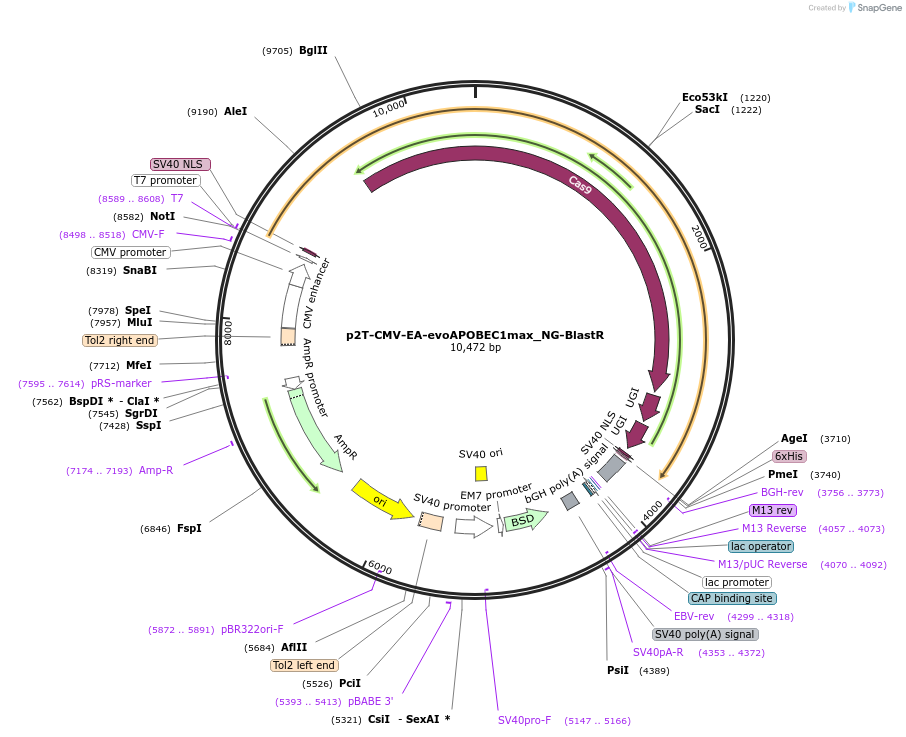
p2T-CMV-EA-evoAPOBEC1max_NG-BlastR
Plasmid#232721PurposeC-to-T base editorDepositorInsertevoAPOBEC1max H47E+S48A-nCas9(NG)-UGI-UGI
ExpressionMammalianMutationwithin evoAPOBEC1: H47E+S48A; within Cas9 D10AAvailable SinceMarch 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
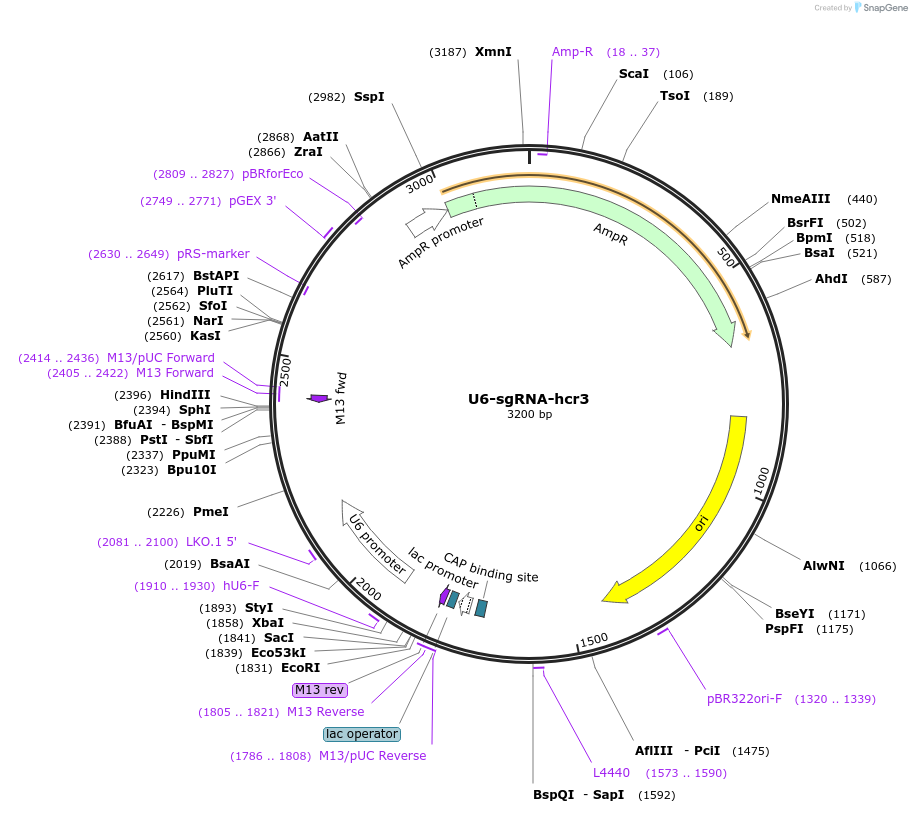
U6-sgRNA-hcr3
Plasmid#193660PurposeExpression of tandem pre-sgRNA array hcr3 for LbCas12aDepositorInsertU6-CFTR-sgRNA-KLF4-sgRNA-TET1-sgRNA-tRNA
ExpressionMammalianPromoterU6Available SinceFeb. 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
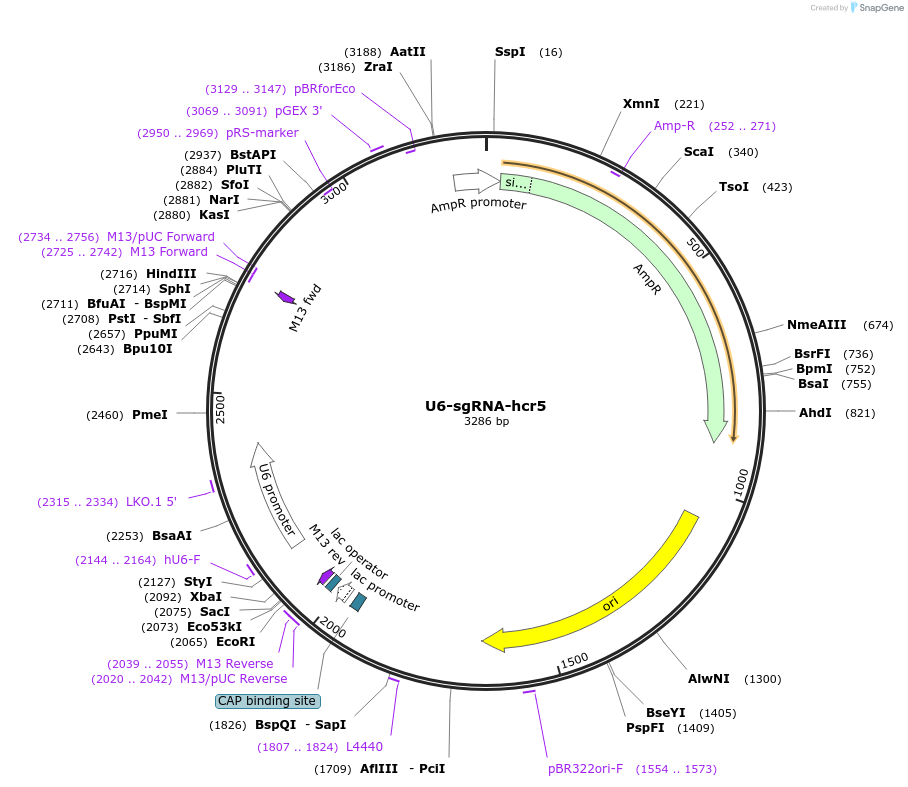
U6-sgRNA-hcr5
Plasmid#193661PurposeExpression of tandem pre-sgRNA array hcr5 for LbCas12aDepositorInsertU6-DNMT3B-sgRNA-KLF4-sgRNA-TET1-sgRNA-PRR5L-sgRNA-CFTR-sgRNA-tRNA
ExpressionMammalianPromoterU6Available SinceFeb. 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
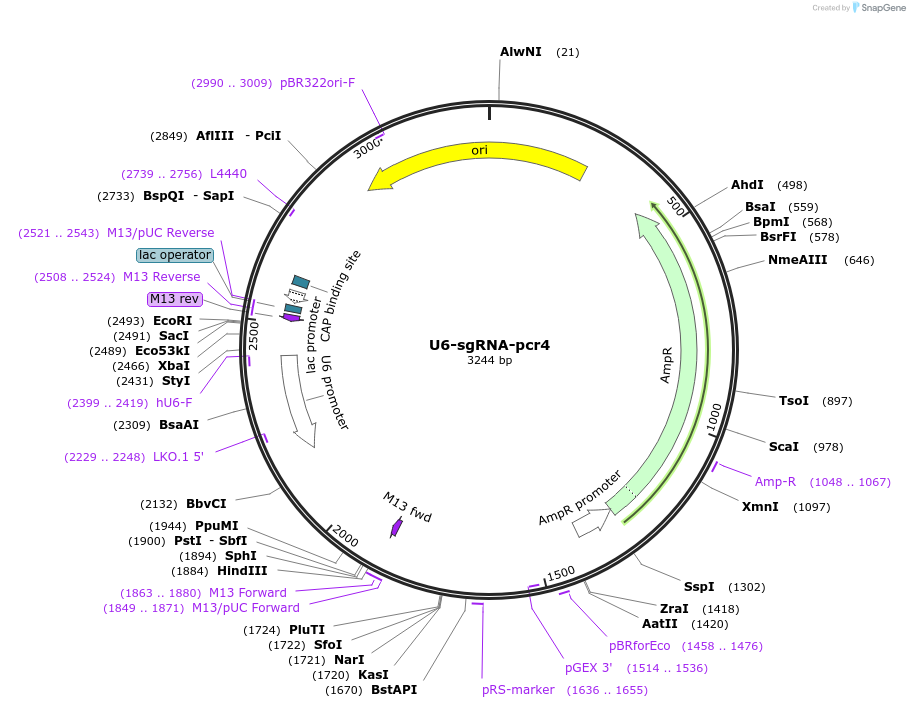
U6-sgRNA-pcr4
Plasmid#193662PurposeExpression of tandem pre-sgRNA array pcr4 for LbCas12aDepositorInsertU6-PUM1-sgRNA-GHR-sgRNA-HMGA2-sgRNA-PUM2-sgRNA-tRNA
ExpressionMammalianPromoterU6Available SinceFeb. 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
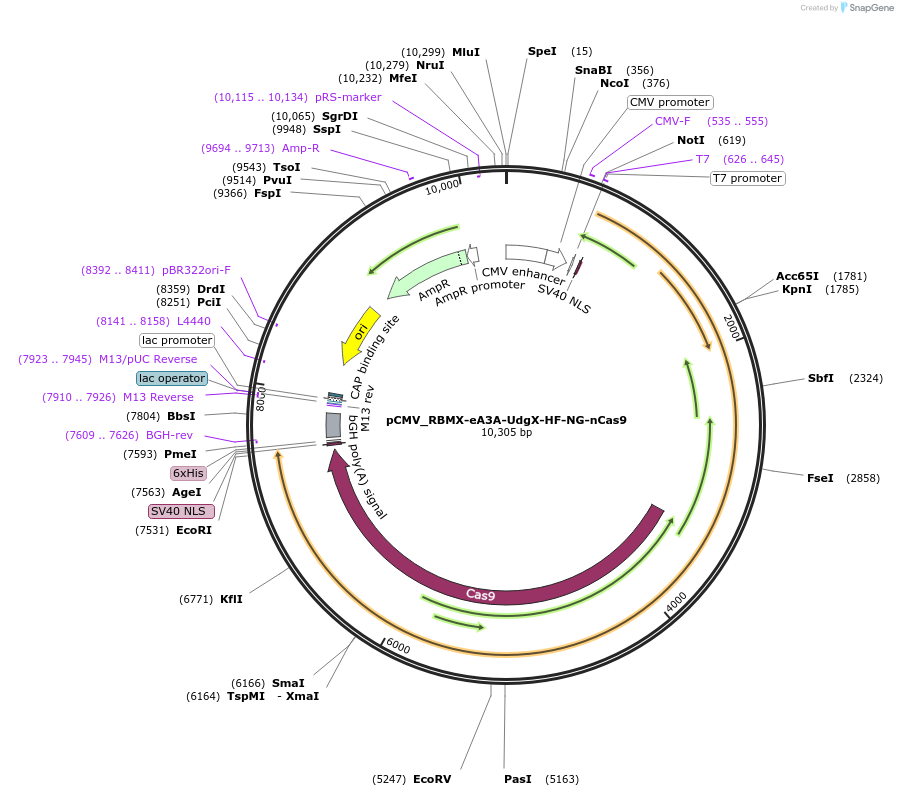
pCMV_RBMX-eA3A-UdgX-HF-NG-nCas9
Plasmid#163550PurposeMammalian CG-to-GC base editingDepositorInsertRBMX-eA3A-UdgX-HF-NG-nCas9
UseCRISPRExpressionMammalianMutationnCas9 (D10A)N497A, R661A, Q695A, Q926A//L111R, D1…Available SinceDec. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
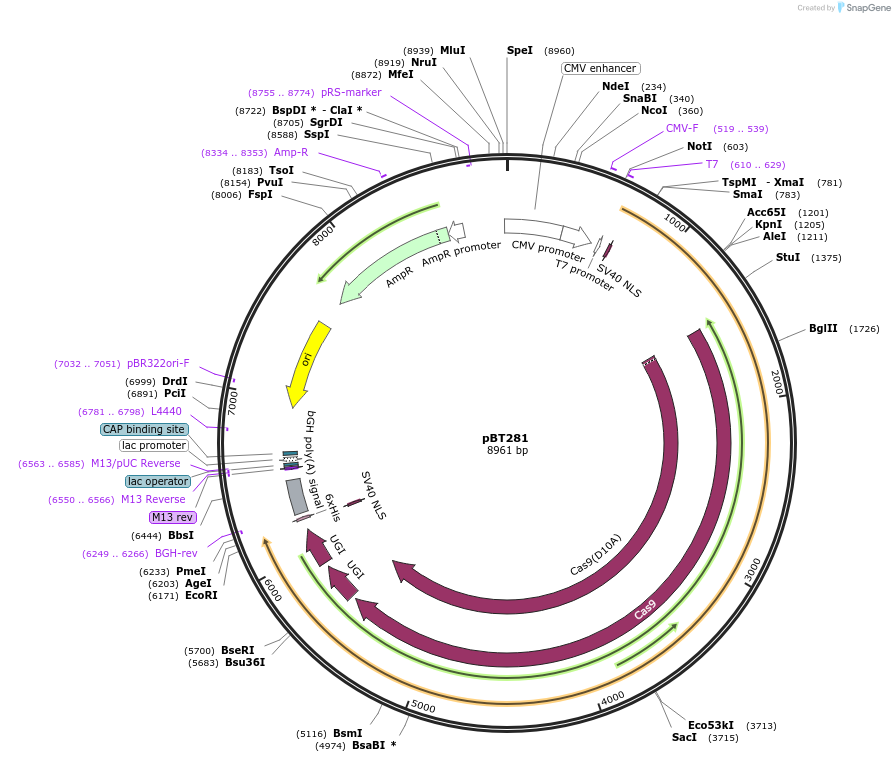
pBT281
Plasmid#122611Purposeexpress evoAPOBEC1-BE4max in mammalian cellsDepositorInsertevoAPOBEC1-BE4max
ExpressionMammalianMutationE4K H109N H122L D124N R154H A165S P201S F205SAvailable SinceJuly 29, 2019AvailabilityAcademic Institutions and Nonprofits only