We narrowed to 13,971 results for: EGFP
-
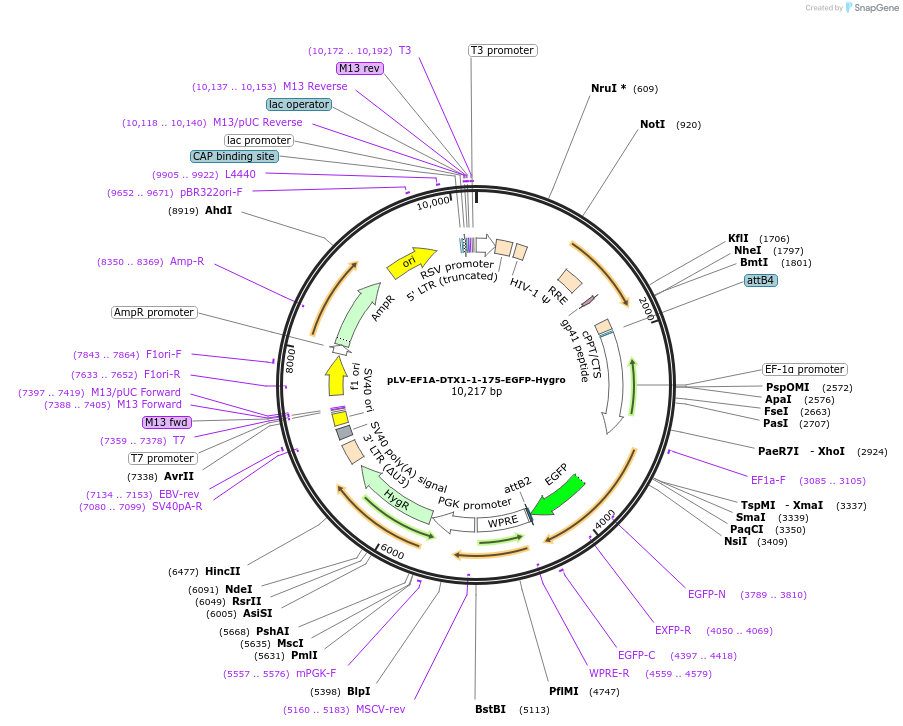
Plasmid#238093PurposeExpression of structurally predicted WWE domain of DTX1 with EGFP on C terminalDepositorInsertDTX1 (DTX1 Human)
UseLentiviralTagsEGFPExpressionMammalianMutationaa 1-175 onlyPromoterEF1alphaAvailable SinceAug. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
pLV-EF1A-DTX4-1-166-EGFP-Hygro
Plasmid#238095PurposeExpression of structurally predicted WWE domain of DTX4 with EGFP on C terminalDepositorInsertDTX4 (DTX4 Human)
UseLentiviralTagsEGFPExpressionMammalianMutationaa 1-166 onlyPromoterEF1alphaAvailable SinceAug. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
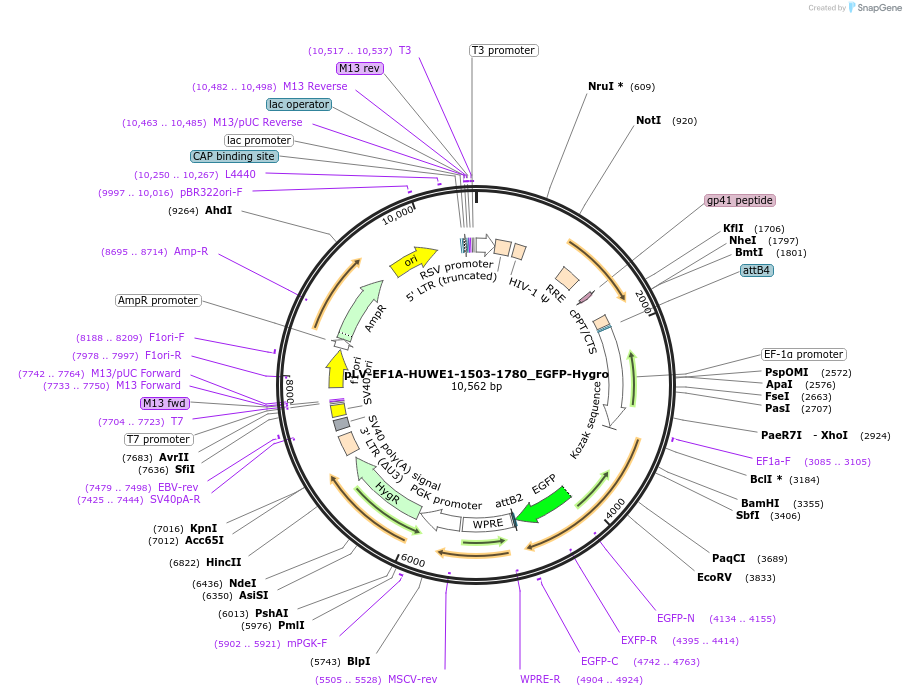
pLV-EF1A-HUWE1-1503-1780_EGFP-Hygro
Plasmid#238096PurposeExpression of structurally predicted WWE domain of HUWE1 with EGFP on C terminalDepositorInsertHUWE1 (HUWE1 Human)
UseLentiviralTagsEGFPExpressionMammalianMutationaa 1-166 onlyPromoterEF1alphaAvailable SinceAug. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
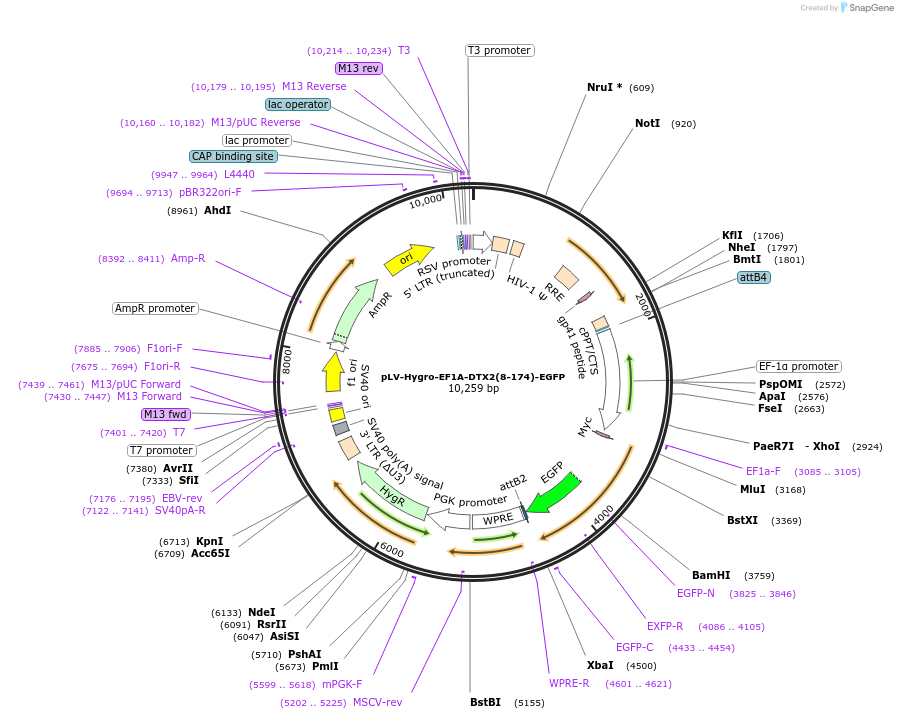
pLV-Hygro-EF1A-DTX2(8-174)-EGFP
Plasmid#196232PurposeEGFP fused to the C-terminus of tandem WWE domains within Deltex2 & a hygromycin resistance cassetteDepositorInsertDTX2(8-174)-EGFP
UseLentiviralTagsEGFP and Myc tagExpressionMammalianPromoterEF1AAvailable SinceAug. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
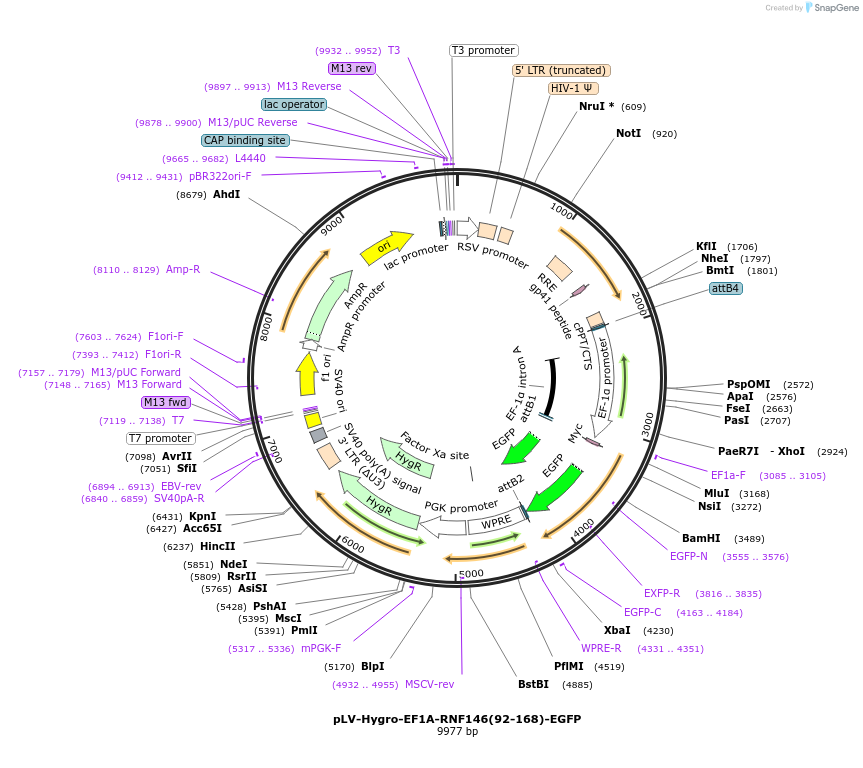
pLV-Hygro-EF1A-RNF146(92-168)-EGFP
Plasmid#196228PurposeEGFP fused to the C-terminus of a WWE domain within RNF146 & a hygromycin resistance cassetteDepositorInsertRNF146(92-168)-EGFP
UseLentiviralTagsEGFP and Myc tagExpressionMammalianPromoterEF1AAvailable SinceAug. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
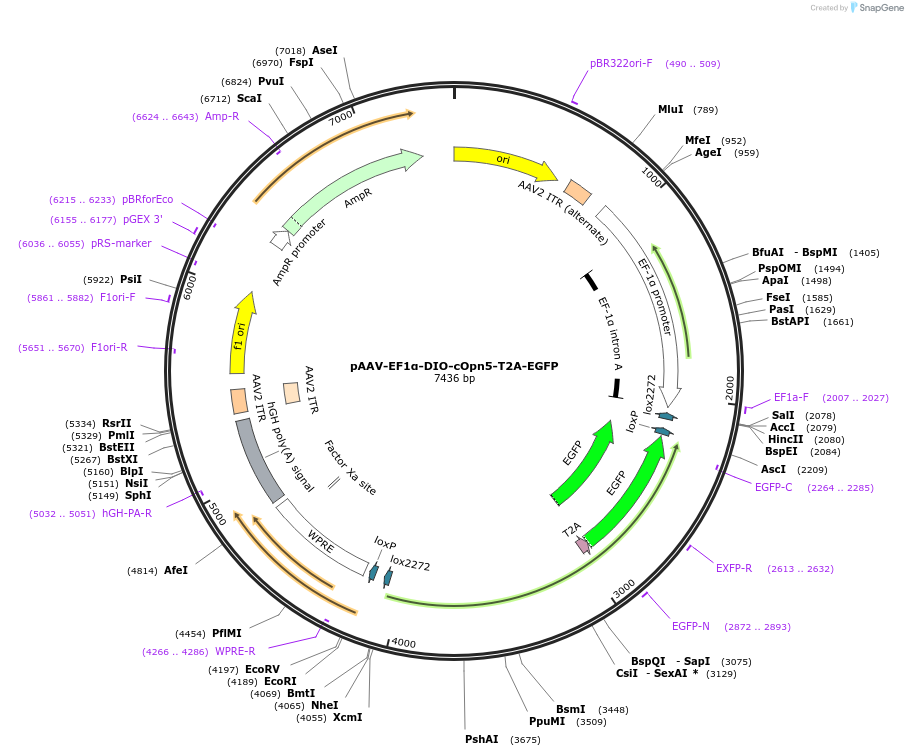
pAAV-EF1α-DIO-cOpn5-T2A-EGFP
Plasmid#237858PurposeThe transfer plasmid for packaging AAV expressing Cre-dependent chicken OPN5 was constructed by inserting the OPN5 coding sequence into the multiple cloning site under the control of EF1α promoterDepositorAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
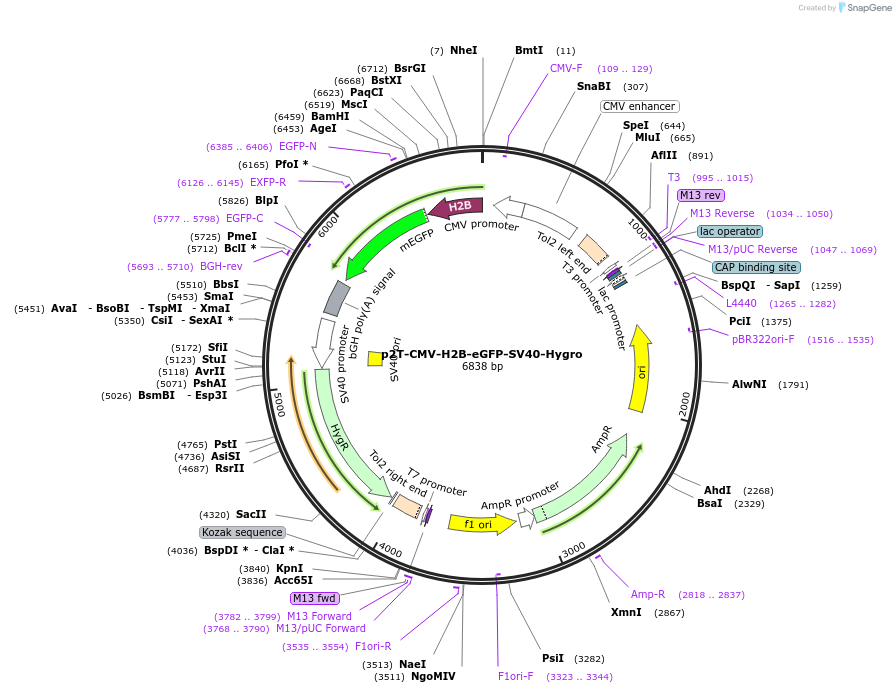
p2T-CMV-H2B-eGFP-SV40-Hygro
Plasmid#240801PurposeConstitutive expression of H2B-eGFPDepositorInsertH2B-eGFP
ExpressionMammalianMutationnonePromoterCMVAvailable SinceAug. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
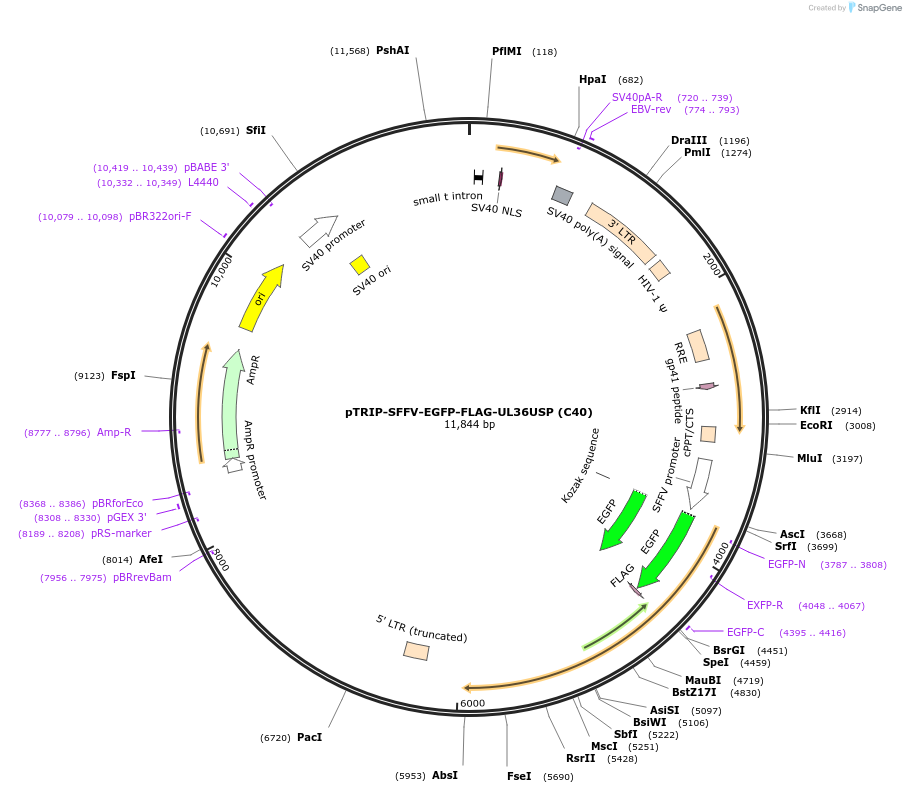
pTRIP-SFFV-EGFP-FLAG-UL36USP (C40)
Plasmid#235548PurposeTo express GFP-FLAG-UL36USP (C40) - region 25-520 of UL36DepositorInsertUL36USP (C40)
UseLentiviralTagsEGFP-FLAGMutationN-terminal part of UL36 starting at the second AT…Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
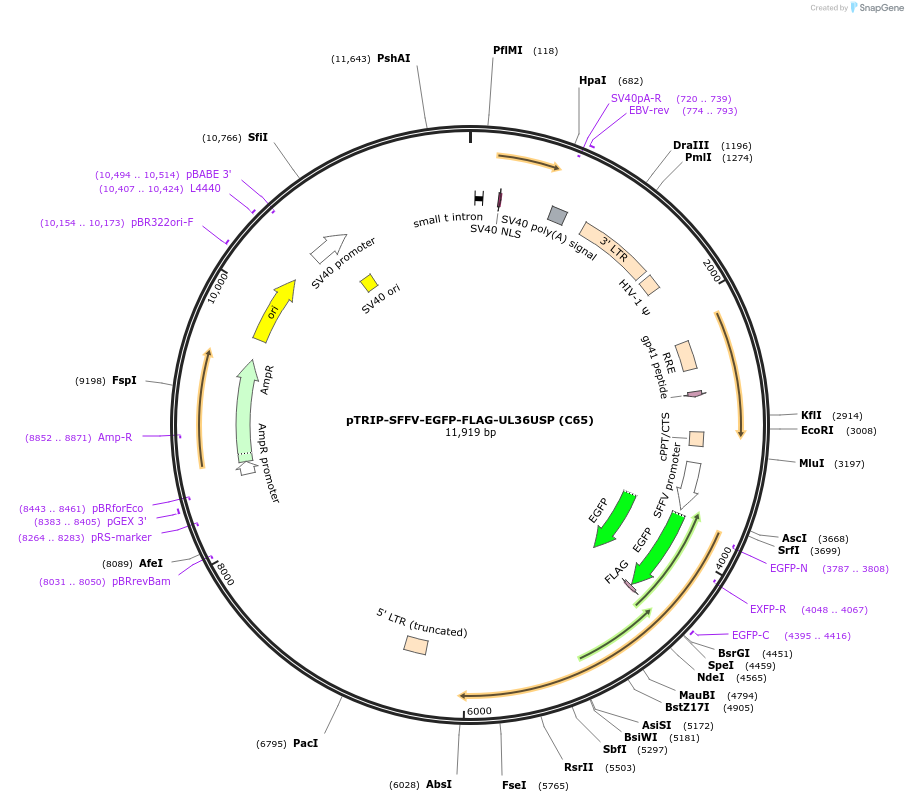
pTRIP-SFFV-EGFP-FLAG-UL36USP (C65)
Plasmid#235549PurposeTo express GFP-FLAG-UL36USP (C65) - region 1-520 of UL36DepositorInsertUL36USP (C65)
UseLentiviralTagsEGFP-FLAGMutationN-terminal part of UL36 starting at the first ATG…Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
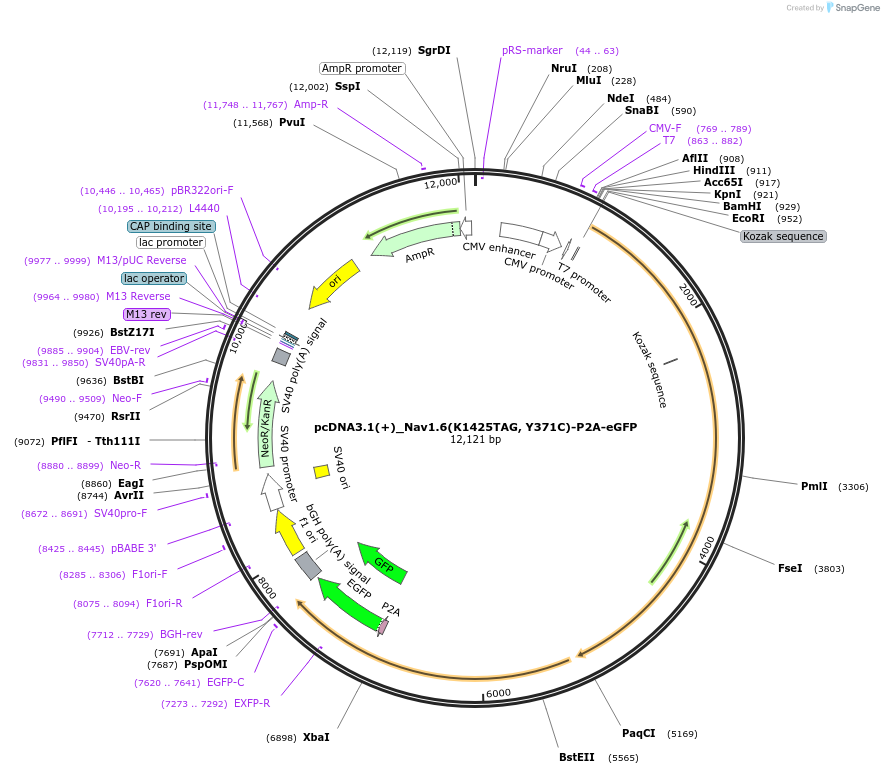
pcDNA3.1(+)_Nav1.6(K1425TAG, Y371C)-P2A-eGFP
Plasmid#239769PurposeExpresses independantly TTX-resistant (Y371C) variant of mouse Nav1.6 with a TAG codon at position 1425 and eGFP in mammalian cells.DepositorAvailable SinceJuly 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
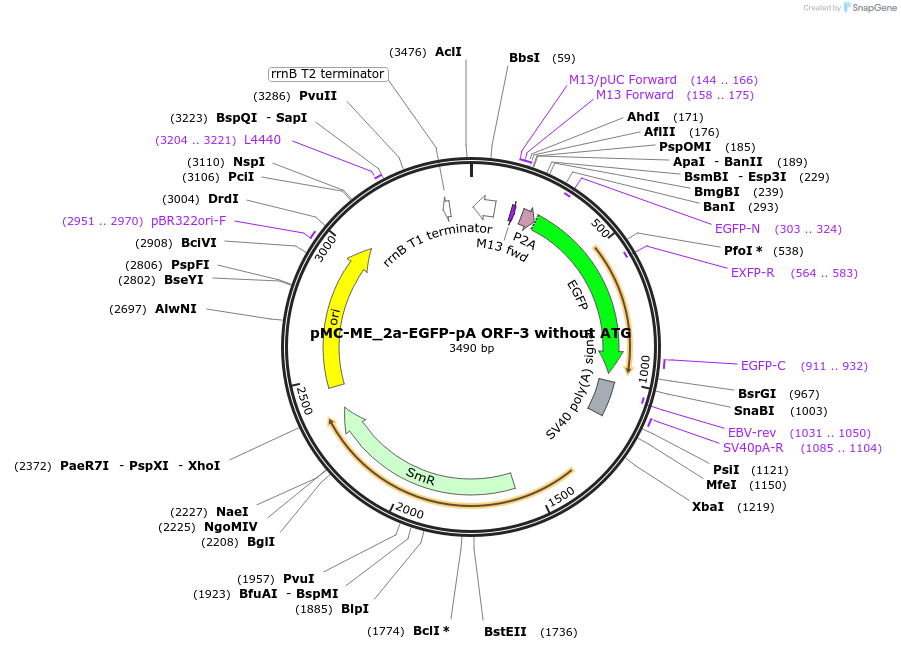
pMC-ME_2a-EGFP-pA ORF-3 without ATG
Plasmid#200519Purposemini circle middle entry plasmid for 2a-eGFP-pA ORF3 (without ATG) with flanking BsaI cut sitesDepositorInserteGFP
UseMini-goldenAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
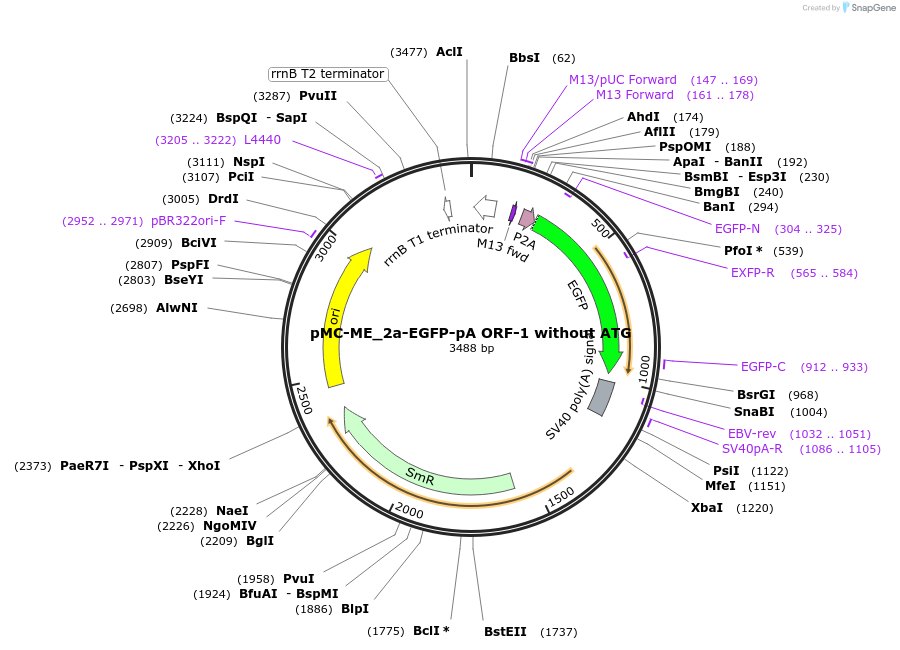
pMC-ME_2a-EGFP-pA ORF-1 without ATG
Plasmid#200517Purposemini circle middle entry plasmid for 2a-eGFP-pA ORF1 (without ATG) with flanking BsaI cut sitesDepositorInserteGFP
UseMini-goldenAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
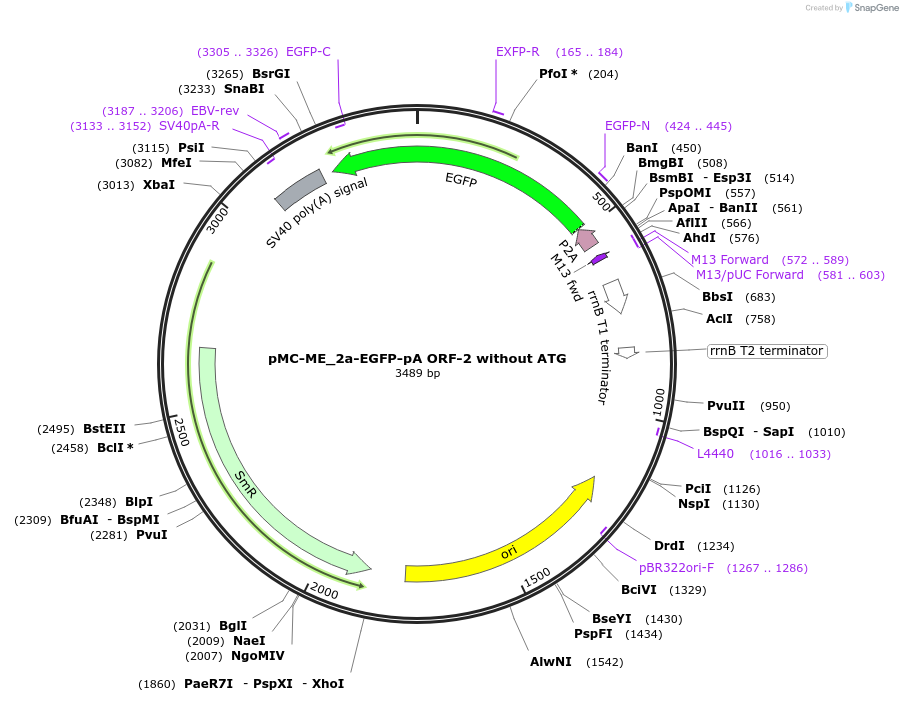
pMC-ME_2a-EGFP-pA ORF-2 without ATG
Plasmid#200518Purposemini circle middle entry plasmid for 2a-eGFP-pA ORF2 (without ATG) with flanking BsaI cut sitesDepositorInserteGFP
UseMini-goldenAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
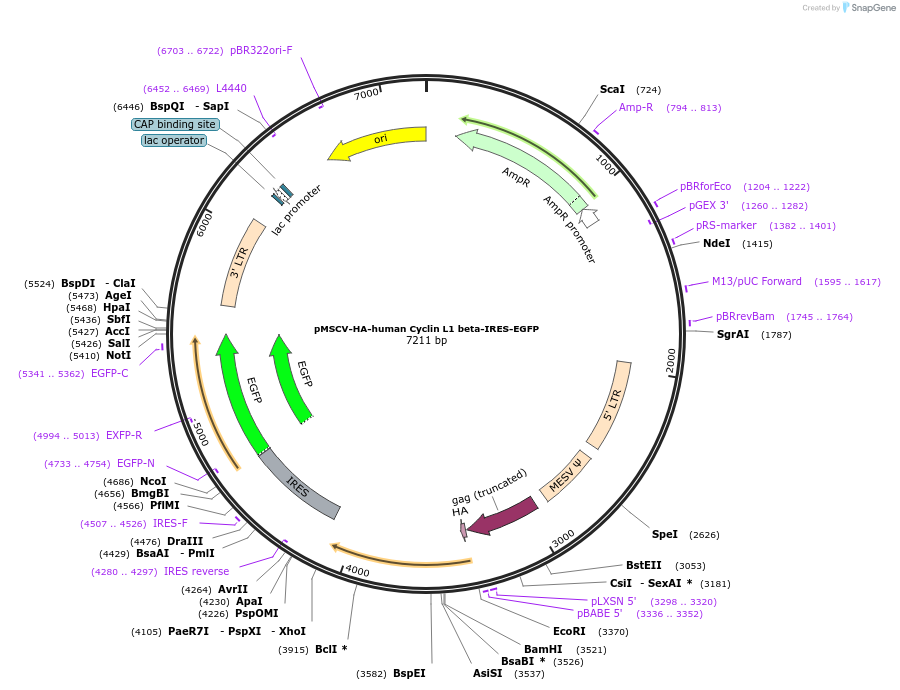
pMSCV-HA-human Cyclin L1 beta-IRES-EGFP
Plasmid#240141PurposeExpress Cyclin L1 betaDepositorAvailable SinceJune 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
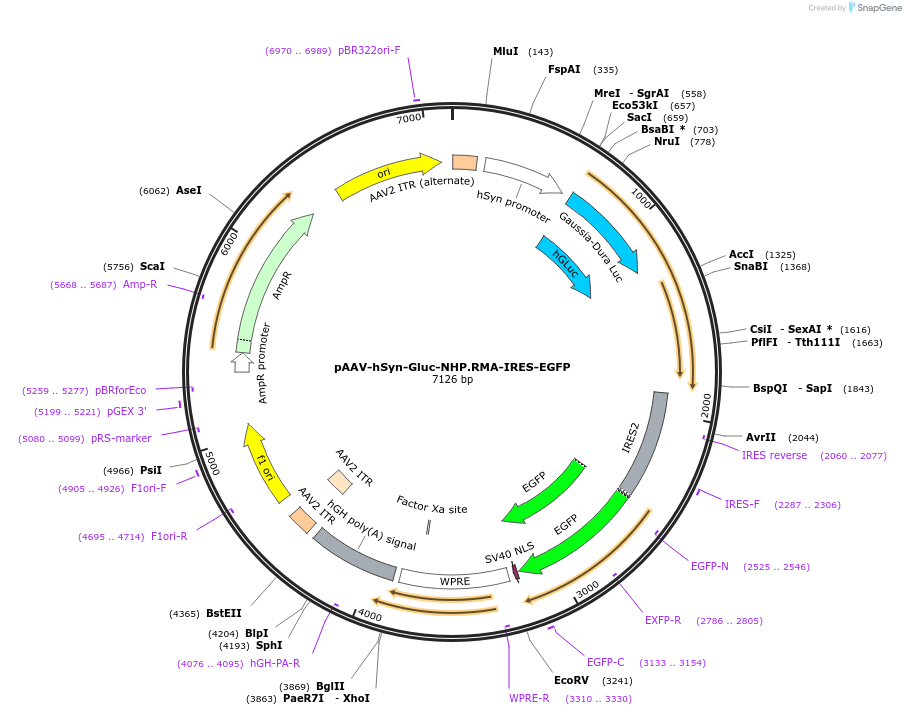
pAAV-hSyn-Gluc-NHP.RMA-IRES-EGFP
Plasmid#227886PurposeExpresses Gluc-NHP.RMA and EGFP under the hSyn promoter. Gluc-NHP.RMA for monitoring neuronal transduction.DepositorInsertsGaussia luciferase fused to macaque Fc
IRES-EGFP
UseAAV and LuciferaseTagsGluc and IgG1 FcExpressionMammalianAvailable SinceJune 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
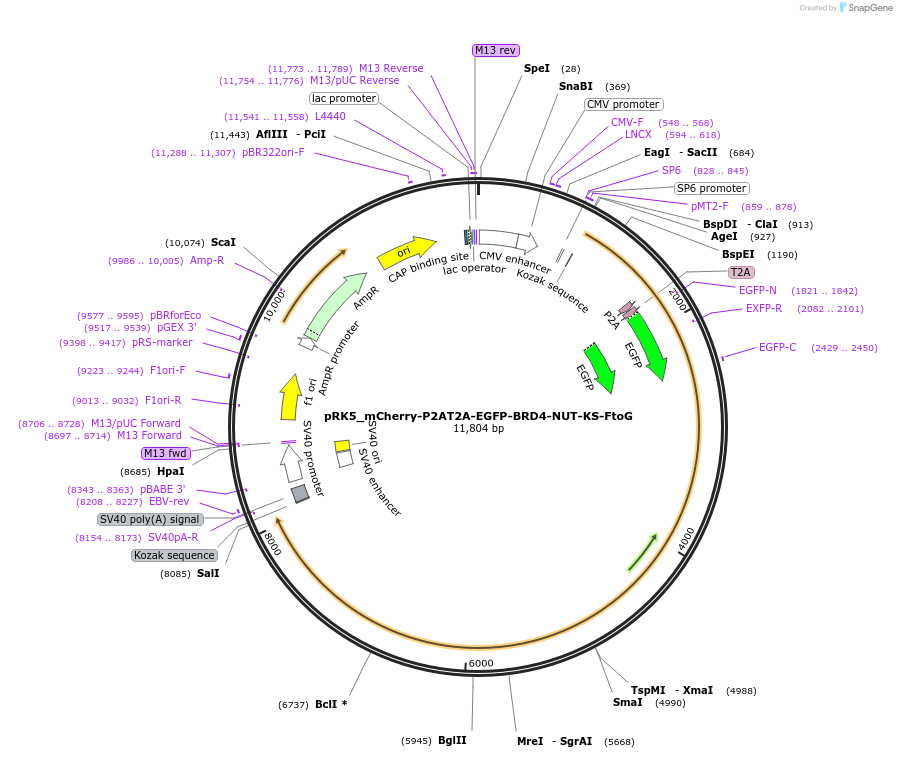
pRK5_mCherry-P2AT2A-EGFP-BRD4-NUT-KS-FtoG
Plasmid#238283PurposeFor overexpression of mCherry-P2AT2A-EGFP-BRD4-NUT-KS-FtoGDepositorInsertmCherry-P2AT2A-EGFP-BRD4-NUT-KS-FtoG
ExpressionMammalianAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
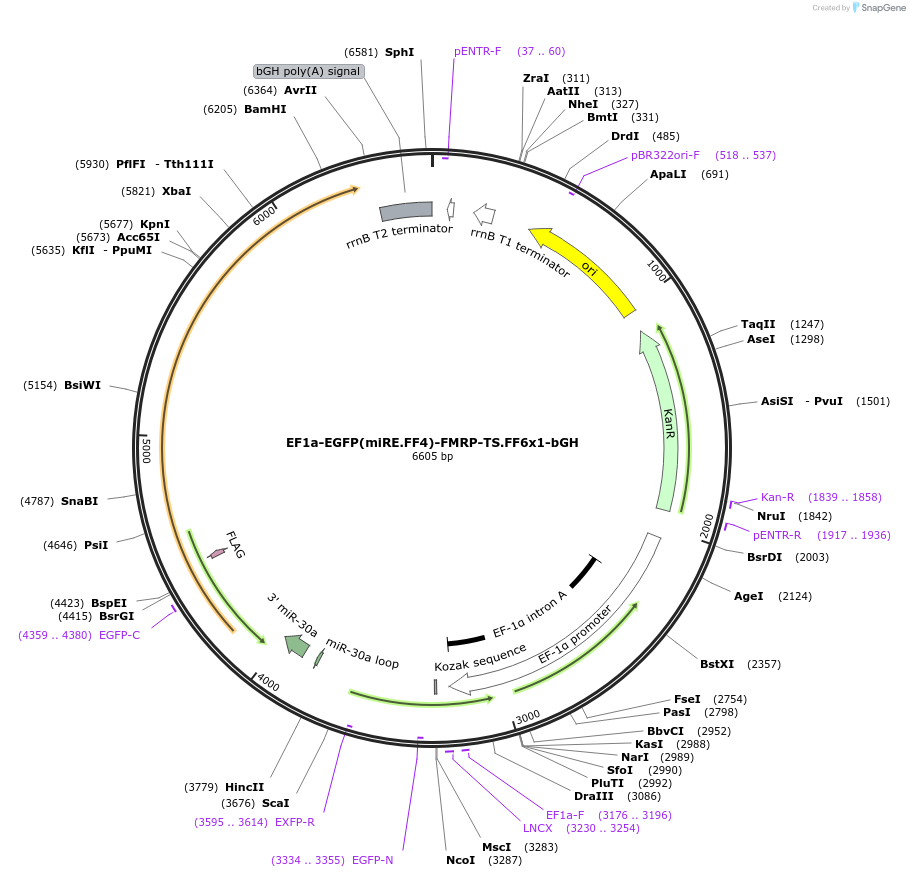
EF1a-EGFP(miRE.FF4)-FMRP-TS.FF6x1-bGH
Plasmid#235266PurposeComMAND EGFP-FMRP open-loop circuitDepositorInsertEGFP-Fmr1 (Fmr1 Mouse)
UseSynthetic BiologyTagsFLAGExpressionMammalianPromoterEF1a (human EF1a)Available SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
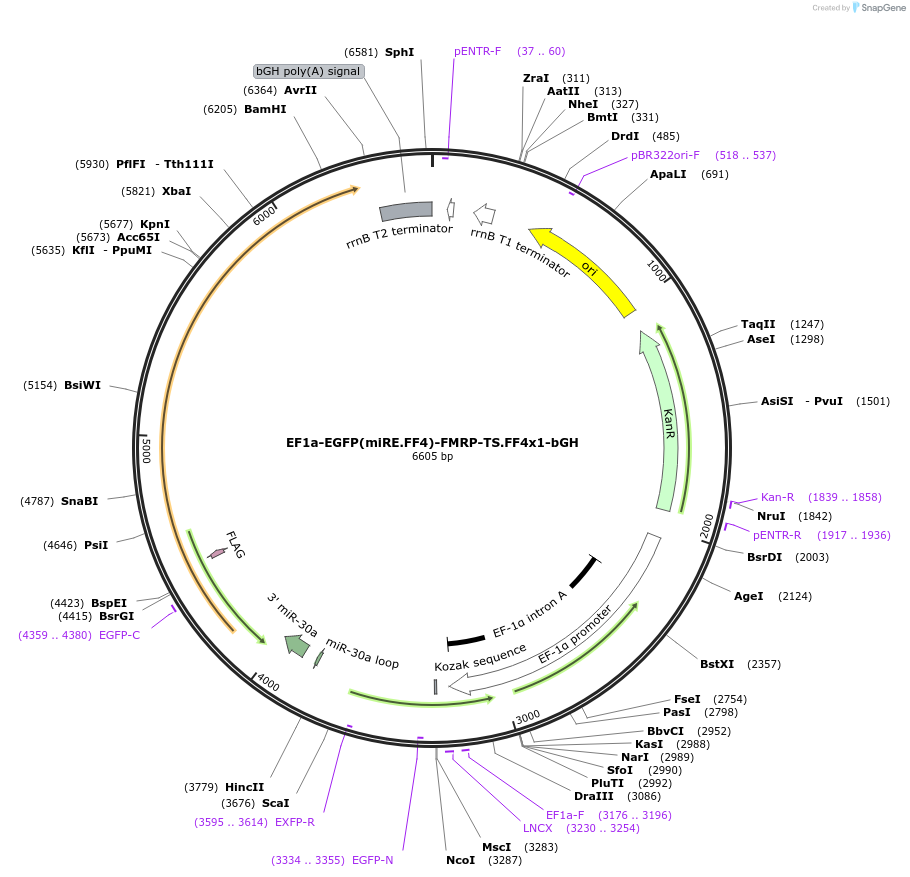
EF1a-EGFP(miRE.FF4)-FMRP-TS.FF4x1-bGH
Plasmid#235267PurposeComMAND EGFP-FMRP closed-loop circuitDepositorInsertEGFP-Fmr1 (Fmr1 Mouse)
UseSynthetic BiologyTagsFLAGExpressionMammalianPromoterEF1a (human EF1a)Available SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
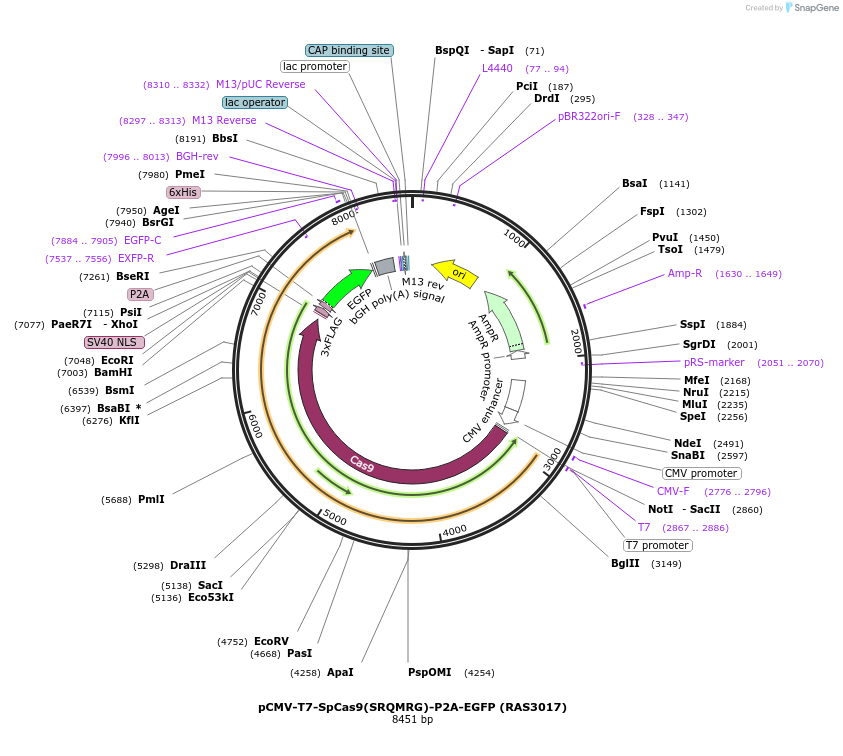
pCMV-T7-SpCas9(SRQMRG)-P2A-EGFP (RAS3017)
Plasmid#223103PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with SRQMRG amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(SRQMRG)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(SRQMRG)=D1135S, S1136R, G1218Q, E1219M, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
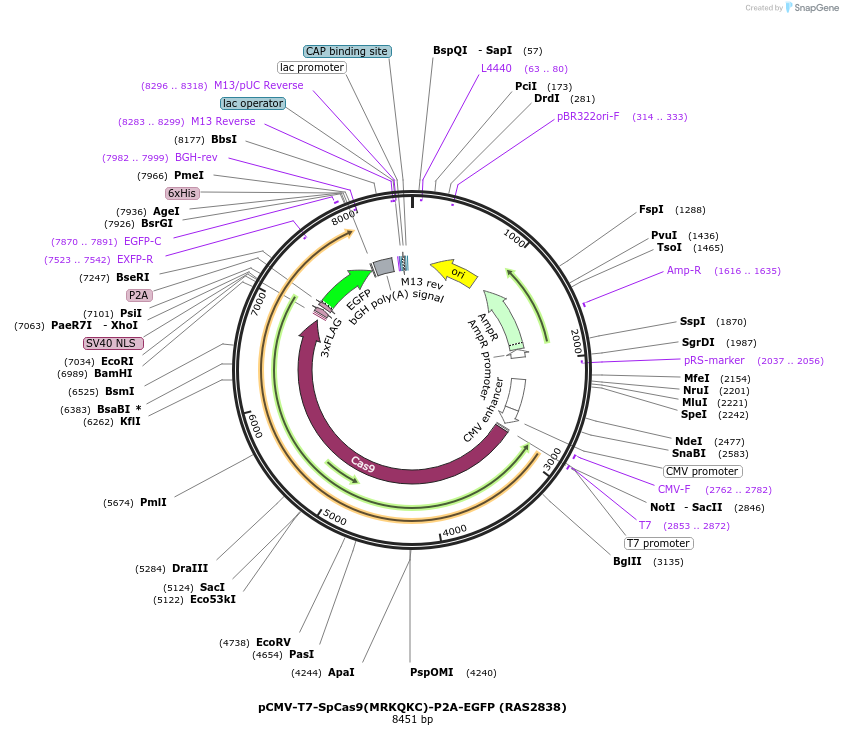
pCMV-T7-SpCas9(MRKQKC)-P2A-EGFP (RAS2838)
Plasmid#223104PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MRKQKC amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MRKQKC)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MRKQKC)=D1135M, S1136R, G1218K, E1219Q, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
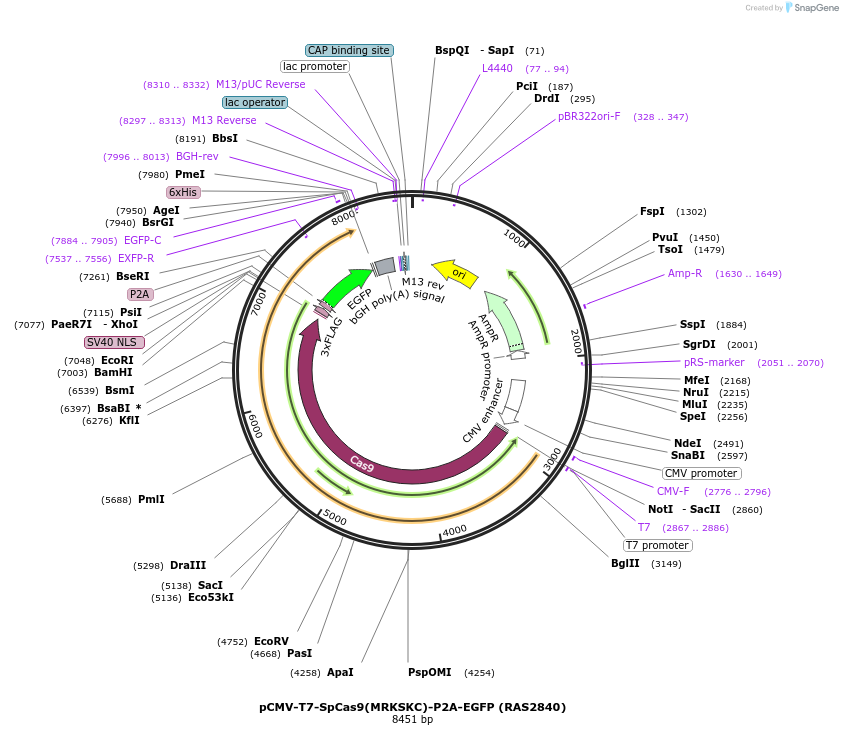
pCMV-T7-SpCas9(MRKSKC)-P2A-EGFP (RAS2840)
Plasmid#223105PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MRKSKC amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MRKSKC)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MRKSKC)=D1135M, S1136R, G1218K, E1219S, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
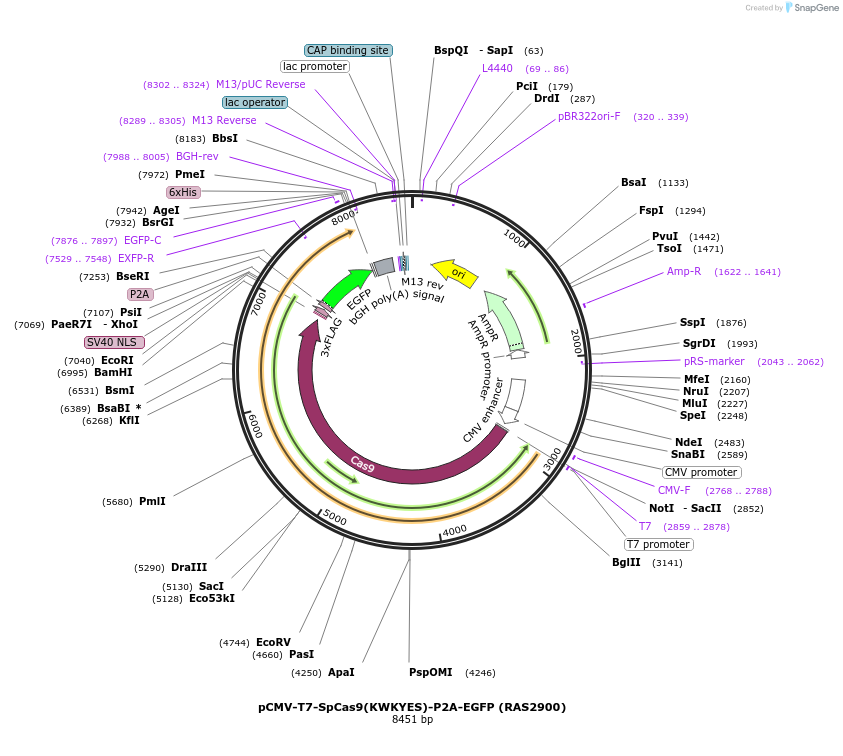
pCMV-T7-SpCas9(KWKYES)-P2A-EGFP (RAS2900)
Plasmid#223095PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KWKYES amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KWKYES)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KWKYES)=D1135K, S1136W, G1218K, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
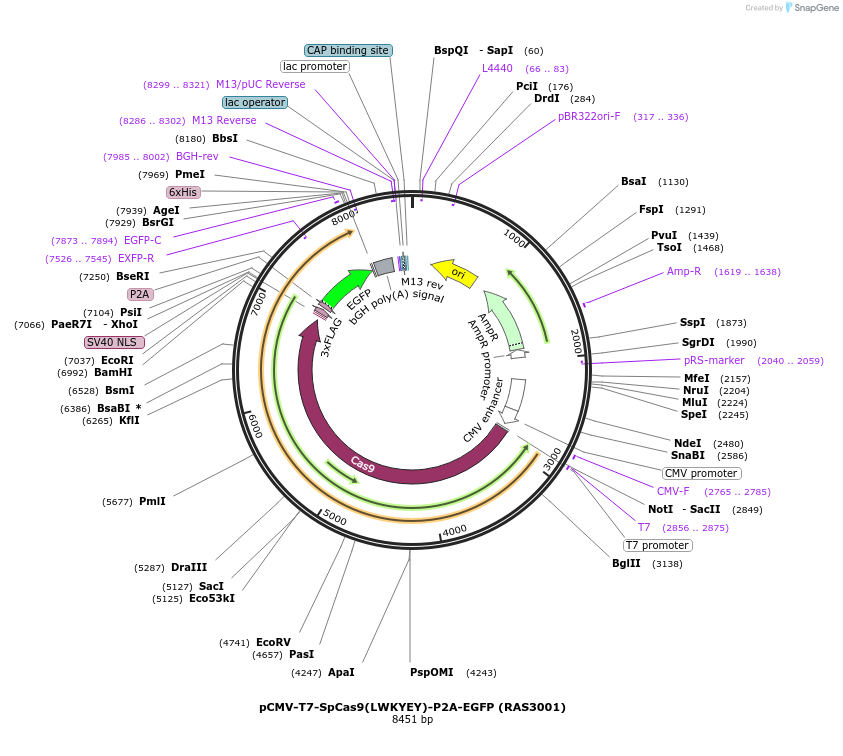
pCMV-T7-SpCas9(LWKYEY)-P2A-EGFP (RAS3001)
Plasmid#223096PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with LWKYEY amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(LWKYEY)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(LWKYEY)=D1135L, S1136W, G1218K, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
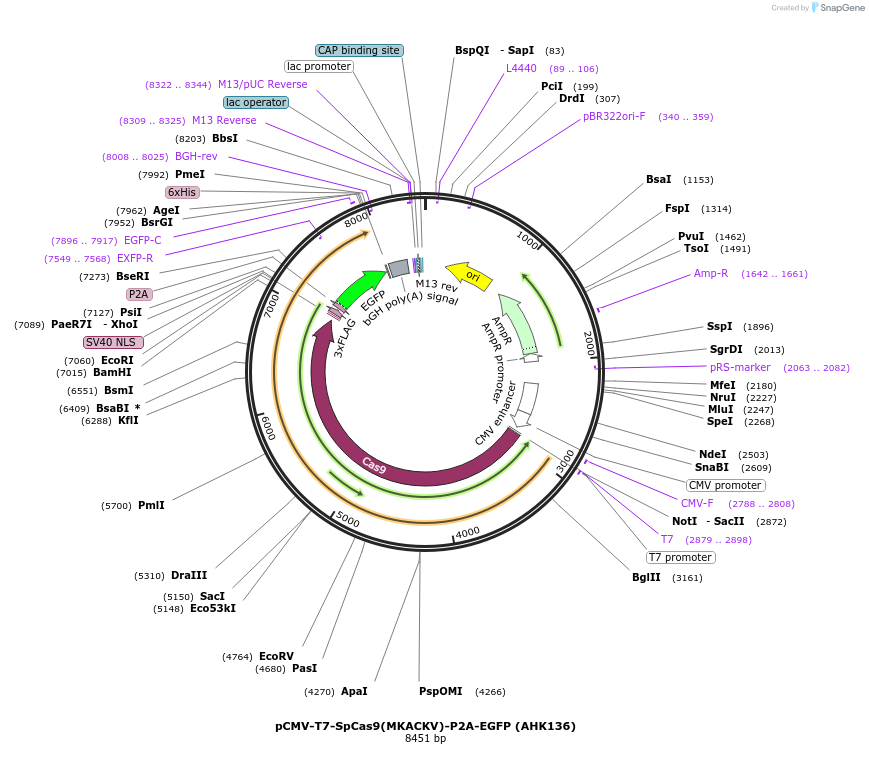
pCMV-T7-SpCas9(MKACKV)-P2A-EGFP (AHK136)
Plasmid#223097PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MKACKV amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MKACKV)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MKACKV)=D1135M, S1136K, G1218A, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
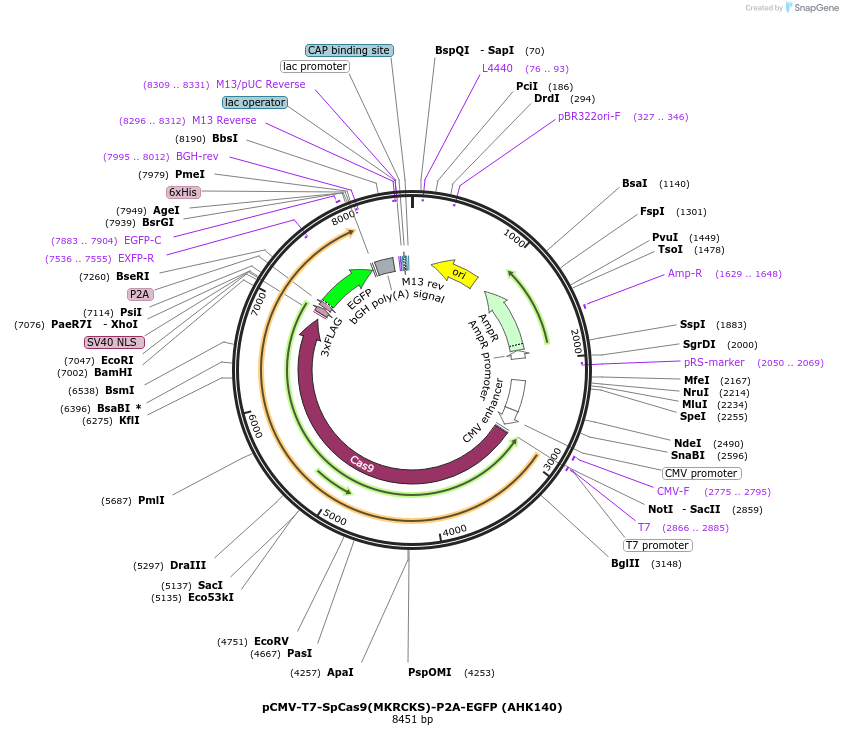
pCMV-T7-SpCas9(MKRCKS)-P2A-EGFP (AHK140)
Plasmid#223098PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MKRCKS amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MKRCKS)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MKRCKS)=D1135M, S1136K, G1218R, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
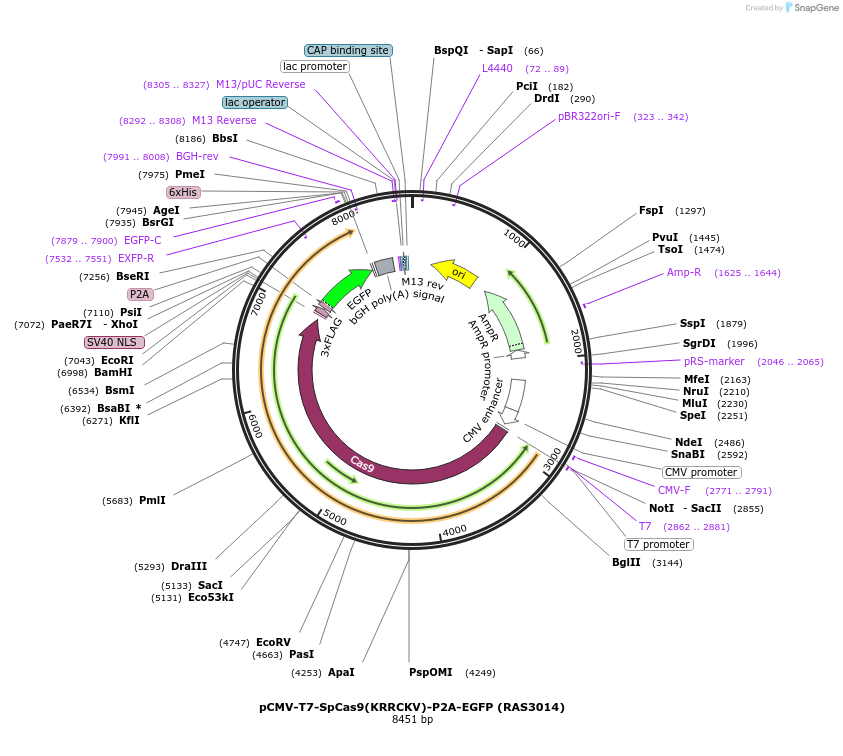
pCMV-T7-SpCas9(KRRCKV)-P2A-EGFP (RAS3014)
Plasmid#223099PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KRRCKV amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KRRCKV)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KRRCKV)=D1135K, S1136R, G1218R, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
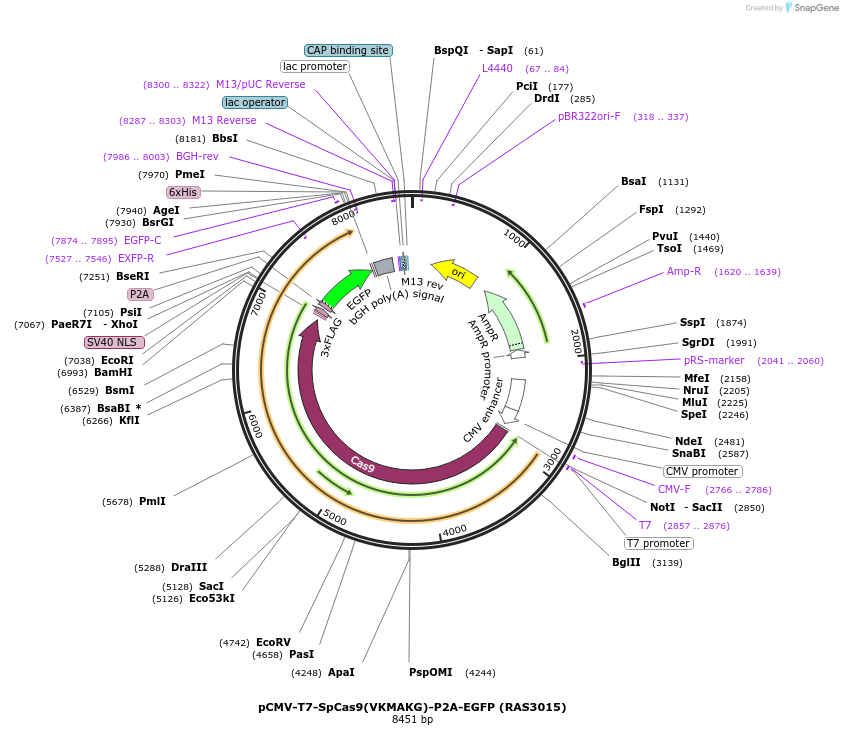
pCMV-T7-SpCas9(VKMAKG)-P2A-EGFP (RAS3015)
Plasmid#223100PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with VKMAKG amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(VKMAKG)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(VKMAKG)=D1135V, S1136K, G1218M, E1219A, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
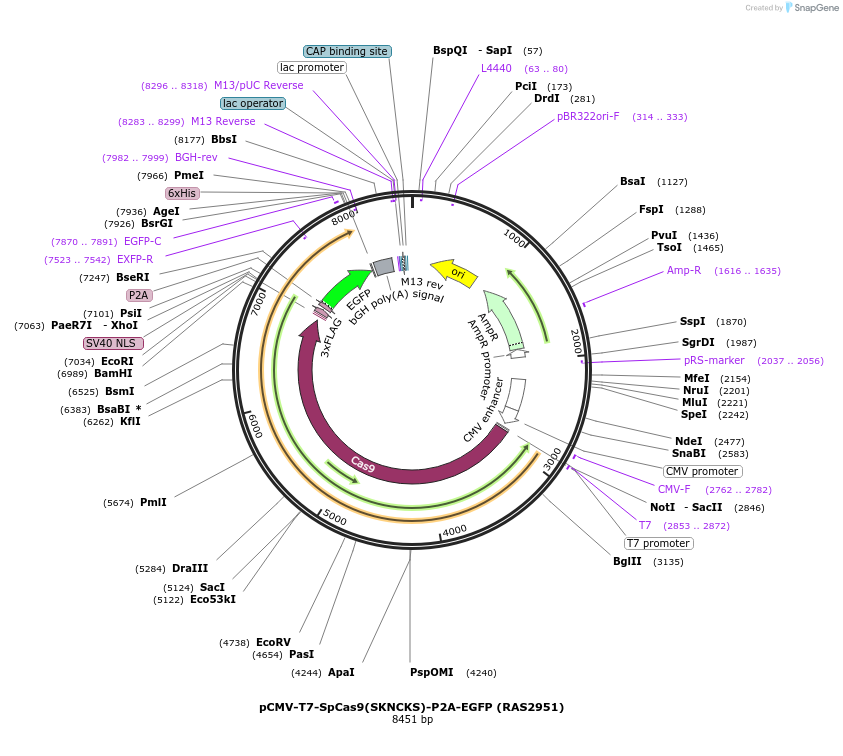
pCMV-T7-SpCas9(SKNCKS)-P2A-EGFP (RAS2951)
Plasmid#223101PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with SKNCKS amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(SKNCKS)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(SKNCKS)=D1135S, S1136K, G1218N, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
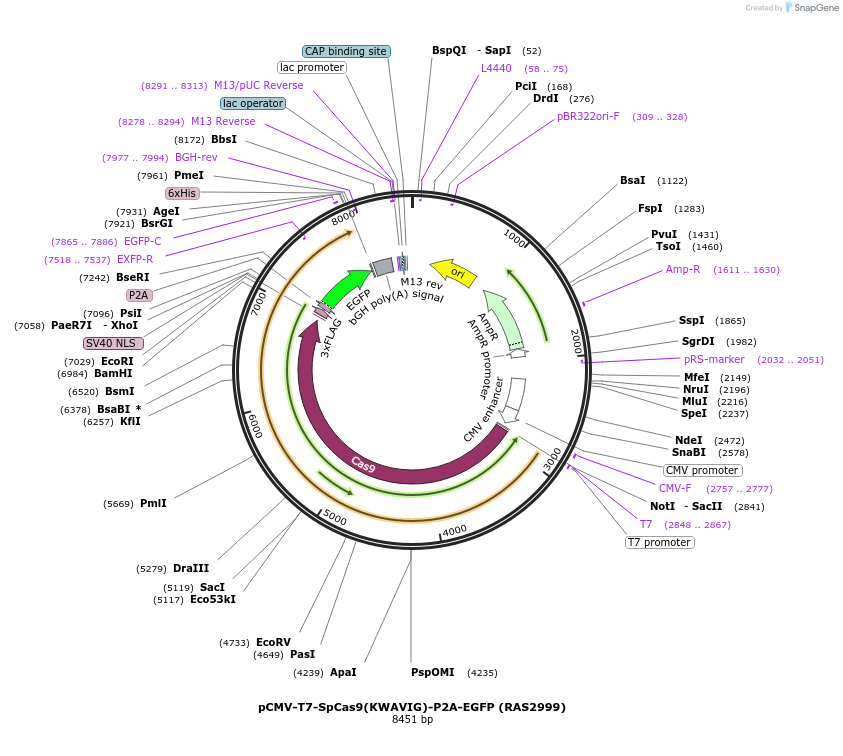
pCMV-T7-SpCas9(KWAVIG)-P2A-EGFP (RAS2999)
Plasmid#223088PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KWAVIG amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KWAVIG)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KWAVIG)=D1135K, S1136W, G1218A, E1219V, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
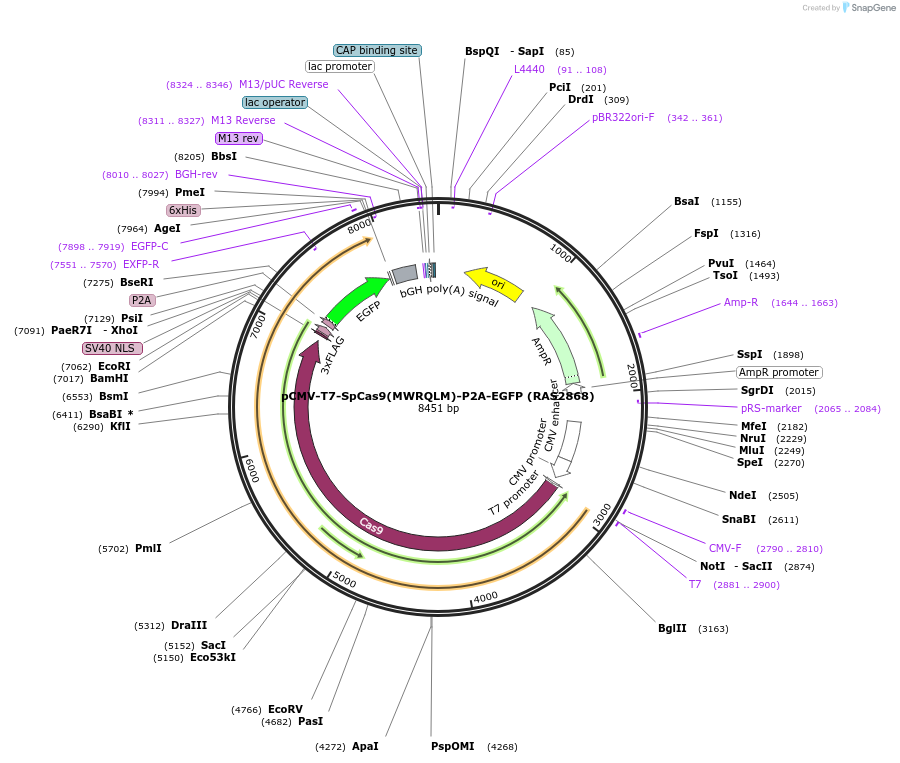
pCMV-T7-SpCas9(MWRQLM)-P2A-EGFP (RAS2868)
Plasmid#223089PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MWRQLM amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MWRQLM)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MWRQLM)=D1135M, S1136W, G1218R, E1219Q, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
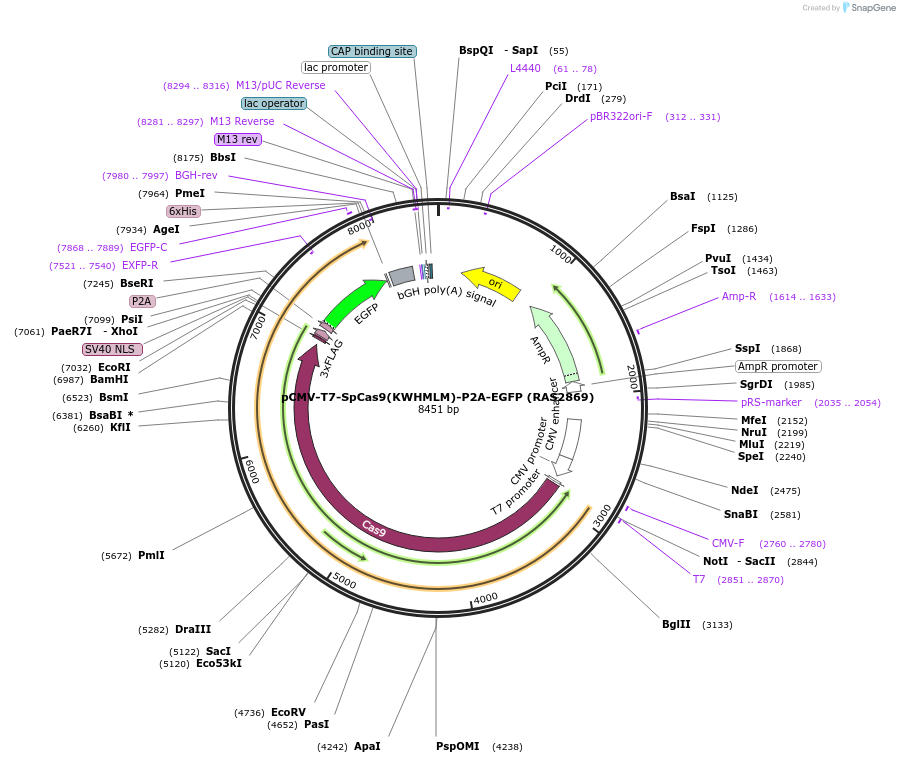
pCMV-T7-SpCas9(KWHMLM)-P2A-EGFP (RAS2869)
Plasmid#223090PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KWHMLM amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KWHMLM)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KWHMLM)=D1135K, S1136W, G1218H, E1219M, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
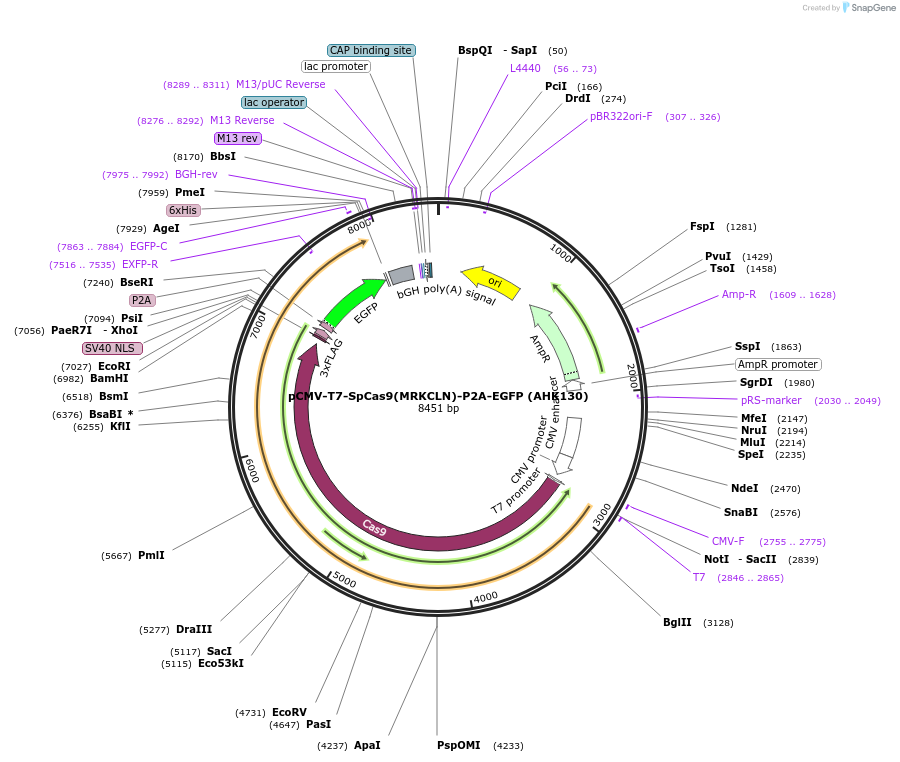
pCMV-T7-SpCas9(MRKCLN)-P2A-EGFP (AHK130)
Plasmid#223091PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MRKCLN amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MRKCLN)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MRKCLN)=D1135M, S1136R, G1218K, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
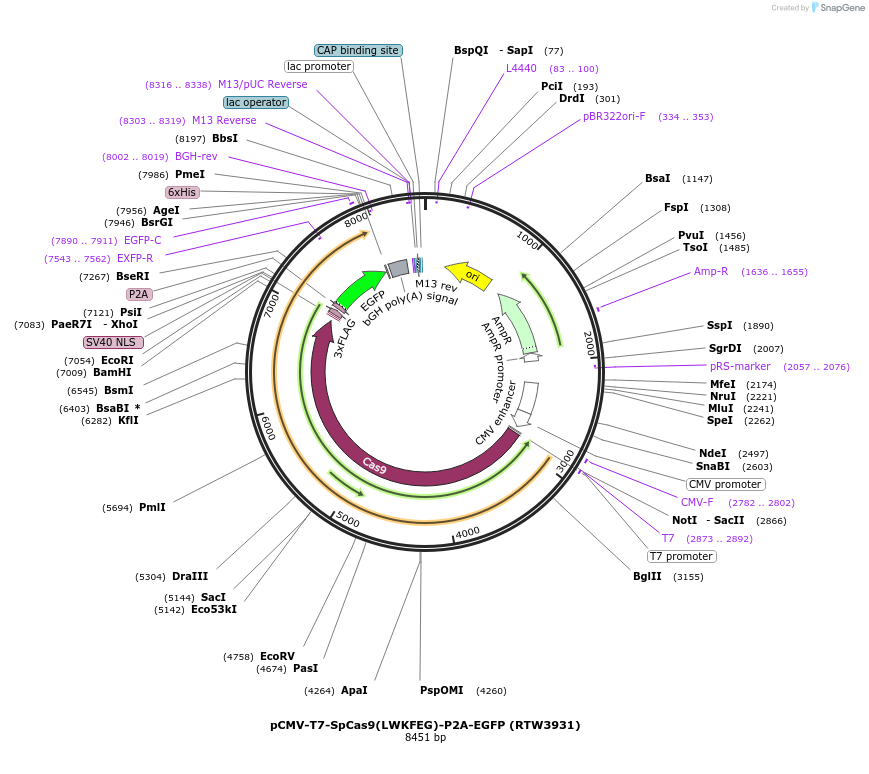
pCMV-T7-SpCas9(LWKFEG)-P2A-EGFP (RTW3931)
Plasmid#223092PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with LWKFEG amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(LWKFEG)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(LWKFEG)=D1135L, S1136W, G1218K, E1219F, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
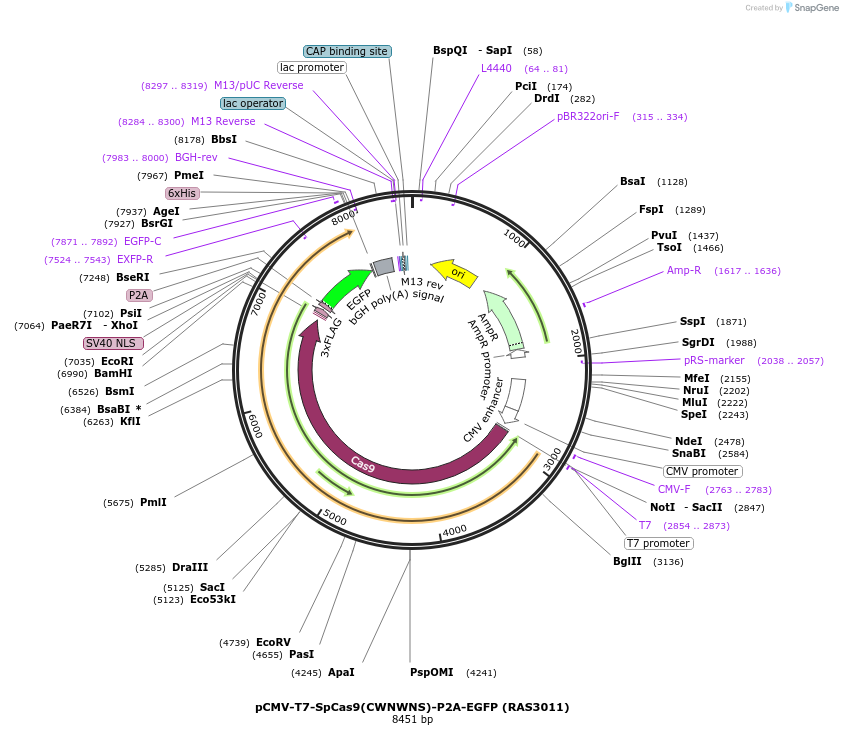
pCMV-T7-SpCas9(CWNWNS)-P2A-EGFP (RAS3011)
Plasmid#223093PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with CWNWNS amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(CWNWNS)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(CWNWNS)=D1135C, S1136W, G1218N, E1219W, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
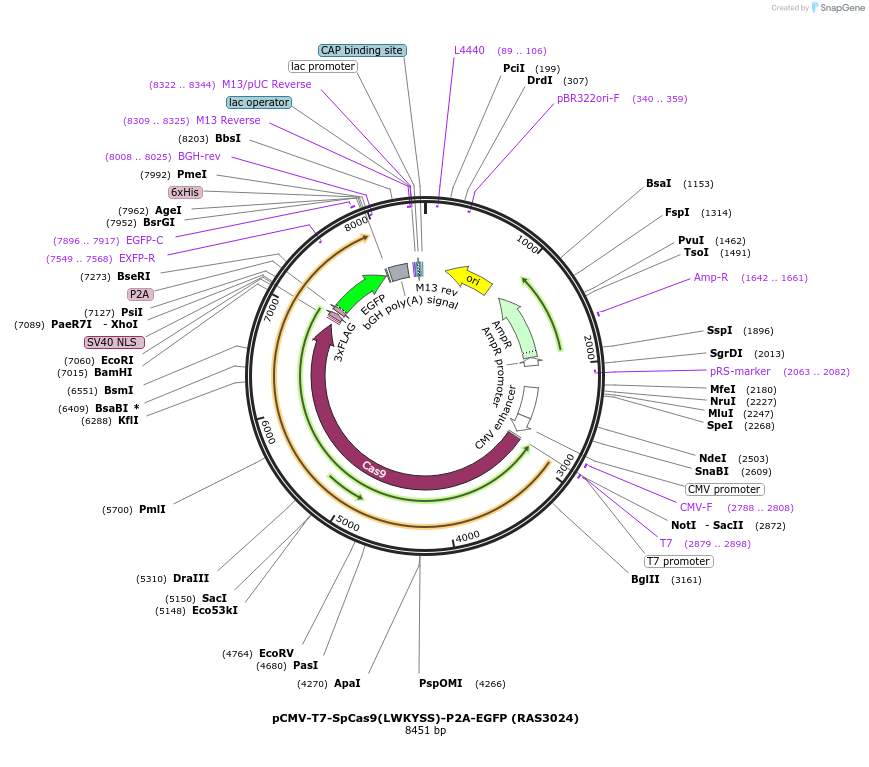
pCMV-T7-SpCas9(LWKYSS)-P2A-EGFP (RAS3024)
Plasmid#223080PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with LWKYSS amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(LWKYSS)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(LWKYSS)=D1135L, S1136W, G1218K, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
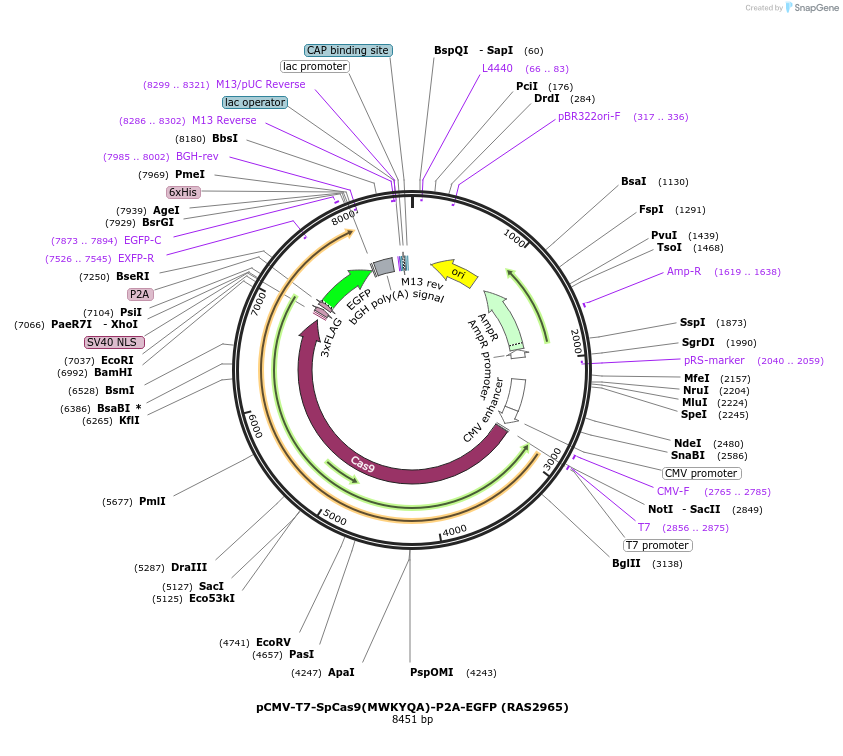
pCMV-T7-SpCas9(MWKYQA)-P2A-EGFP (RAS2965)
Plasmid#223081PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MWKYQA amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MWKYQA)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MWKYQA)=D1135M, S1136W, G1218K, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
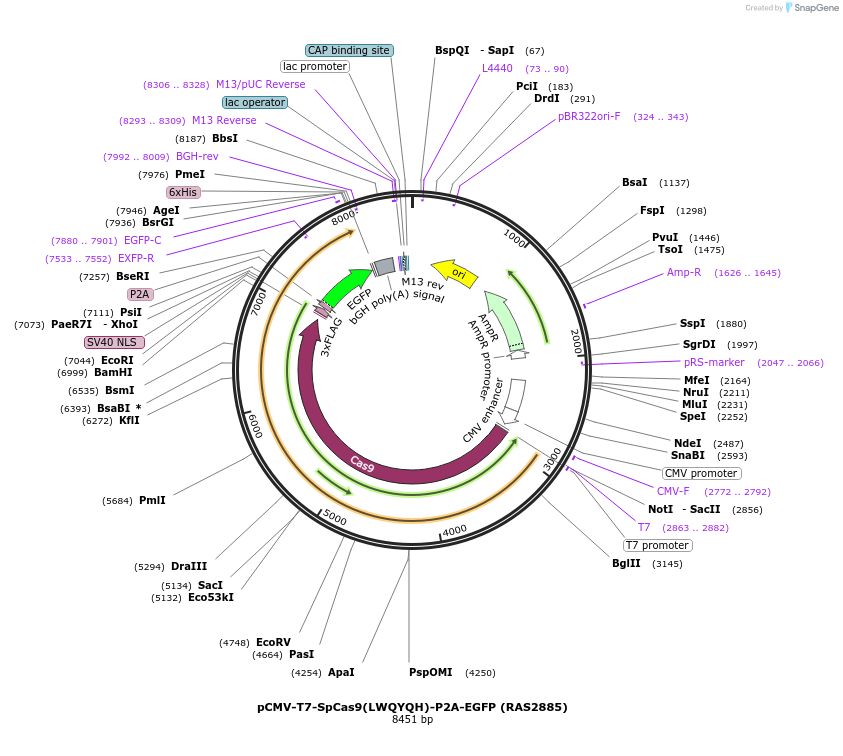
pCMV-T7-SpCas9(LWQYQH)-P2A-EGFP (RAS2885)
Plasmid#223082PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with LWQYQH amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(LWQYQH)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(LWQYQH)=D1135L, S1136W, G1218Q, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
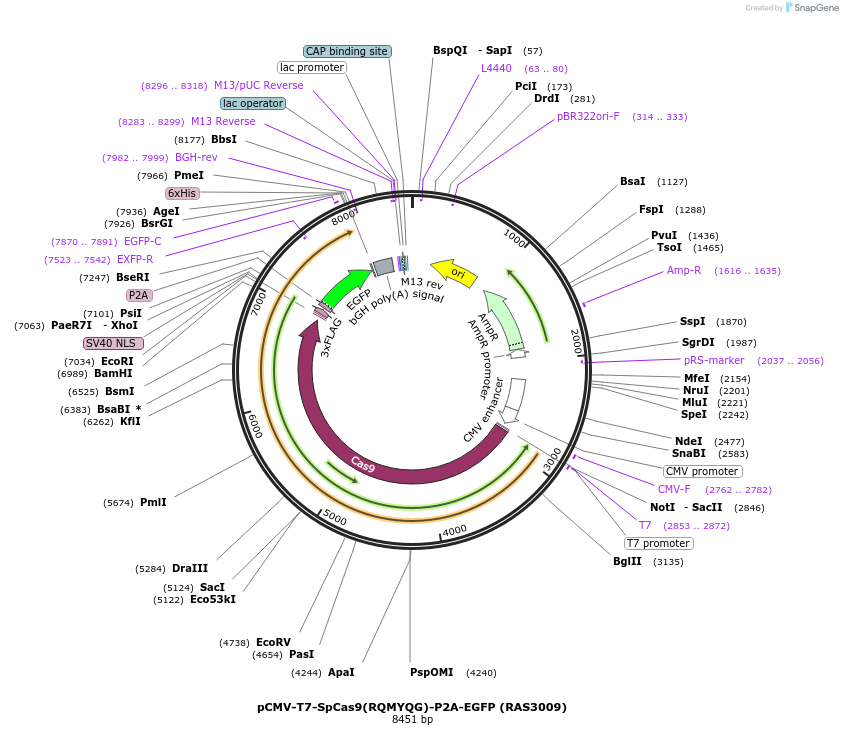
pCMV-T7-SpCas9(RQMYQG)-P2A-EGFP (RAS3009)
Plasmid#223083PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with RQMYQG amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(RQMYQG)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(RQMYQG)=D1135R, S1136Q, G1218M, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
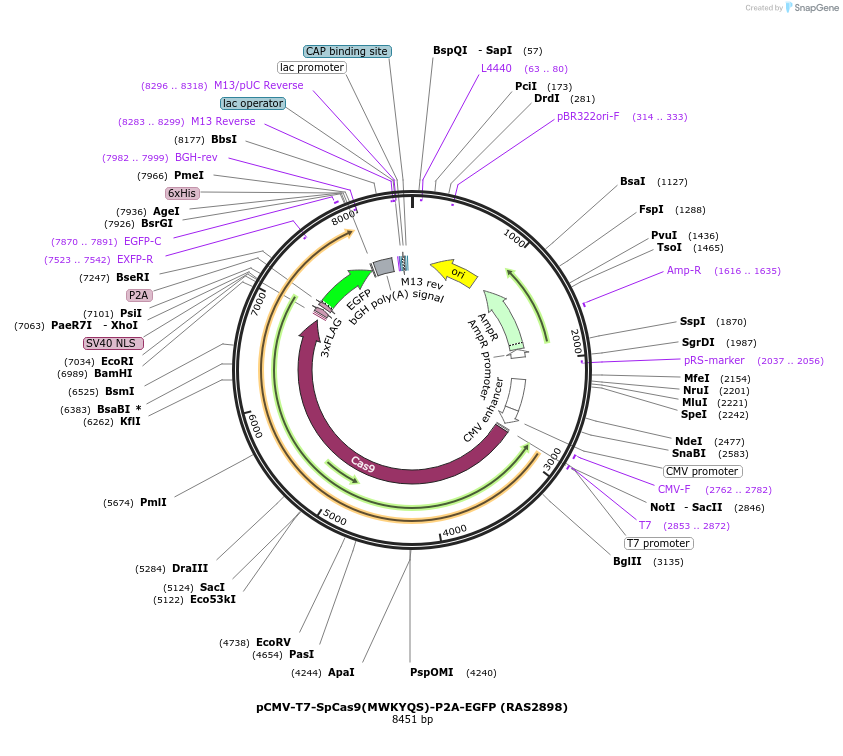
pCMV-T7-SpCas9(MWKYQS)-P2A-EGFP (RAS2898)
Plasmid#223084PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MWKYQS amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MWKYQS)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MWKYQS)=D1135M, S1136W, G1218K, E1219Y, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
pCMV-T7-SpCas9(LWSSLY)-P2A-EGFP (RAS2997)
Plasmid#223085PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with LWSSLY amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(LWSSLY)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(LWSSLY)=D1135L, S1136W, G1218S, E1219S, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
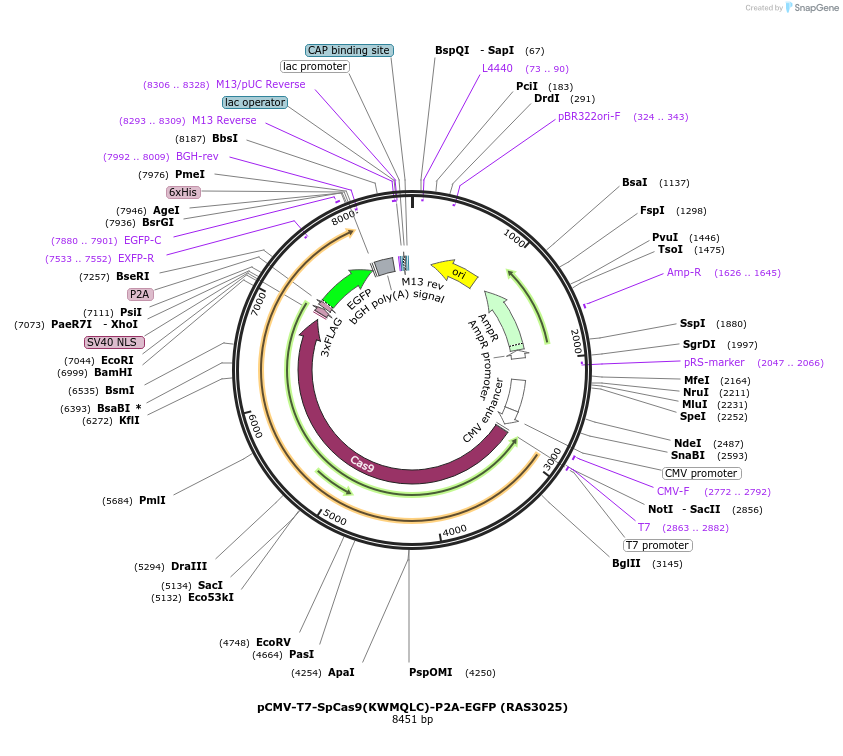
pCMV-T7-SpCas9(KWMQLC)-P2A-EGFP (RAS3025)
Plasmid#223086PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KWMQLC amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KWMQLC)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KWMQLC)=D1135K, S1136W, G1218M, E1219Q, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
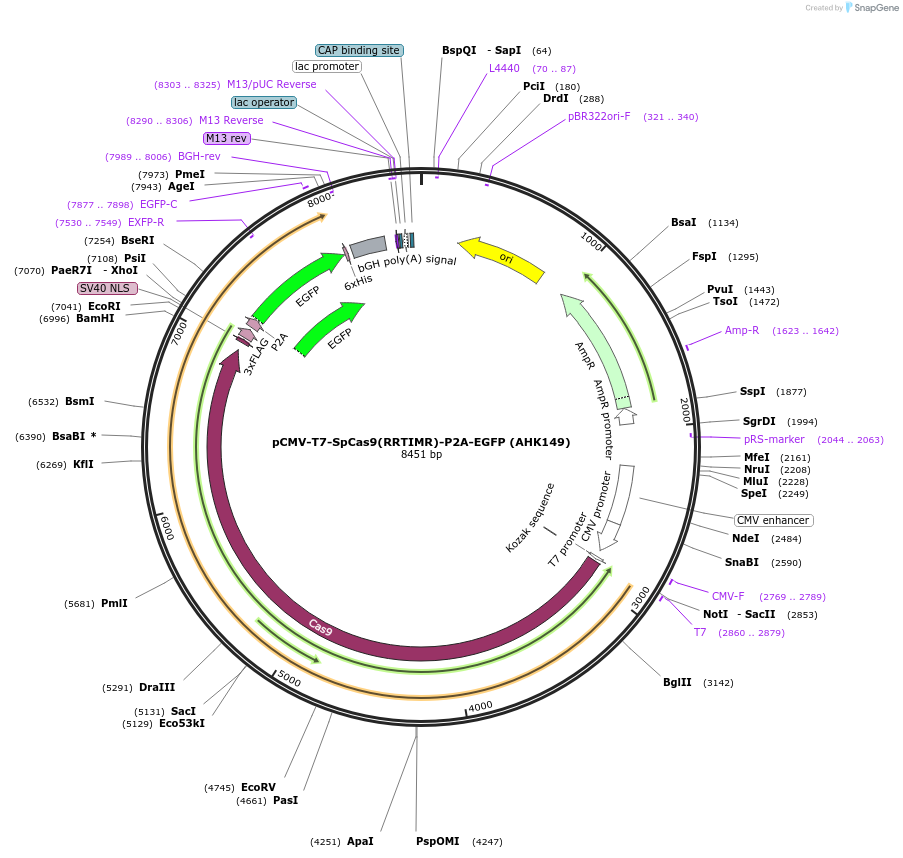
pCMV-T7-SpCas9(RRTIMR)-P2A-EGFP (AHK149)
Plasmid#223071PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with RRTIMR amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(RRTIMR)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(RRTIMR)=D1135R, S1136R, G1218T, E1219I, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
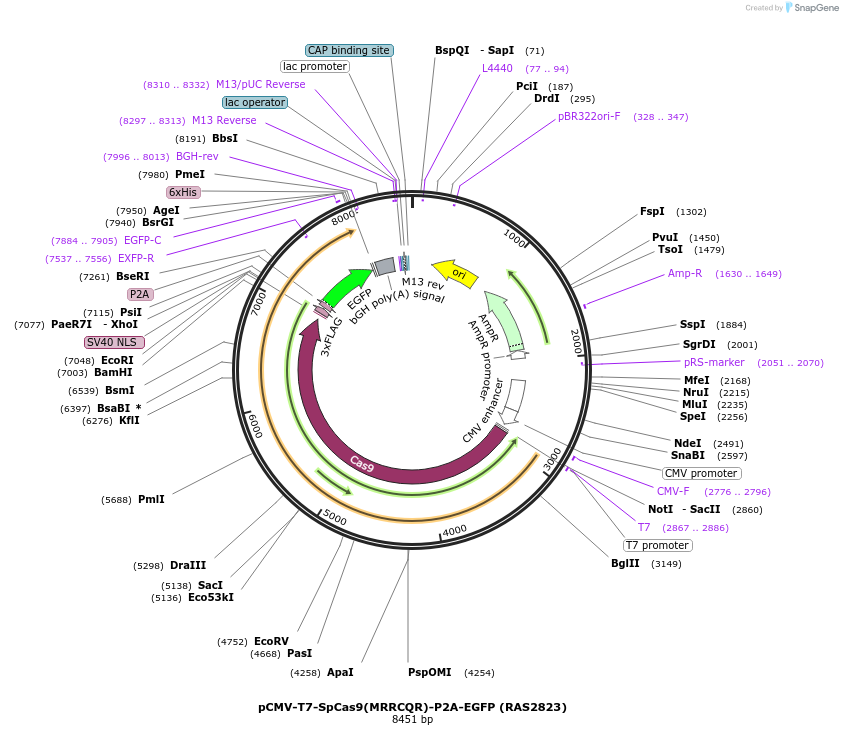
pCMV-T7-SpCas9(MRRCQR)-P2A-EGFP (RAS2823)
Plasmid#223072PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MRRCQR amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MRRCQR)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MRRCQR)=D1135M, S1136R, G1218R, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
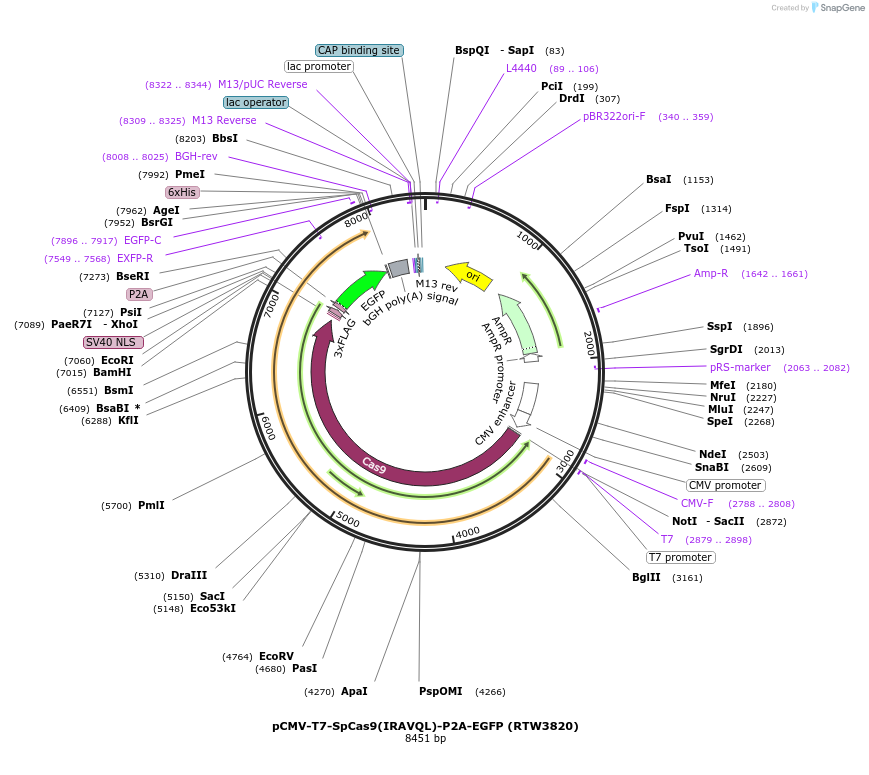
pCMV-T7-SpCas9(IRAVQL)-P2A-EGFP (RTW3820)
Plasmid#223073PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with IRAVQL amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(IRAVQL)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(IRAVQL)=D1135I, S1136R, G1218A, E1219V, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
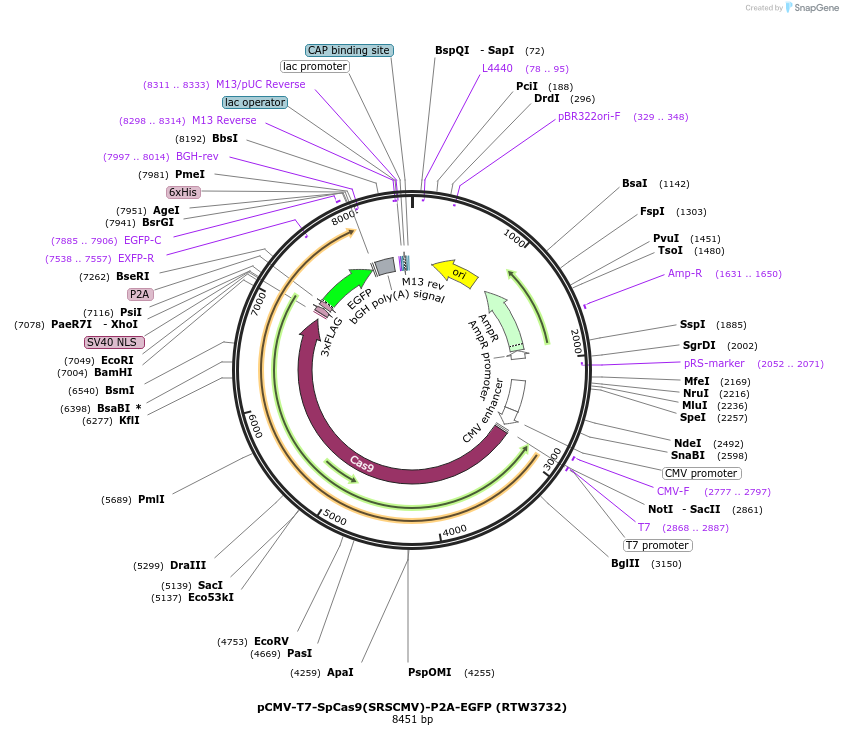
pCMV-T7-SpCas9(SRSCMV)-P2A-EGFP (RTW3732)
Plasmid#223074PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with SRSCMV amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(SRSCMV)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(SRSCMV)=D1135S, S1136R, G1218S, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
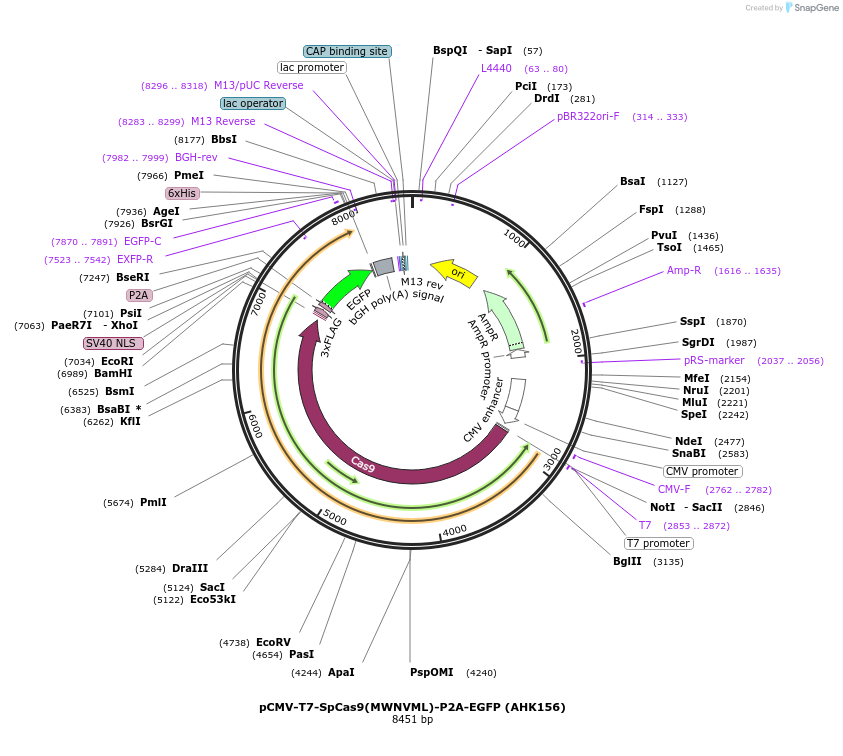
pCMV-T7-SpCas9(MWNVML)-P2A-EGFP (AHK156)
Plasmid#223076PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MWNVML amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MWNVML)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MWNVML)=D1135M, S1136W, G1218N, E1219V, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
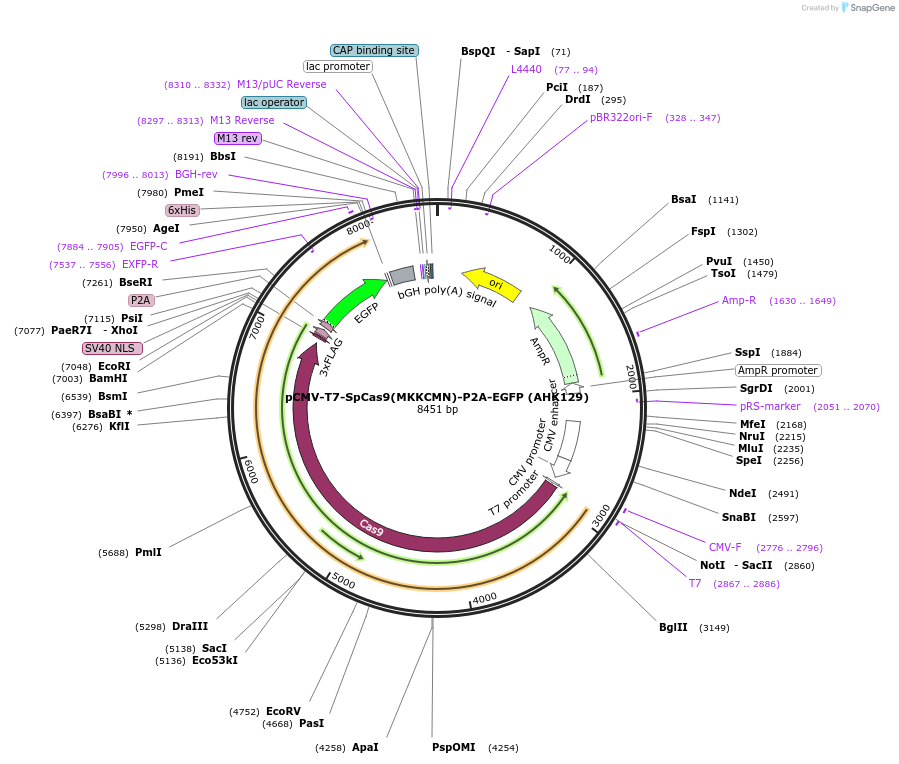
pCMV-T7-SpCas9(MKKCMN)-P2A-EGFP (AHK129)
Plasmid#223077PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with MKKCMN amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(MKKCMN)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(MKKCMN)=D1135M, S1136K, G1218K, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
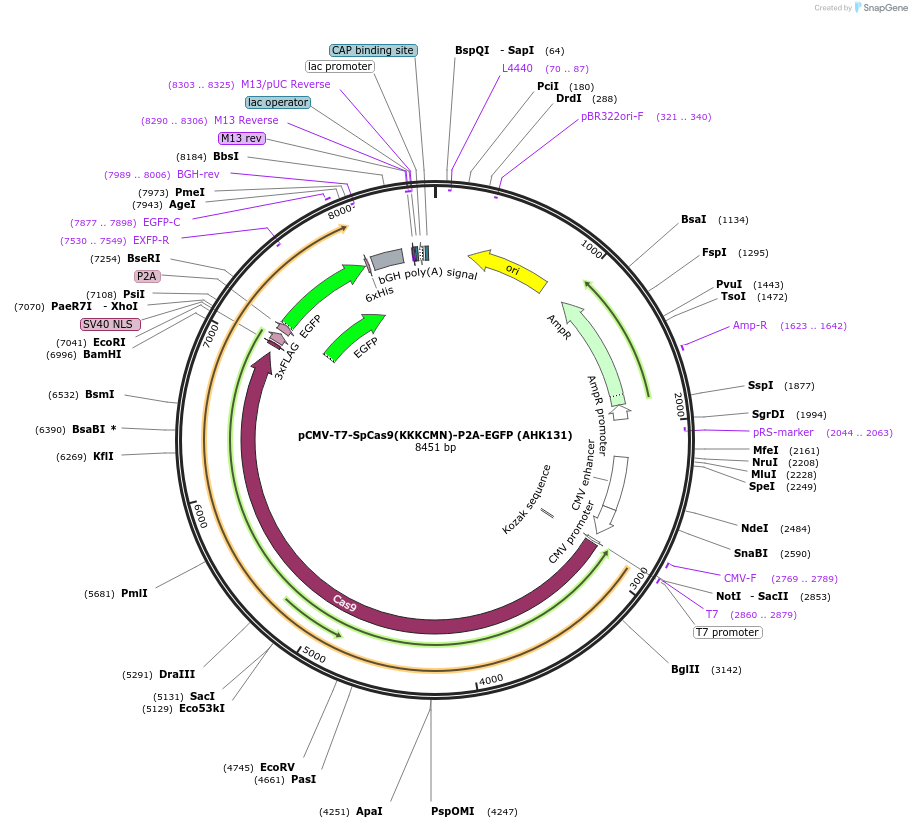
pCMV-T7-SpCas9(KKKCMN)-P2A-EGFP (AHK131)
Plasmid#223078PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with KKKCMN amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(KKKCMN)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(KKKCMN)=D1135K, S1136K, G1218K, E1219C, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
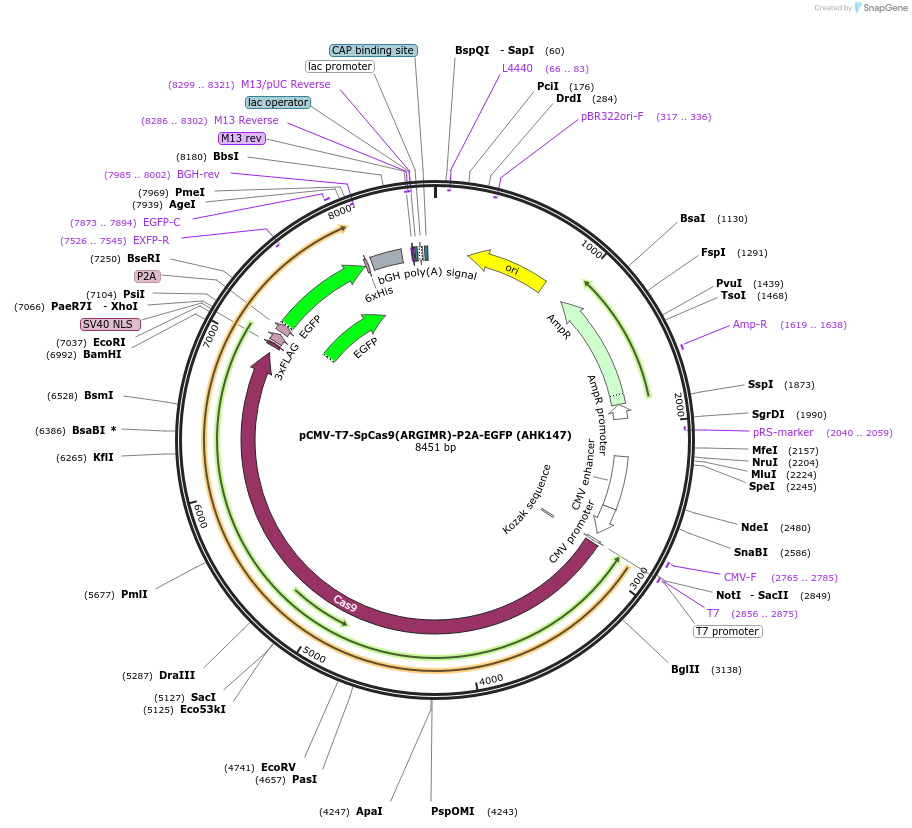
pCMV-T7-SpCas9(ARGIMR)-P2A-EGFP (AHK147)
Plasmid#223069PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with ARGIMR amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(ARGIMR)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(ARGIMR)=D1135A, S1136R, G1218G, E1219I, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
pCMV-T7-SpCas9(ARTWGR)-P2A-EGFP (RAS2995)
Plasmid#223070PurposepCMV and pT7 Human expression plasmid for SpCas9 enzyme with ARTWGR amino acids at positions 1135,1136,1218,1219,1335,T1337 with C-term BPNLS-P2A-EGFPDepositorInserthuman codon optimized nuclease SpCas9(ARTWGR)-P2A-EGFP
UseCRISPRTagsBPNLS-3xFLAG-P2A-EGFPExpressionMammalianMutationSpCas9(ARTWGR)=D1135A, S1136R, G1218T, E1219W, R1…PromoterCMV and T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only