We narrowed to 8,199 results for: Gal
-
Plasmid#237520PurposeRecombinant protein expressionDepositorInsertRwt4_kinase
UseTagsExpressionBacterialMutationKinase region of RWT4PromoterAvailable SinceMay 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
pUC19::ZmUBI-RWT4Ca_Reg1
Plasmid#237512PurposeProtoplast expressionDepositorInsertRwt4Ca_Reg1
UseTagsExpressionPlantMutationRemoved binding region 1PromoterAvailable SinceMay 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
pUC19::ZmUBI-RWT4Ca+C-Tail
Plasmid#237514PurposeProtoplast expressionDepositorInsertRwt4Ca+C-Tail
UseTagsExpressionPlantMutationAdd C-terminal extensionPromoterAvailable SinceApril 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
MPS1 Malassezia globosa
Plasmid#232151PurposeMPS1 from Malassezia globosa codon optimized for S. cerevisiae in pUCIDTDepositorInsertMPS1
UseTagsExpressionYeastMutationLikely not full proteinPromoterAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
MPS1 Schizosaccharomyces pombe
Plasmid#232147PurposeMPS1 from Schizosaccharomyces pombe codon optimized for S. cerevisiae in pUCIDTDepositorAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
PAF23
Plasmid#223171PurposeExpresses yeGFP under control of CTR1 promoter and terminatorDepositorInsertgreen fluorescent protein
UseTagsExpressionYeastMutationPromoterSaccharomyces cerevisiae CTR1Available SinceFeb. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
PAF24
Plasmid#223172PurposeExpresses VC83_00191-yeGFP under control of CTR1 promoter and terminatorDepositorInsertCopper Transporter
UseTagsStreptagII and yeGFPExpressionYeastMutationPromotersaccharomyces cerevisiae CTR1Available SinceFeb. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
pET28a-hfYFP-TEV-mScarlet-i
Plasmid#191196PurposeFusion of hfYFP and mScarlet-I separated by a TEV cleavage site for fluorescence-assisted protein purificationDepositorInserthfYFP & mScarlet-I fusion
UseTags6xHis and mScarlet-IExpressionBacterialMutationPromoterT7 promoterAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET28a-LSSmGFP-TEV-mScarlet-i
Plasmid#191199PurposeFusion of LSSmGFP and mScarlet-I separated by a TEV cleavage site for fluorescence-assisted protein purificationDepositorInsertLSSmGFP & mScarlet-I fusion
UseTags6xHis and mScarlet-IExpressionBacterialMutationPromoterT7 promoterAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
Dm_λHA-FLuc100_CSC
Plasmid#210592PurposeExpresses lambdaHA-optimal Firefly (CSC metric; Drosophila)DepositorInsertLambdaHA-optimal Firefly luciferase (Dm optimization, CSC metric)
UseTagslambda HAExpressionInsectMutationPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
Dm_Opt_Renilla_CSC
Plasmid#210593PurposeExpresses Optimal Renilla luciferase (CSC metric; Drosophila)DepositorInsertOptimal Renilla luciferase (Optimized for Drosophila, CSC metric)
UseTagsExpressionInsectMutationPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUB_OptλHAramp-FLuc100
Plasmid#210577PurposeExpresses Optimal lambda HA-Optimal Firefly luciferaseDepositorInsertOptimal lambda HA-Optimal Firefly luciferase
UseTagslambda HAExpressionMammalianMutationPromoterUBC humanAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUB_NonoptλHAramp-FLuc100
Plasmid#210579PurposeExpresses Nonoptimal lambda HA-Optimal Firefly luciferaseDepositorInsertNonoptimal lambda HA-Optimal Firefly luciferase
UseLentiviral and LuciferaseTagslambda HAExpressionMammalianMutationPromoterUBC humanAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1_SM-FLuc0-pA
Plasmid#210583PurposeExpresses SM(FLAG)-nonoptimal Firefly luciferase- pADepositorInsertSpaghetti monster (FLAG)-nonoptimal FLuc
UseLuciferaseTagsSpaghetti monster (FLAG)ExpressionMammalianMutationPromoterCMVAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1_SM-FLuc100-pA
Plasmid#210584PurposeExpresses SM(FLAG)-optimal Firefly luciferase - pADepositorInsertSpaghetti monster (FLAG)-optimal FLuc
UseLuciferaseTagsSpaghetti monster (FLAG)ExpressionMammalianMutationPromoterCMVAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
Dm_λHA-FLuc0_CSC
Plasmid#210589PurposeExpresses lambdaHA-nonoptimal Firefly (CSC metric; Drosophila)DepositorInsertLambdaHA-nonoptimal Firefly luciferase (Dm optimization, CSC metric)
UseLuciferaseTagslambda HAExpressionInsectMutationPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.425
Plasmid#203944PurposeUp Flank. Up flanking region of RSF1010-Cas12a vector for introducing gRNA expression cassette and swapping out the AbR.DepositorInsertCas12a UP
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.426
Plasmid#203945PurposeDown Flank. Down flanking region of RSF1010-Cas12a vector for introducing gRNA expression cassette and swapping out the AbR.DepositorInsertCas12a DOWN
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.391
Plasmid#203948PurposeLevel 0 part. Cas12a coding sequence from Francisella novicida in CDS1 positionDepositorInsertFnCas12a
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.385
Plasmid#203936PurposeUp Flank. Neutral site. Up flanking sequence of ω3 acyl-lipid desaturase (desB, FEK30_04840) in Synechococcus sp. PCC 11901.DepositorInsertdesB UP
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.386
Plasmid#203938PurposeUp Flank. Neutral site. Up flanking sequence of glycogen synthase A1 (glgA1, FEK30_14880) in Synechococcus sp. PCC 11901.DepositorInsertglgA1 UP
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.387
Plasmid#203939PurposeDown Flank. Neutral site. Down flanking sequence of glycogen synthase A1 (glgA1, FEK30_14880) in Synechococcus sp. PCC 11901.DepositorInsertglgA1 DOWN
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC0.437
Plasmid#203942PurposeUp Flank. Neutral site. Up flanking sequence of intergenic region (NS1) between FEK30_11550 and FEK30_11555 in Synechococcus sp. PCC 11901.DepositorInsertNS1 UP
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
Fg vector
Plasmid#213464PurposeDestination vector for cloning and transformation into TSI locus 1.DepositorTypeEmpty backboneUseTagsExpressionBacterialMutationPromoterAvailable SinceMarch 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
pHYX143
Plasmid#202816PurposeControl plasmid expressing mScarlet-I fluorescent protein.DepositorInsertmScarlet-I
UseTagsExpressionYeastMutationPromoterScPCK1Available SinceNov. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
PlayBack-CMV-Bamh1
Plasmid#203303PurposeFor expression of eukaryotic genes of interest with CMV promoter with Bamh1 as a PlayBack Restriction EnzymeDepositorTypeEmpty backboneUseTagsExpressionMammalianMutationPromoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
PlayBack-CMV-Nhe1
Plasmid#203304PurposeFor expression of eukaryotic genes of interest with CMV promoter with NheI as a PlayBack Restriction EnzymeDepositorTypeEmpty backboneUseTagsExpressionMammalianMutationPromoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
PlayBack-CMV-Bgl11
Plasmid#203306PurposeFor expression of eukaryotic genes of interest with CMV promoter with BglII as a PlayBack Restriction EnzymeDepositorTypeEmpty backboneUseTagsExpressionMammalianMutationPromoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
PlayBack-Ef1a-Nde1
Plasmid#203309PurposeFor expression of eukaryotic genes of interest with Ef1a promoter with NdeI as a PlayBack Restriction EnzymeDepositorTypeEmpty backboneUseTagsExpressionMammalianMutationPromoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
PlayBack-Ef1a-Nhe1
Plasmid#203310PurposeFor expression of eukaryotic genes of interest with Ef1a promoter with NheI as a PlayBack Restriction EnzymeDepositorTypeEmpty backboneUseTagsExpressionMammalianMutationPromoterAvailable SinceOct. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB406
Plasmid#203627PurposePromoter of the TAKA-amylase A gene from A. oryzae, maltose/starch-inducible.DepositorInsertPamyB
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB407
Plasmid#203628PurposePromoter of the elongation factor 1-alpha gene from P. digitatum (PDIG_59570).DepositorInsertPef1A
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB408
Plasmid#203629PurposePromoter of the ubiquitin ligase gene from P. digitatum (PDIG_07760).DepositorInsertP07760
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
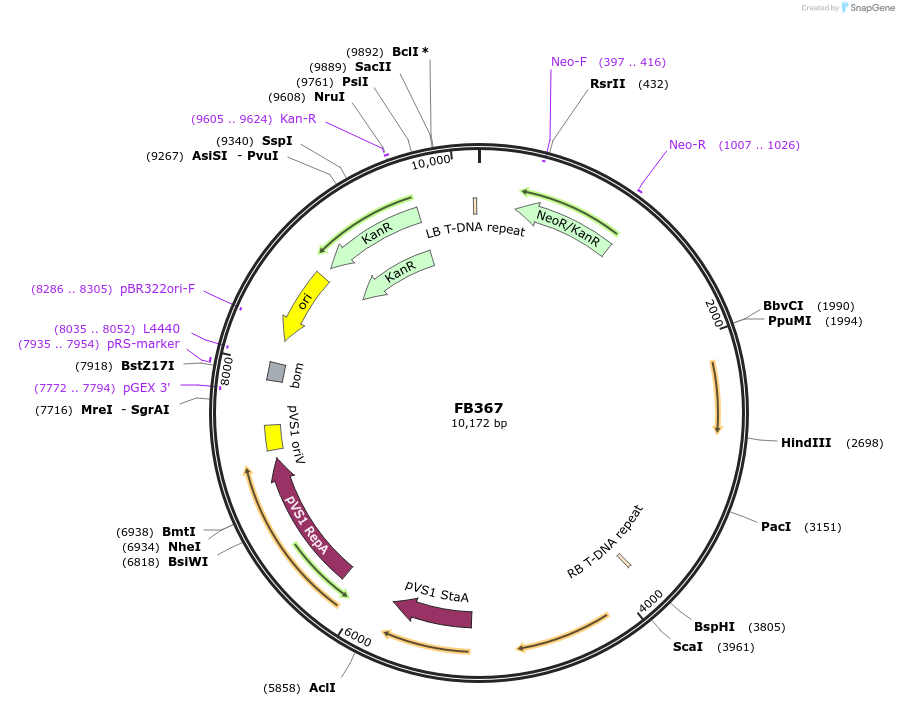
FB367
Plasmid#203630PurposeModule for the expression of geneticin resistance and Nanoluciferase.DepositorInsertPgpdA:Nluc:Ttub::PtrpC:nptII:Ttub
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB273
Plasmid#203612PurposeTerminator of pyr4 gene from T. reesei.DepositorInsertTpyr4
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB293
Plasmid#203613PurposeAssembly of the transcriptional unit for the auxotrophy marker pyr4 from T. reseei.DepositorInsertTU_pyr4
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB359
Plasmid#203614Purpose5' upstream region of pyrG gene in P. digitatum.DepositorInsert5' upstream Pdig pyrG
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB361
Plasmid#203615Purpose3' downstream region of pyrG gene in P. digitatum.DepositorInsert3' downstream Pdig pyrG
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB372
Plasmid#203616PurposeAssembly for pyrG deletion in P. digitatum.DepositorInsertFB359+FB361
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB413
Plasmid#203617PurposeCoding sequence for phleomycin resistance.DepositorInsertble
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only