We narrowed to 20,834 results for: omp
-
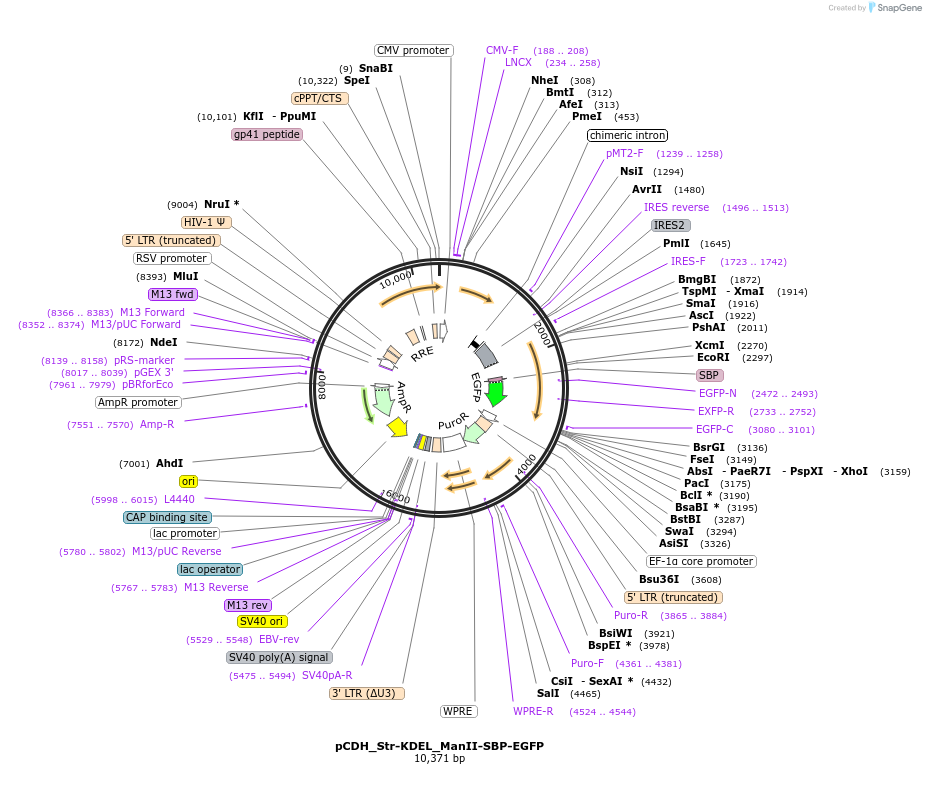
Plasmid#65258Purposesynchronize trafficking of ManII from the ER (RUSH system)DepositorInsertStreptavidin-KDEL and truncated Mannosidase II
UseLentiviralTagsEGFP and Streptavidin Binding Protein (SBP)Mutationcontains only amino acids 1 to 116 of ManIIPromoterCMVAvailable SinceAug. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
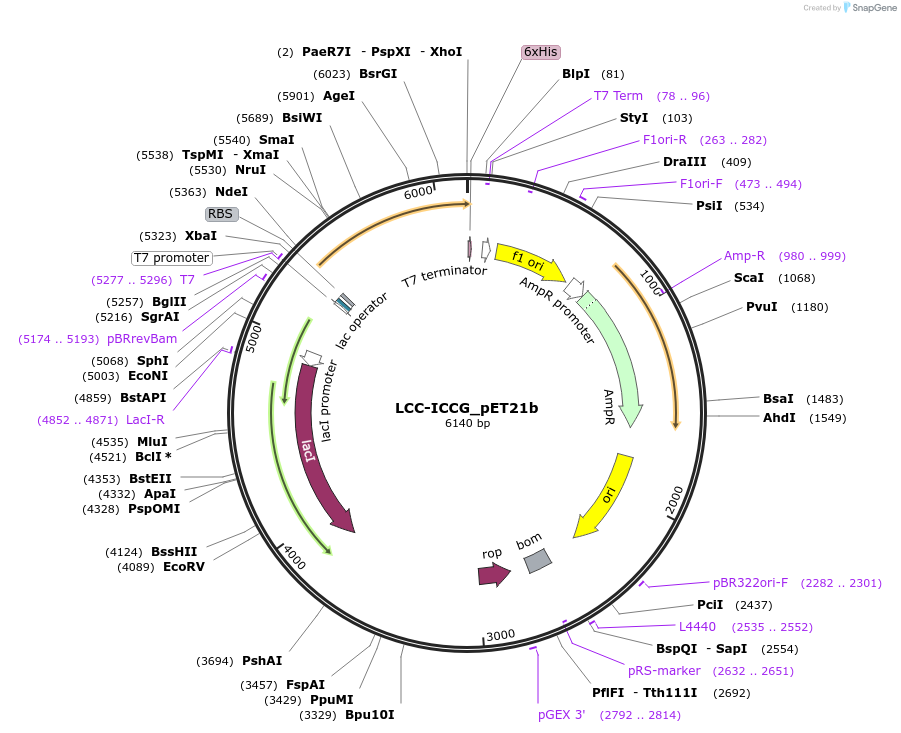
LCC-ICCG_pET21b
Plasmid#215408PurposeExpression construct for LCC-ICCG gene, codon optimized for expression in E. coliDepositorInsertLCC-ICCG
ExpressionBacterialMutationF243I/D238C/S283C/Y127GAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
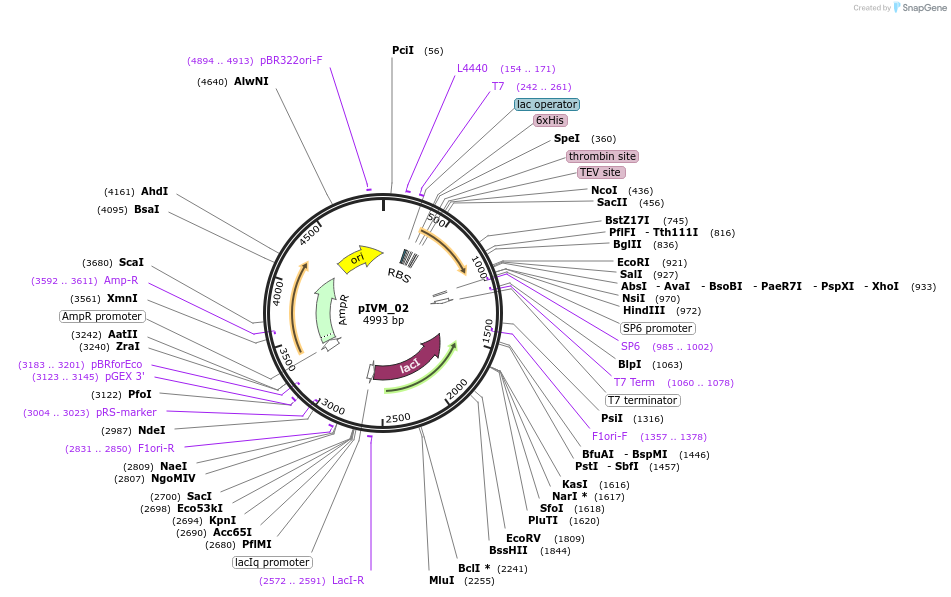
pIVM_02
Plasmid#204500PurposeExpression of VHL (54-213) for production of VCB complexes in conjunction with plasmid pIVM_26 (EloB 1-104, EloC 17-112)DepositorInsertamino acids 54-213 VHL (VHL Human)
Tags6xHis, TEV, and ThrombinExpressionBacterialMutation0Available SinceSept. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
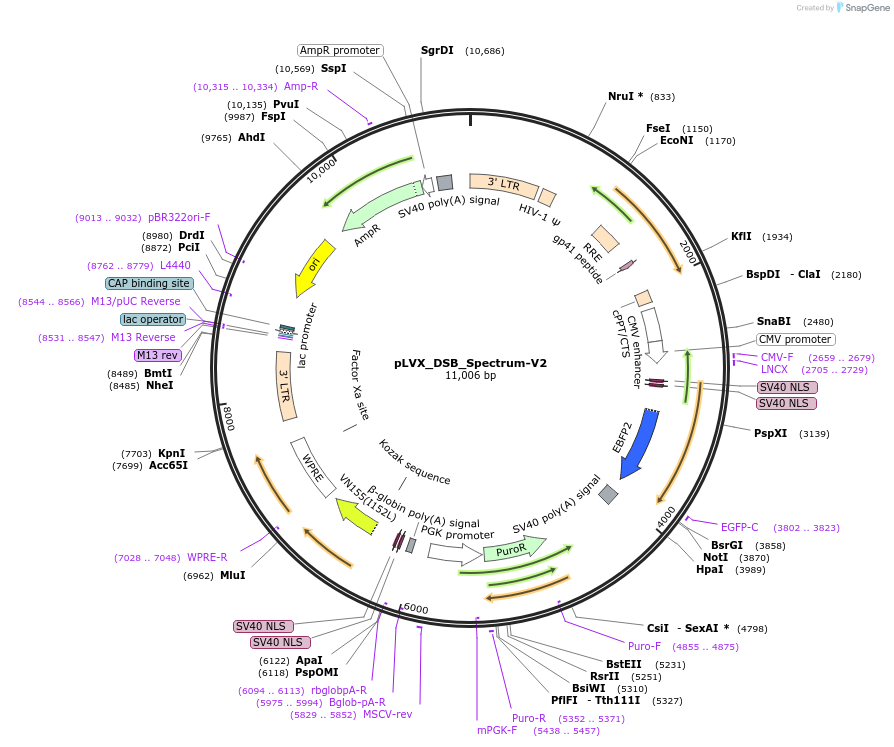
pLVX_DSB_Spectrum-V2
Plasmid#192475PurposeLentiviral DSB Spectrum V2 Reporter SystemDepositorInsertBFP1.2, incompleteGFP
UseLentiviralExpressionMammalianPromoterCMVAvailable SinceDec. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
-
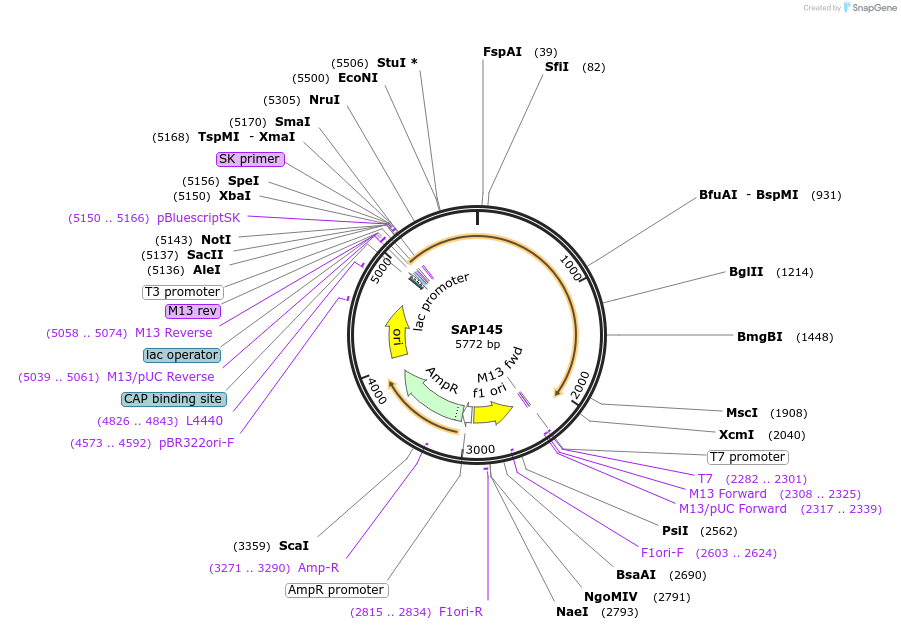
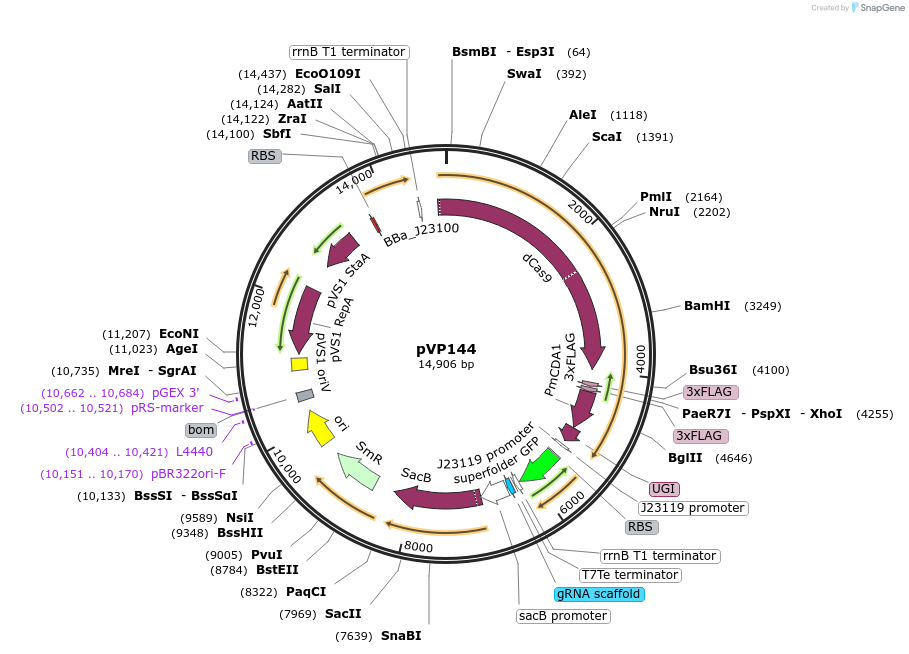
pVP144
Plasmid#222081PurposeSingle component CRISPR-mediated base-editor for Agrobacterium genetic engineering visually marked with sfGFP for guide insertion and eforRED for plasmid eviction.DepositorInsertdCas9-PmCDA1, sfGFP, sacB and eforRED
UseCRISPRExpressionBacterialAvailable SinceJan. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
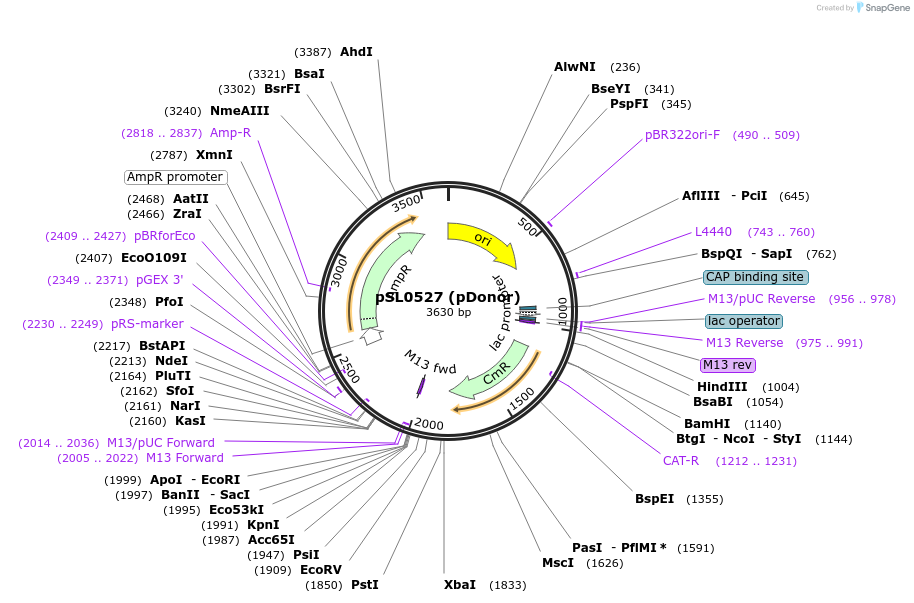
pSL0527 (pDonor)
Plasmid#130634PurposeEncodes a mini-transposon derived from V. cholerae CAST system, with CmR cargo gene. Total transposon size = 977 bp.DepositorInsertVchCAST donor DNA (Mini-Tn)
UseCRISPR; TransposonExpressionBacterialAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
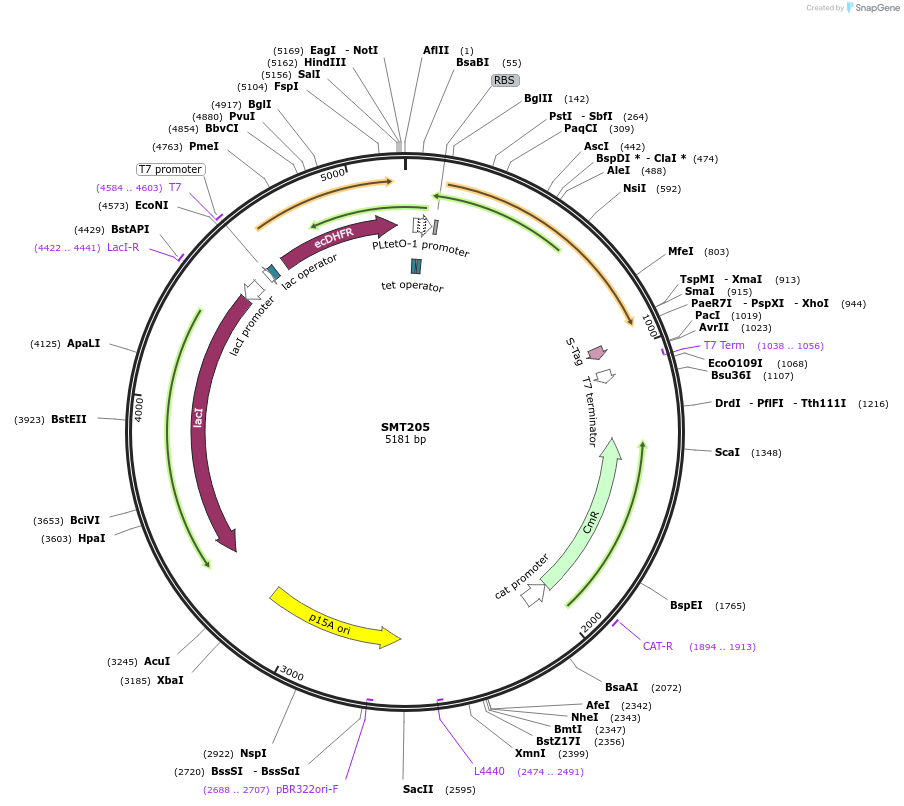
SMT205
Plasmid#134817PurposeSMT201 with mutated RBS for DHFR, in vivo assaysDepositorInsertsDihydrofolate reductase
Thymidylate synthase
ExpressionBacterialMutationSilent mutations to eliminate internal restrictio…PromoterT7 and TETAvailable SinceJuly 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
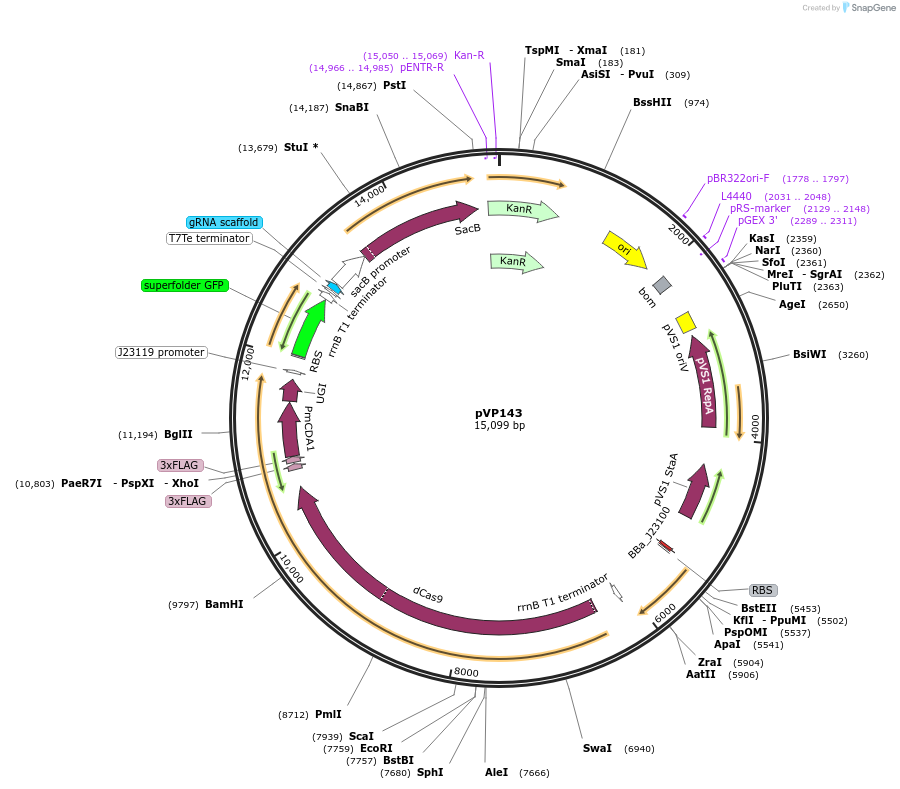
pVP143
Plasmid#222080PurposeSingle component CRISPR-mediated base-editor for Agrobacterium genetic engineering visually marked with sfGFP for guide insertion and AeBlue for plasmid eviction.DepositorInsertdCas9-PmCDA1, sfGFP, sacB and AeBlue
UseCRISPRExpressionBacterialAvailable SinceJan. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
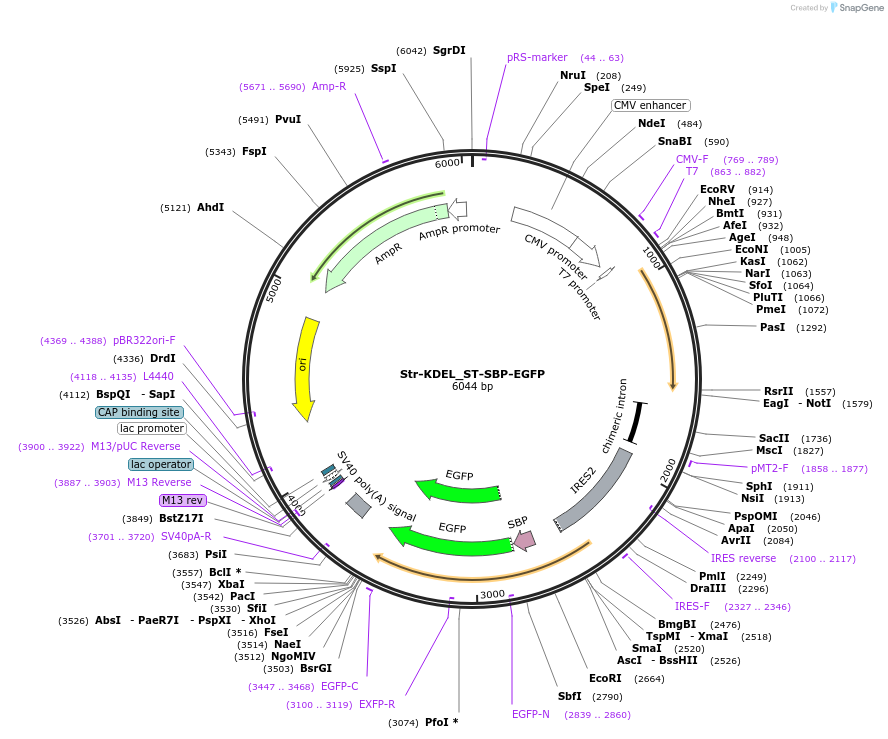
Str-KDEL_ST-SBP-EGFP
Plasmid#65264Purposesynchronize trafficking of ST from the ER (RUSH system)DepositorInsertStr-KDEL and truncated sialyltransferase (ST6GAL1 Human)
TagsEGFP and Streptavidin Binding Protein (SBP)ExpressionMammalianMutationcontains only amino acids 1 to 43 of STPromoterCMVAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
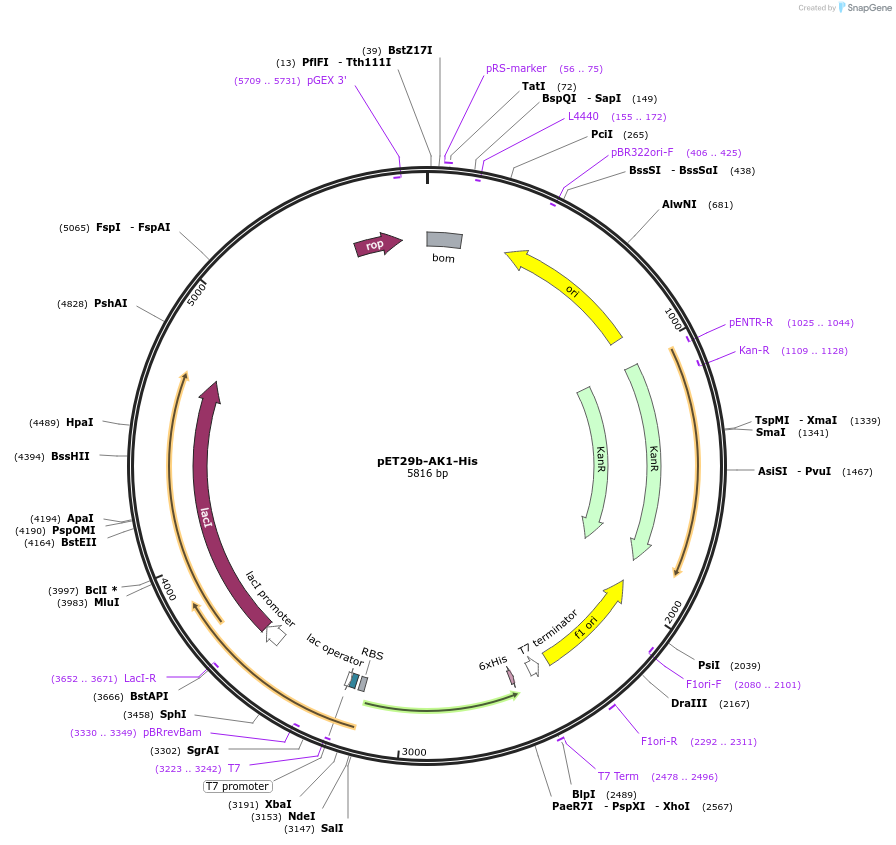
pET29b-AK1-His
Plasmid#124135PurposeBacterial expression plasmid for PURE system componentsDepositorAvailable SinceApril 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
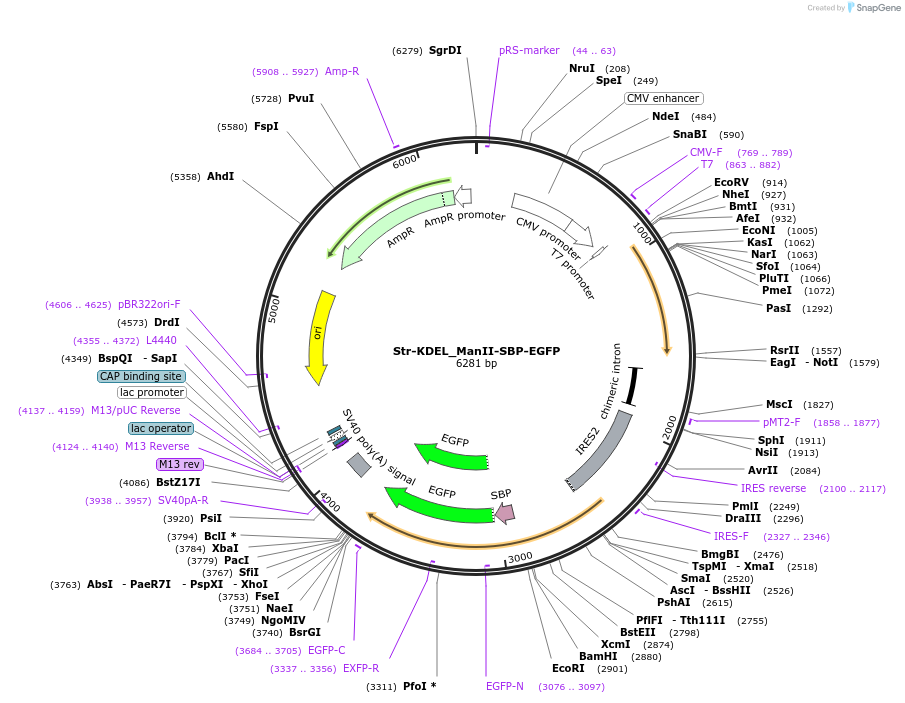
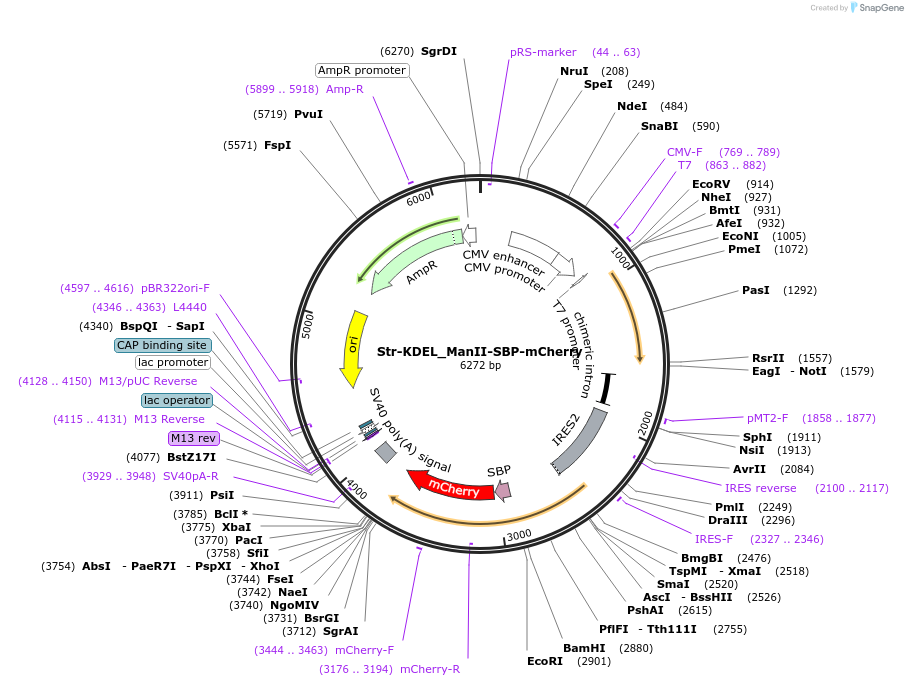
Str-KDEL_ManII-SBP-EGFP
Plasmid#65252Purposesynchronize trafficking of ManII from the ER (RUSH system)DepositorInsertStreptavidin-KDEL and truncated Mannosidase II
TagsEGFP and Streptavidin Binding Protein (SBP)ExpressionMammalianMutationcontains only amino acids 1 to 116 of ManIIPromoterCMVAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
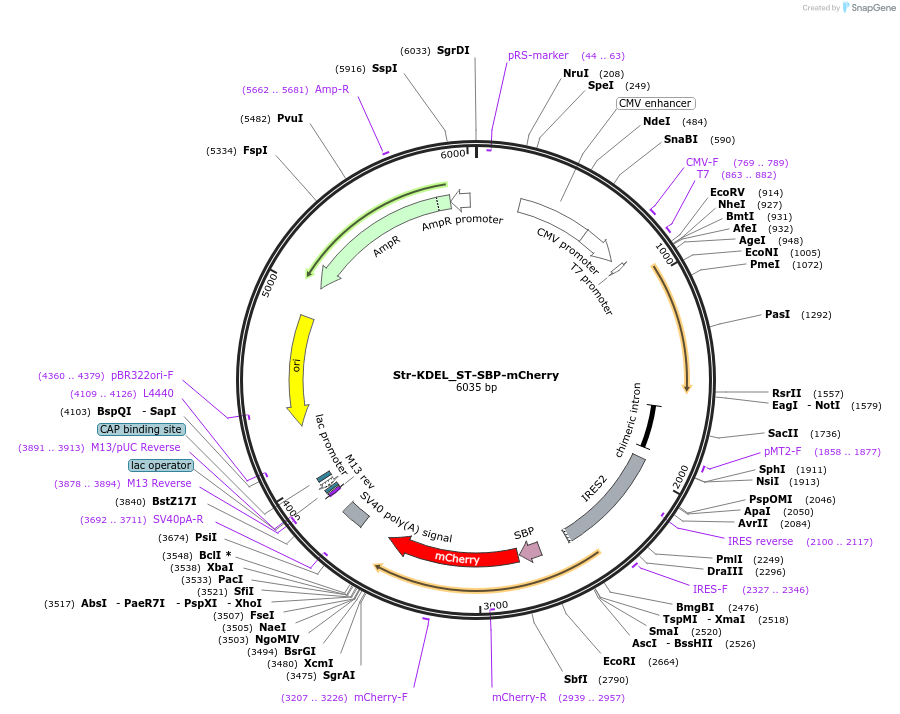
Str-KDEL_ST-SBP-mCherry
Plasmid#65265Purposesynchronize trafficking of ST from the ER (RUSH system)DepositorInsertStr-KDEL and truncated sialyltransferase (ST6GAL1 Human)
TagsStreptavidin Binding Protein (SBP) and mCherryExpressionMammalianMutationcontains only amino acids 1 to 43 of STPromoterCMVAvailable SinceJune 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
Str-KDEL_ManII-SBP-mCherry
Plasmid#65253Purposesynchronize trafficking of ManII from the ER (RUSH system)DepositorInsertStreptavidin-KDEL and truncated Mannosidase II
TagsStreptavidin Binding Protein (SBP) and mCherryExpressionMammalianMutationcontains only amino acids 1 to 116 of ManIIPromoterCMVAvailable SinceJune 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
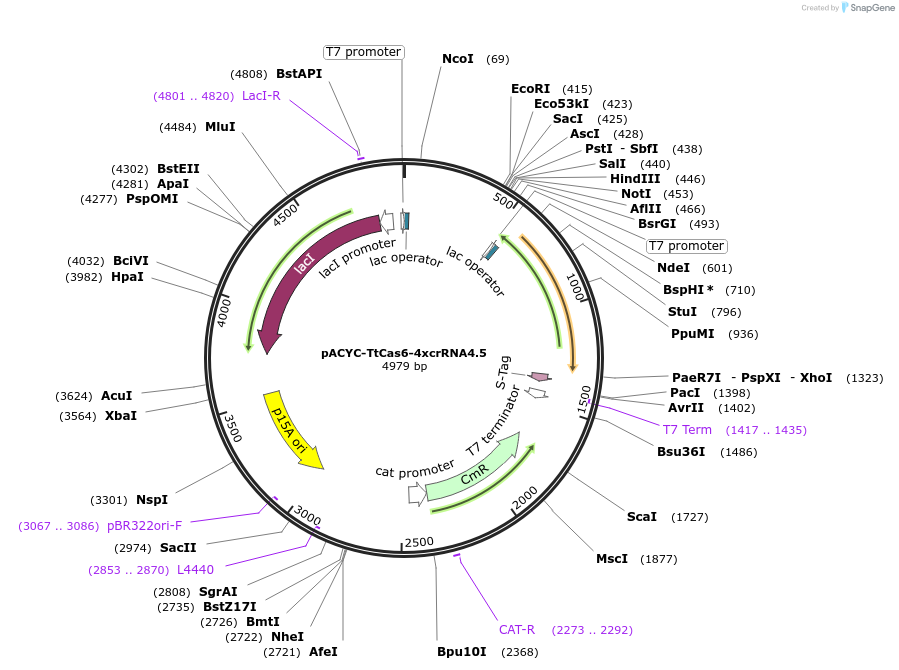
pACYC-TtCas6-4xcrRNA4.5
Plasmid#127764PurposeFor expression of T. thermophilus Csm complex in E. coli (crRNA)DepositorInsertsSynthetic CRISPR array containing four identical T. thermophilus spacers
Cas6A
ExpressionBacterialMutationCodon optimized for expression in E. coliPromoterT7Available SinceAug. 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
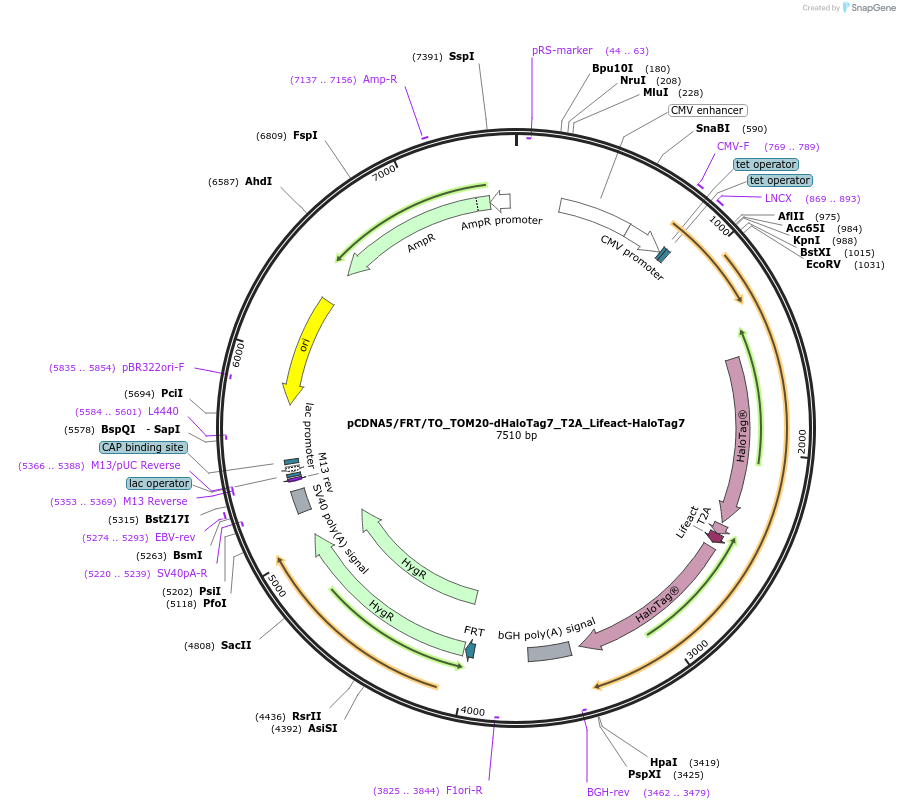
pCDNA5/FRT/TO_TOM20-dHaloTag7_T2A_Lifeact-HaloTag7
Plasmid#187076PurposeCo-expression of dHaloTag7 at the mitochondria surface and HaloTag7 on actin filamentsDepositorInsertTOM20-dHaloTag7_T2A_Lifeact-HaloTag7
ExpressionMammalianMutationdHaloTag7 = HaloTag7-D106AAvailable SinceAug. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
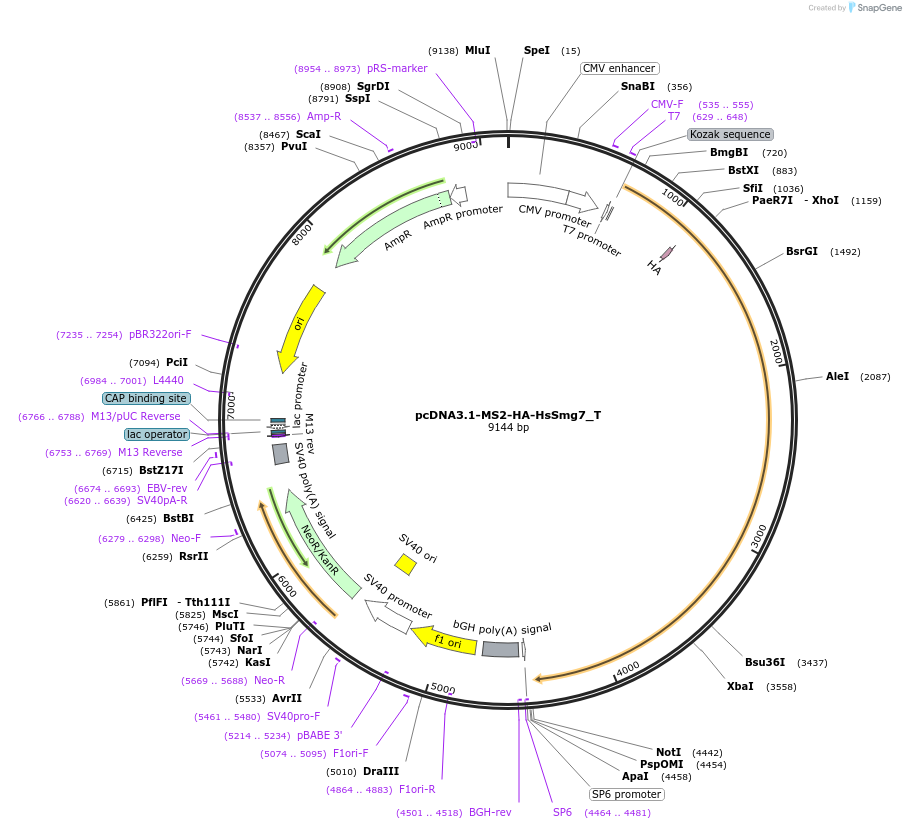
pcDNA3.1-MS2-HA-HsSmg7_T
Plasmid#147556PurposeMammalian Expression of HsSmg7DepositorInsertHsSmg7 (SMG7 Human)
ExpressionMammalianMutationtwo silent mutation G216A, A1521G and one non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
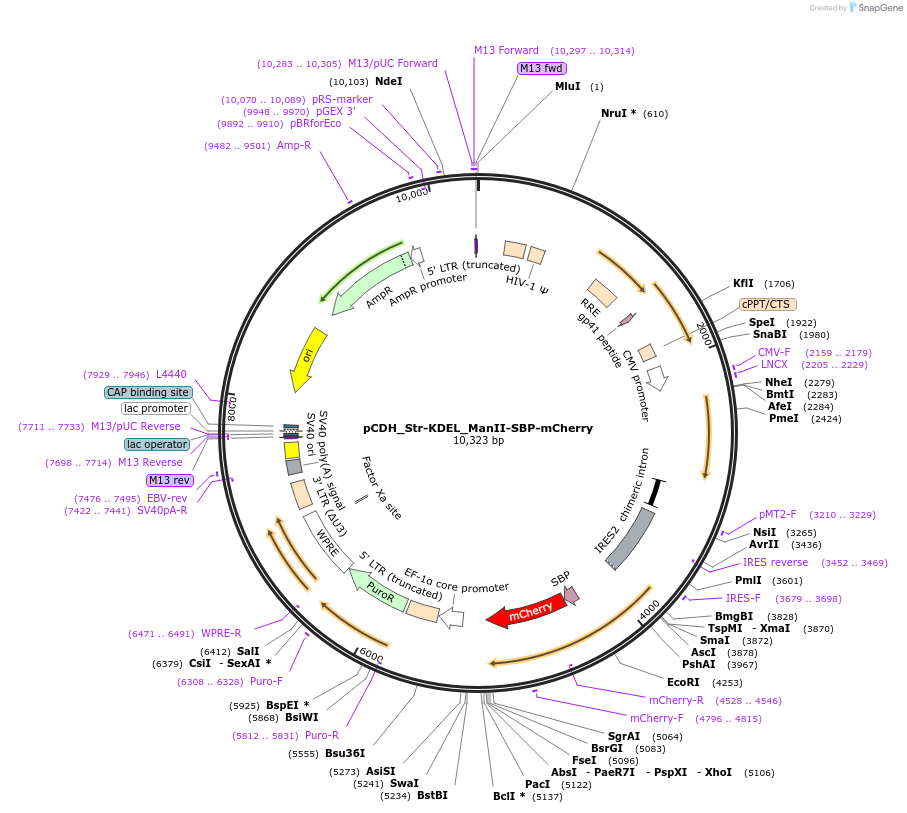
pCDH_Str-KDEL_ManII-SBP-mCherry
Plasmid#65259Purposesynchronize trafficking of ManII from the ER (RUSH system)DepositorInsertStreptavidin-KDEL and truncated Mannosidase II
UseLentiviralTagsStreptavidin Binding Protein (SBP) and mCherryMutationcontains only amino acids 1 to 116 of ManIIPromoterCMVAvailable SinceAug. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
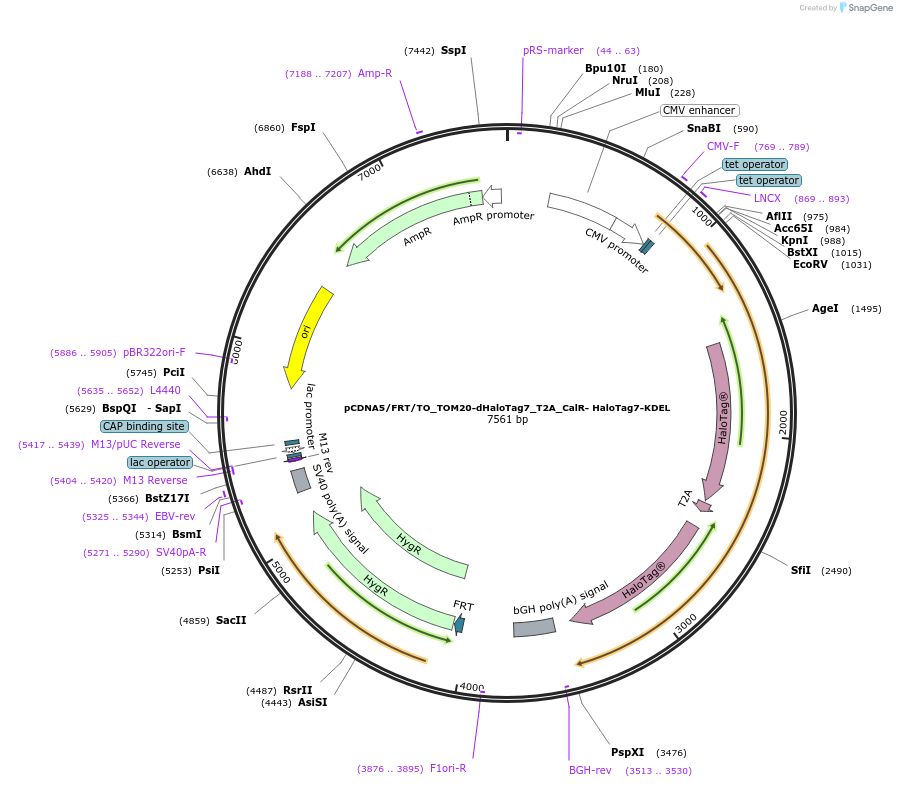
pCDNA5/FRT/TO_TOM20-dHaloTag7_T2A_CalR- HaloTag7-KDEL
Plasmid#187079PurposeCo-expression of dHaloTag7 at the mitochondria surface and HaloTag7 in endoplasmic reticulumDepositorInsertTOM20-dHaloTag7_T2A_CalR- HaloTag7-KDEL
ExpressionMammalianMutationdHaloTag7 = HaloTag7-D106AAvailable SinceAug. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
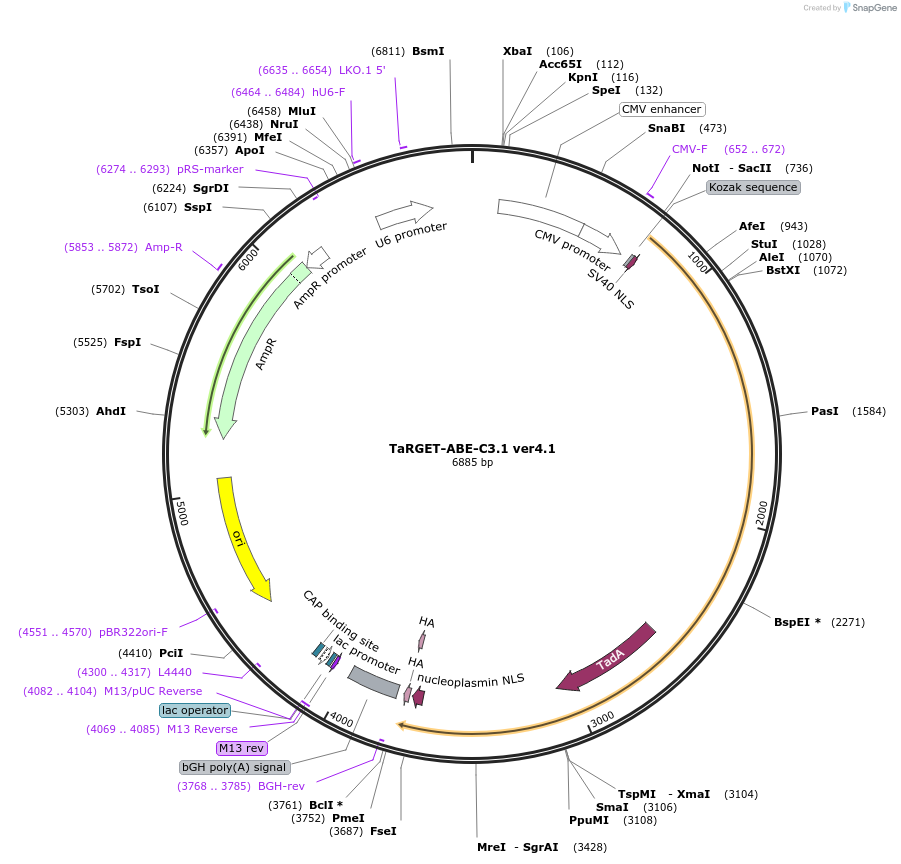
TaRGET-ABE-C3.1 ver4.1
Plasmid#188251PurposeHypercompact adenine base editorsDepositorInsertTaRGET-ABE3.1
UseCRISPRExpressionMammalianAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only