We narrowed to 3,133 results for: eed
-
Plasmid#47587DepositorInsertPMCA2x/b delta6 (ATP2B2 Human)
TagsEGFP-HA-HisExpressionMammalianMutation6 C-terminal AA deleted (needed for PDZ binding)PromoterCMVAvailable SinceOct. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
MDH1-miR-196a-1SM-PGK-GFP-Blast
Plasmid#31510DepositorInsertmiR-196a1 (Mir196a-1 Mouse)
UseRetroviralTagsGFPExpressionMammalianMutationseed mutant controlAvailable SinceSept. 19, 2011AvailabilityAcademic Institutions and Nonprofits only -
MDH1-miR-324SM-PGK-GFP-Blast
Plasmid#31518DepositorInsertmiR-324 (Mir324 Mouse)
UseRetroviralTagsGFPExpressionMammalianMutationseed mutant controlAvailable SinceSept. 19, 2011AvailabilityAcademic Institutions and Nonprofits only -
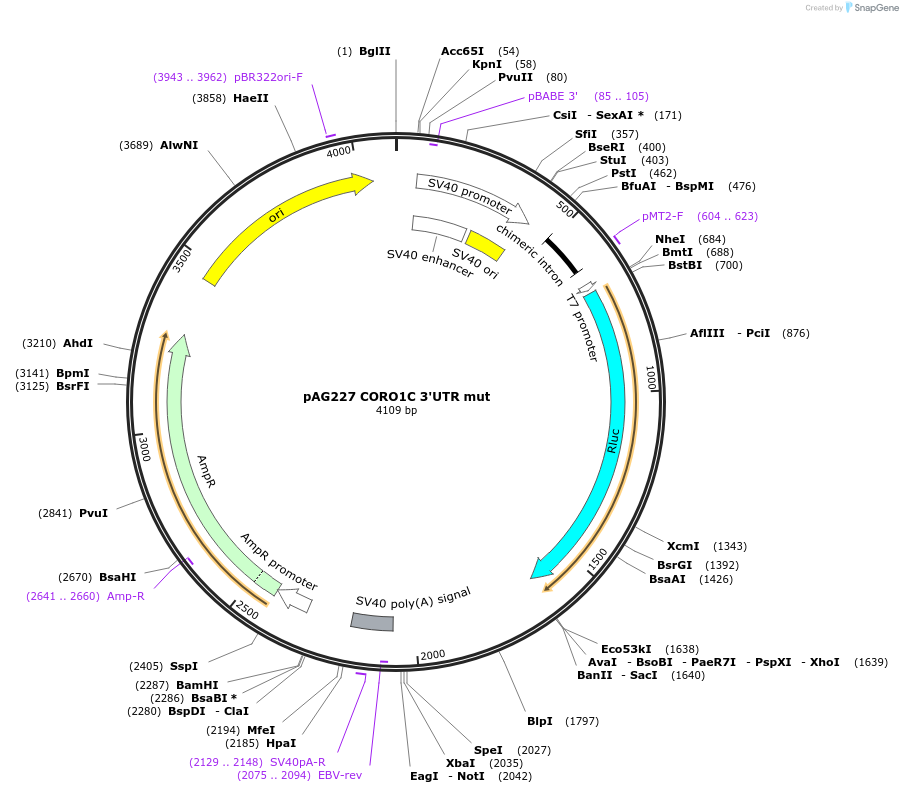
pAG227 CORO1C 3'UTR mut
Plasmid#12030DepositorInsertCORO1C 3'UTR mut (CORO1C Human)
UseLuciferaseExpressionMammalianMutationMutations in microRNA recognition sites (seed mat…Available SinceJune 2, 2006AvailabilityAcademic Institutions and Nonprofits only -
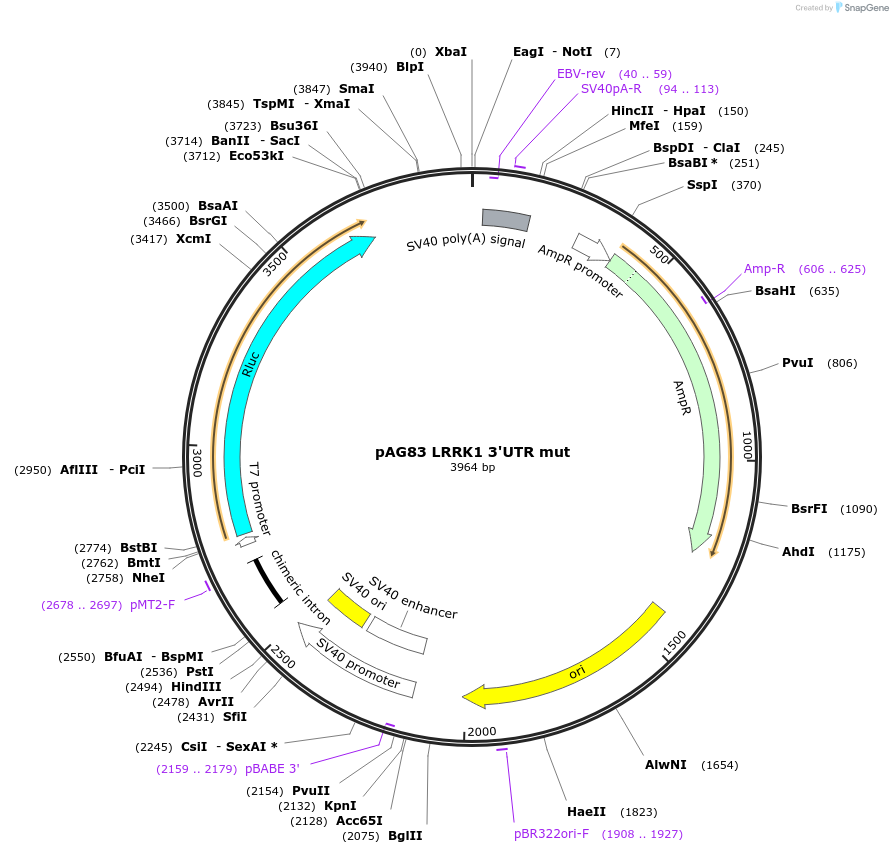
pAG83 LRRK1 3'UTR mut
Plasmid#12058DepositorInsertN4 3'UTR mut (LRRK1 Human)
UseLuciferaseExpressionMammalianMutationMutations in microRNA recognition sites (seed mat…Available SinceJune 2, 2006AvailabilityAcademic Institutions and Nonprofits only -
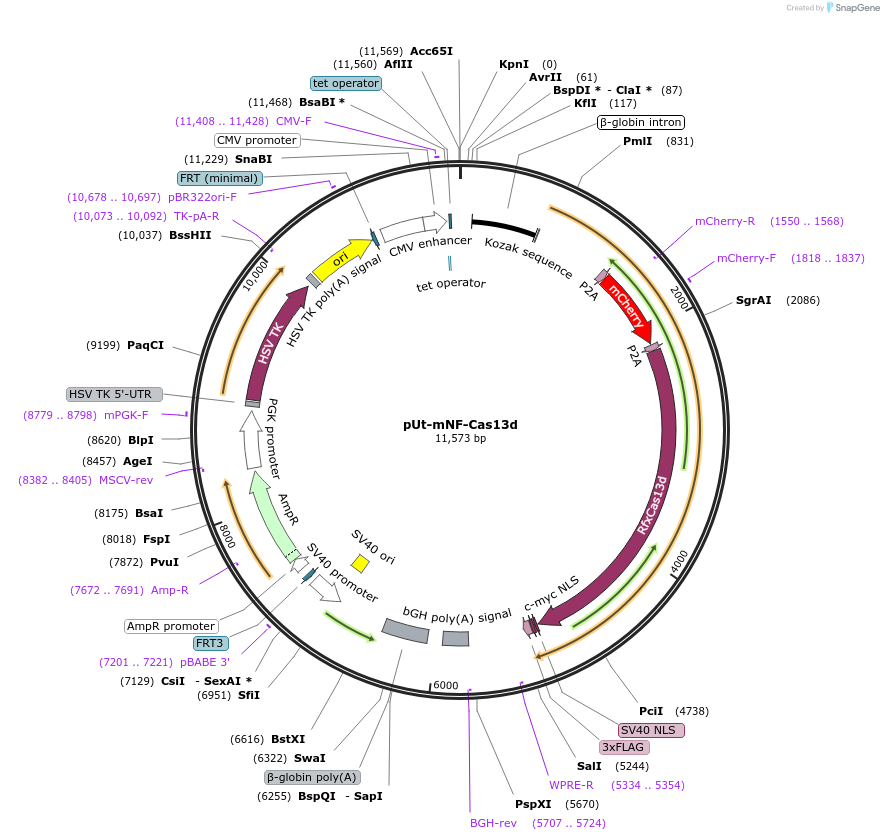
pUt-mNF-Cas13d
Plasmid#224789PurposeLPUtopia matching RMCE donor plasmid with mCherry-P2A-RfxCas13d Negative-Feedback circuit. Use BlastR for positive selection and HSV-TK for negative selection.DepositorInsertmCherry-P2A-RfxCas13d
UseCRISPR and Synthetic BiologyExpressionMammalianPromoterCMV-d2iAvailable SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
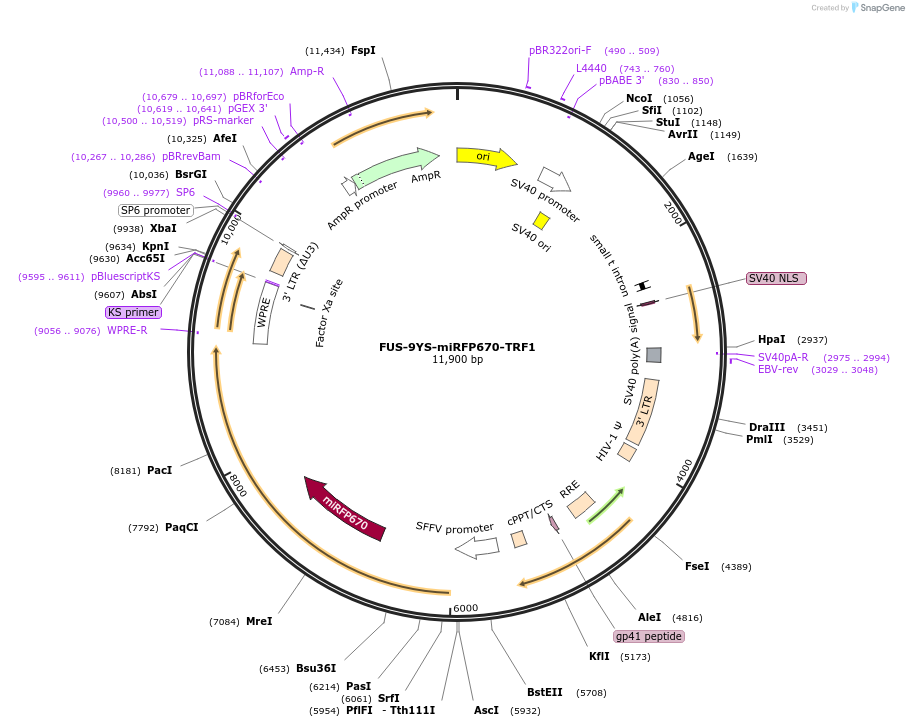
FUS-9YS-miRFP670-TRF1
Plasmid#227381PurposeAdhesion module, creates 'seed' at telomereDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
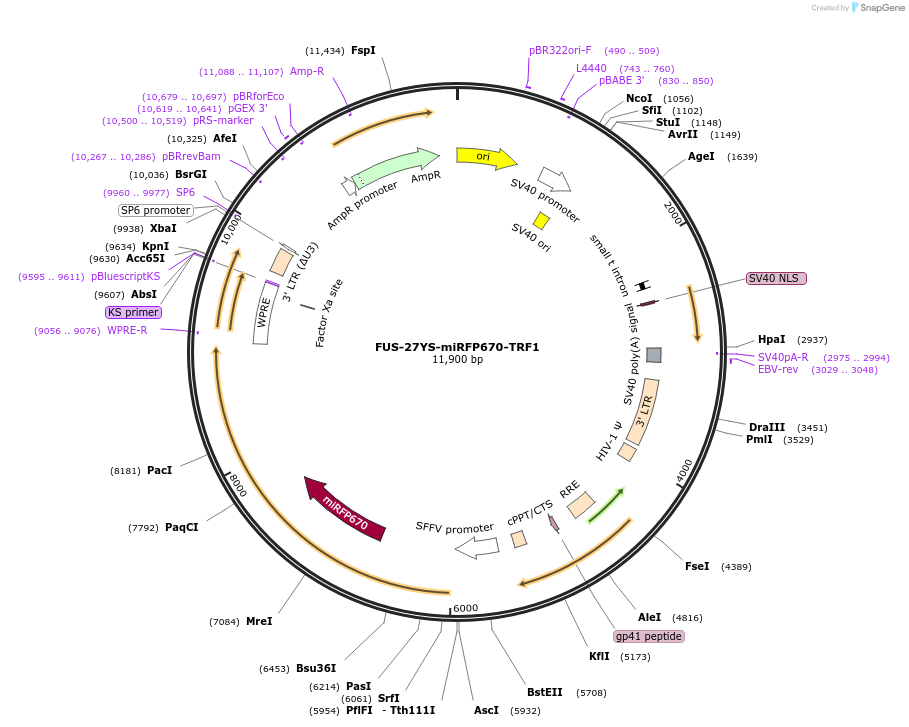
FUS-27YS-miRFP670-TRF1
Plasmid#227383PurposeAdhesion module, creates 'seed' at telomereDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
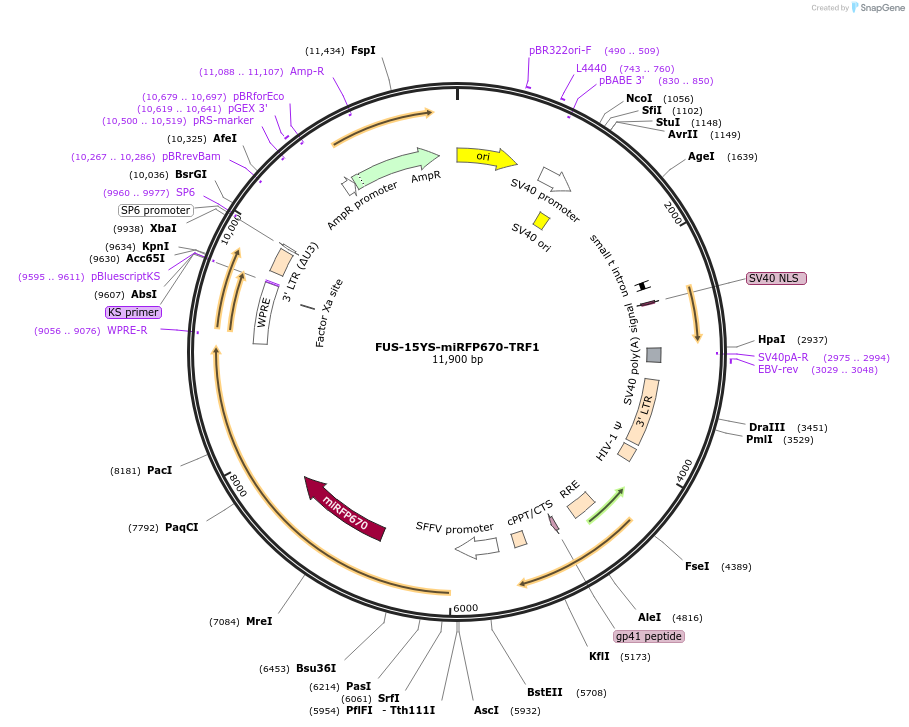
FUS-15YS-miRFP670-TRF1
Plasmid#227382PurposeAdhesion module, creates 'seed' at telomereDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
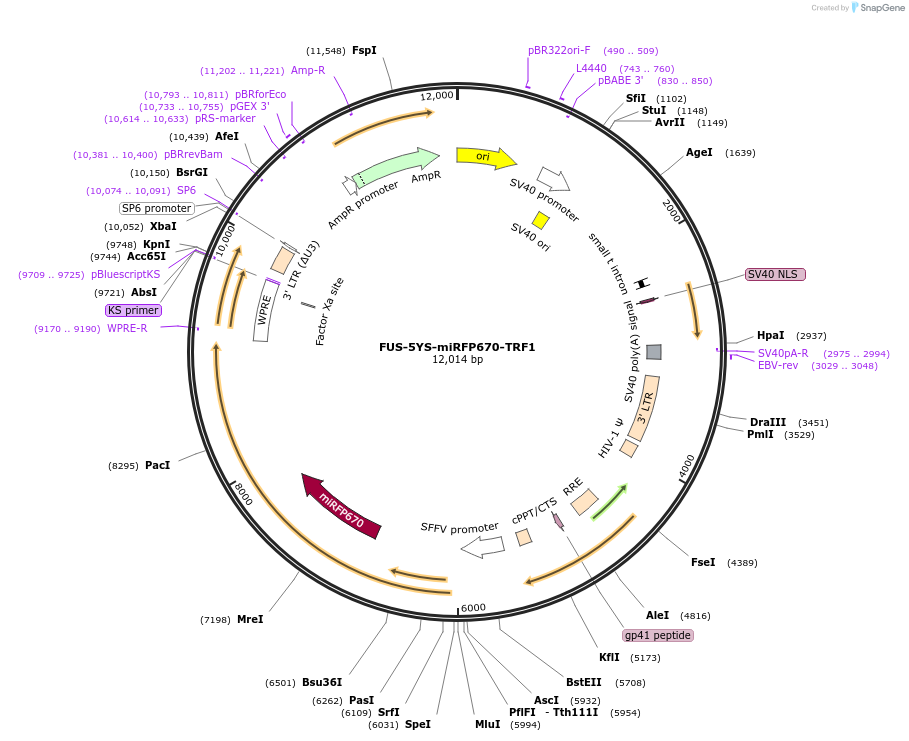
FUS-5YS-miRFP670-TRF1
Plasmid#227380PurposeAdhesion module, creates 'seed' at telomereDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
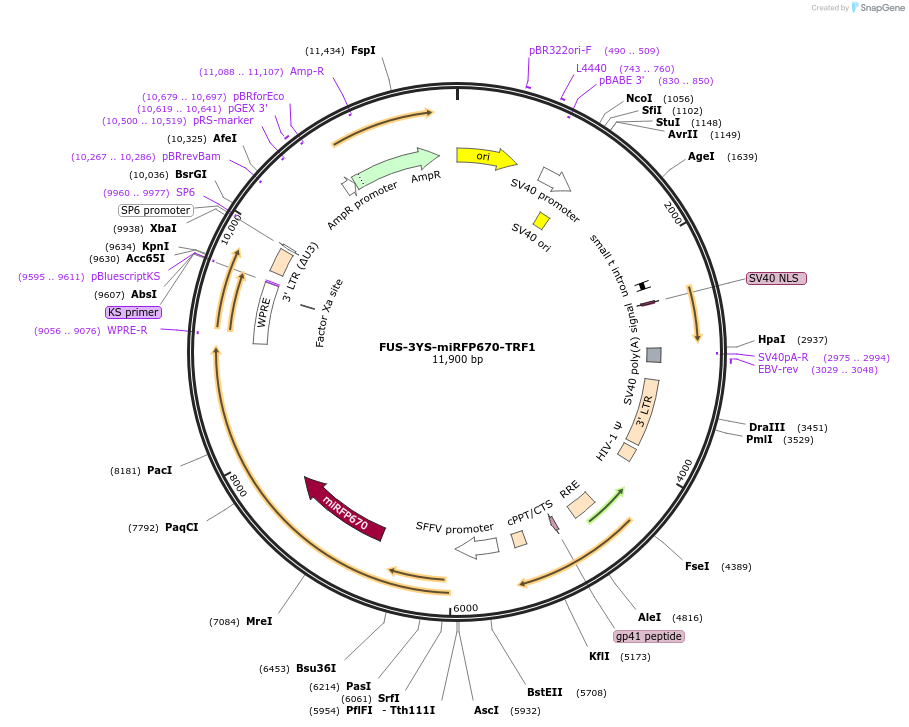
FUS-3YS-miRFP670-TRF1
Plasmid#227379PurposeAdhesion module, creates 'seed' at telomereDepositorAvailable SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
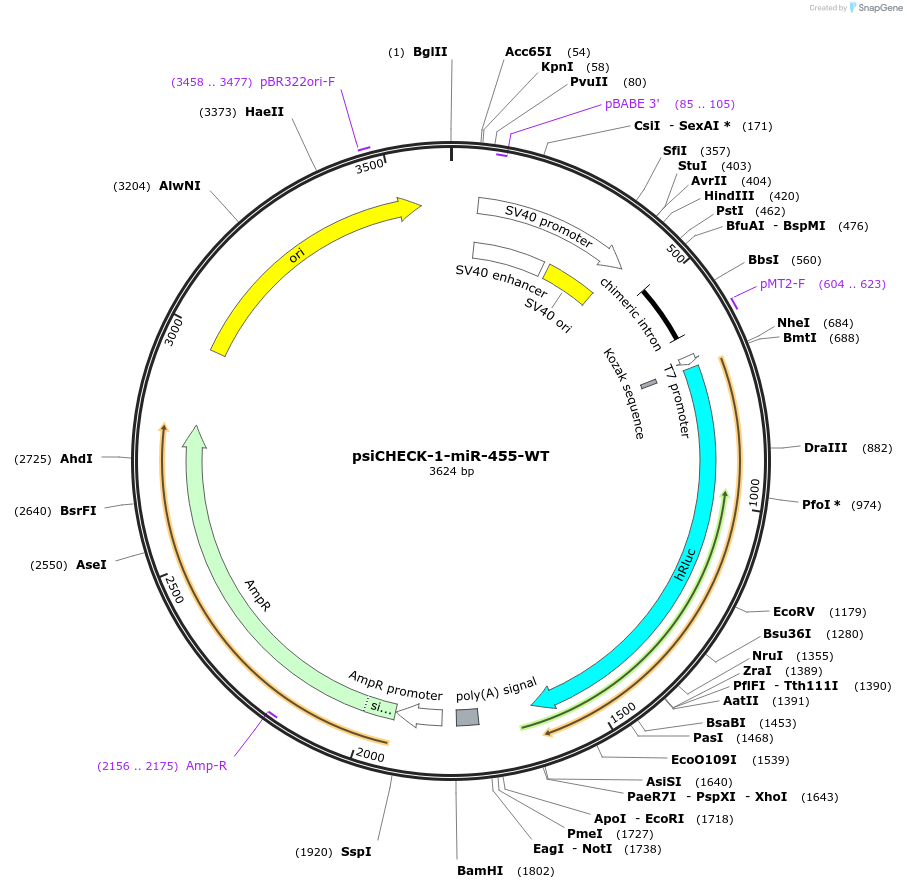
psiCHECK-1-miR-455-WT
Plasmid#215881PurposeThis plasmid encodes luciferase which has microRNA binding sites in 3'UTR for detecting the microRNA silencing activity.DepositorInsertthree tandem repeats of seed-matched (SM) sequence of miR-455-WT (MIR455 Human)
ExpressionMammalianAvailable SinceOct. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
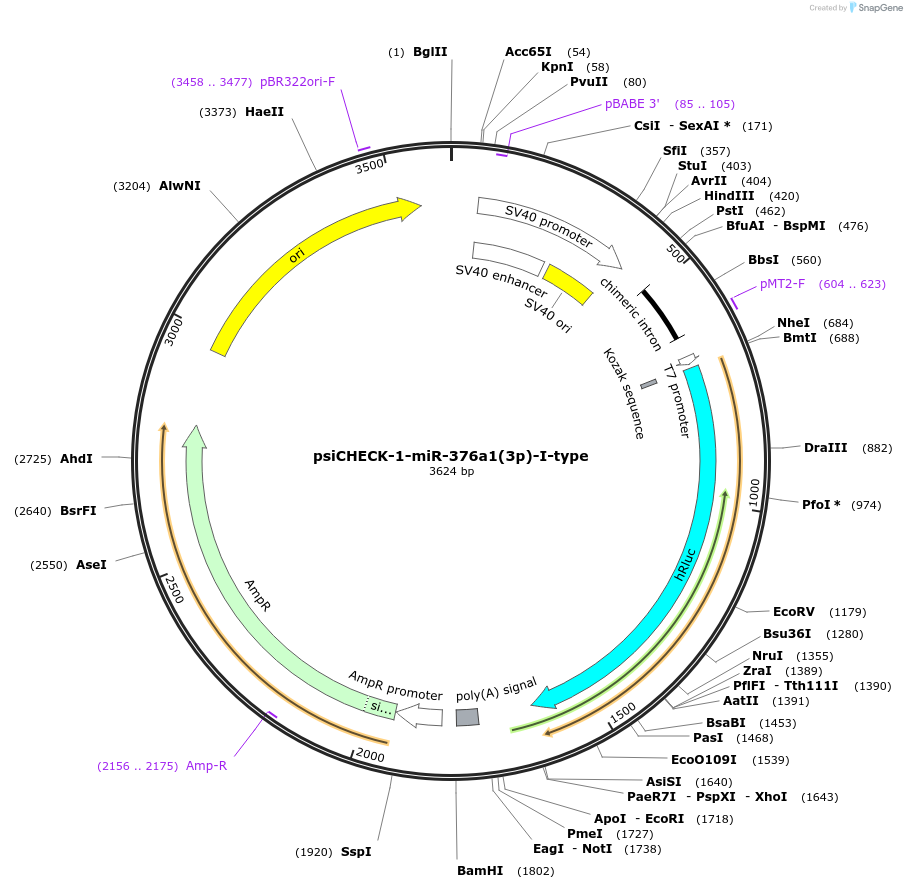
psiCHECK-1-miR-376a1(3p)-I-type
Plasmid#215883PurposeThis plasmid encodes luciferase which has microRNA binding sites in 3'UTR for detecting the microRNA silencing activity.DepositorInsertthree tandem repeats of seed-matched (SM) sequence of miR-376a1(3p)-I-type (MIR376A1 Human)
ExpressionMammalianAvailable SinceOct. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
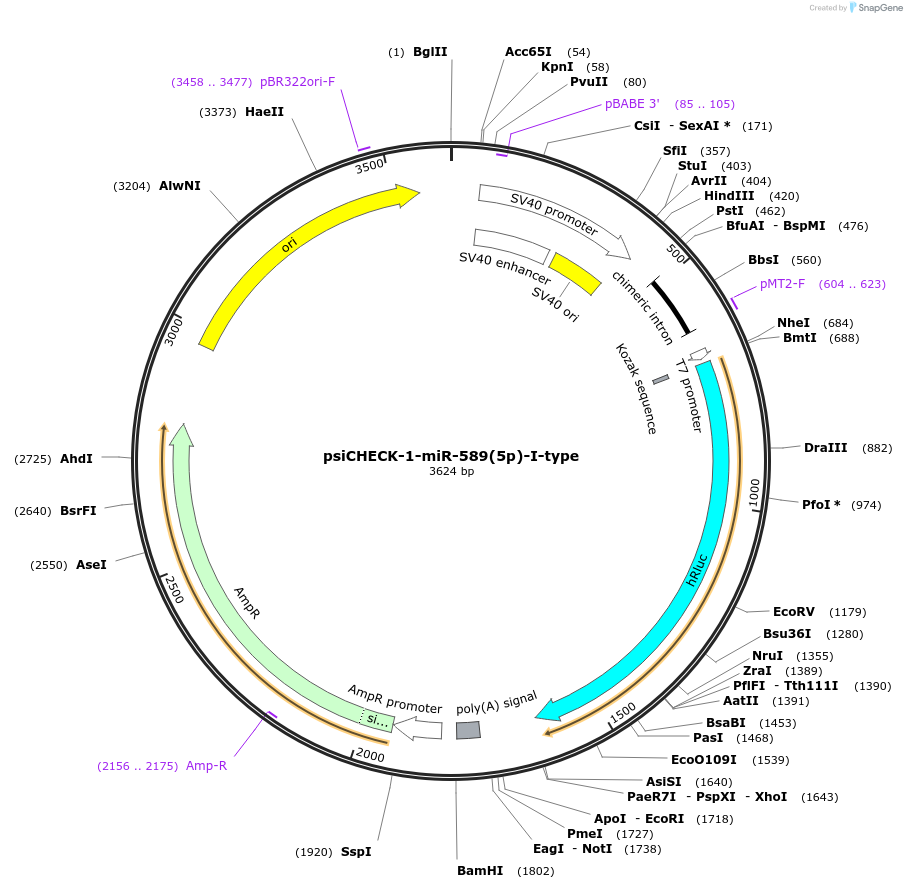
psiCHECK-1-miR-589(5p)-I-type
Plasmid#215884PurposeThis plasmid encodes luciferase which has microRNA binding sites in 3'UTR for detecting the microRNA silencing activity.DepositorInsertthree tandem repeats of seed-matched (SM) sequence of miR-589(5p)-I-type (MIR589 Human)
ExpressionMammalianAvailable SinceOct. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
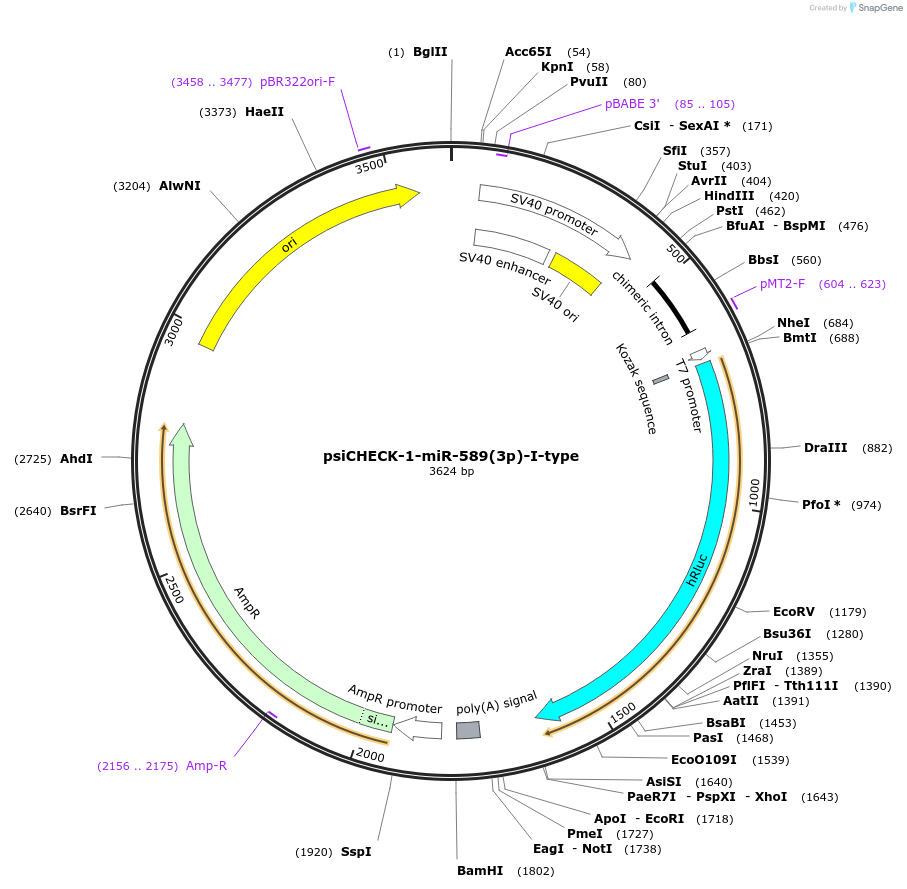
psiCHECK-1-miR-589(3p)-I-type
Plasmid#215885PurposeThis plasmid encodes luciferase which has microRNA binding sites in 3'UTR for detecting the microRNA silencing activity.DepositorInsertthree tandem repeats of seed-matched (SM) sequence of miR-589(3p)-I-type (MIR589 Human)
ExpressionMammalianAvailable SinceOct. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
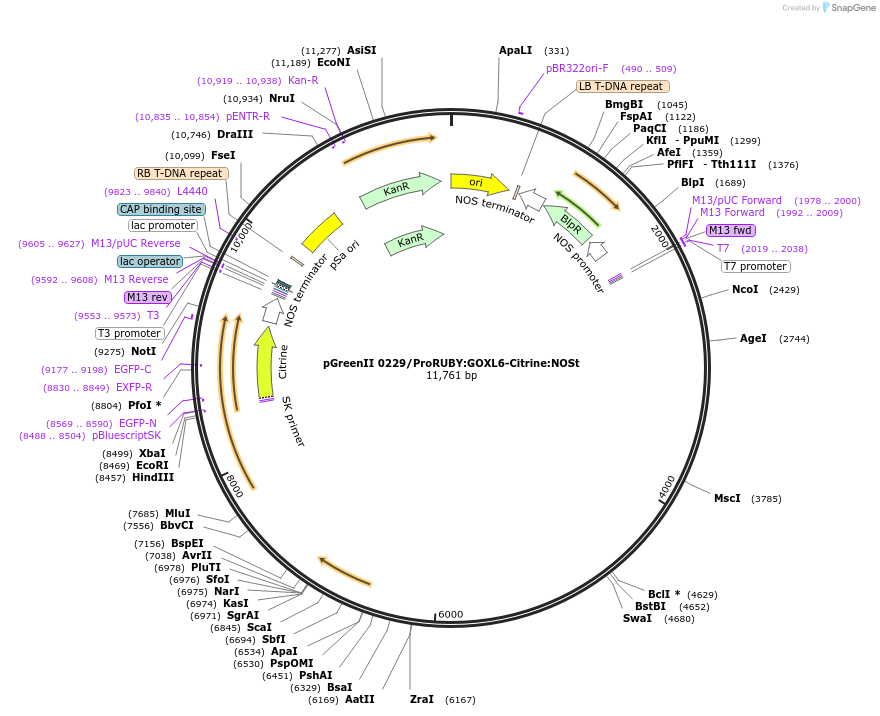
pGreenII 0229/ProRUBY:GOXL6-Citrine:NOSt
Plasmid#175568PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-6DepositorInsertGOXL6
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
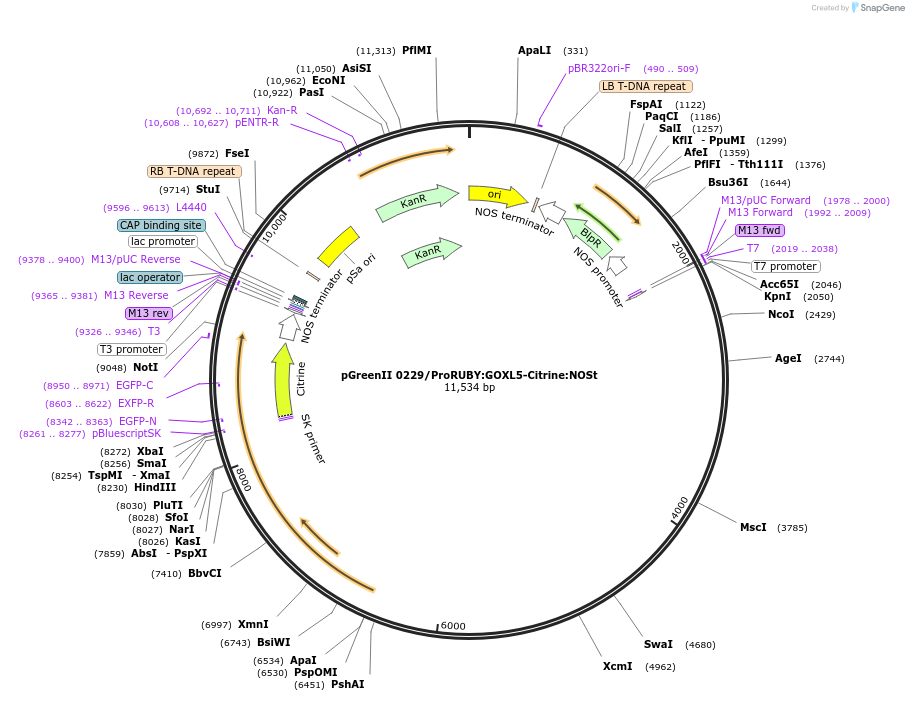
pGreenII 0229/ProRUBY:GOXL5-Citrine:NOSt
Plasmid#175567PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-5DepositorInsertGOXL5
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
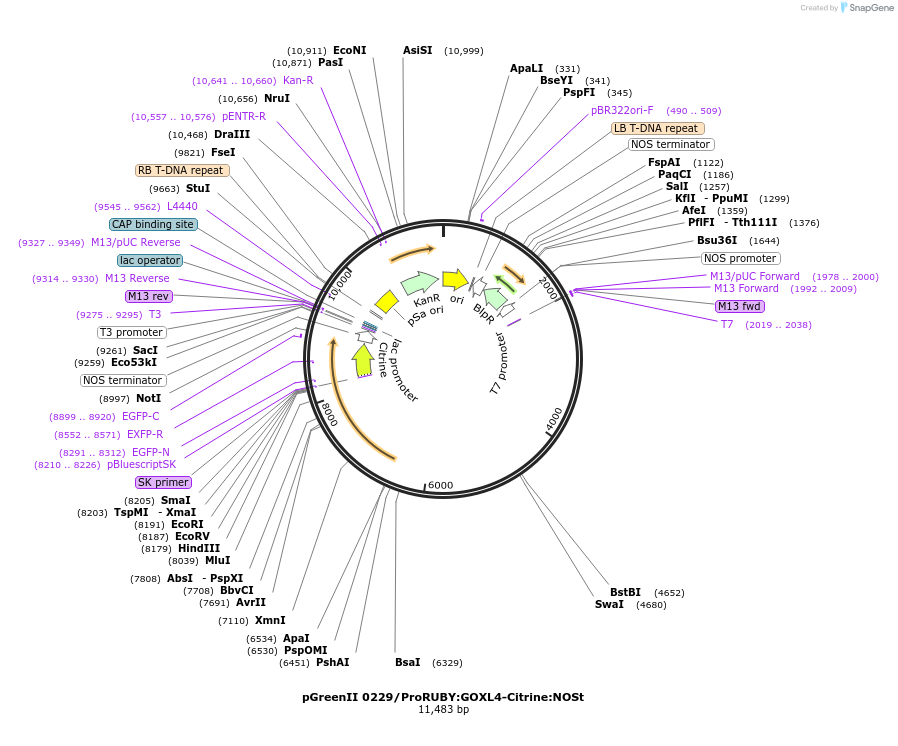
pGreenII 0229/ProRUBY:GOXL4-Citrine:NOSt
Plasmid#175566PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-4DepositorInsertGOXL4
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
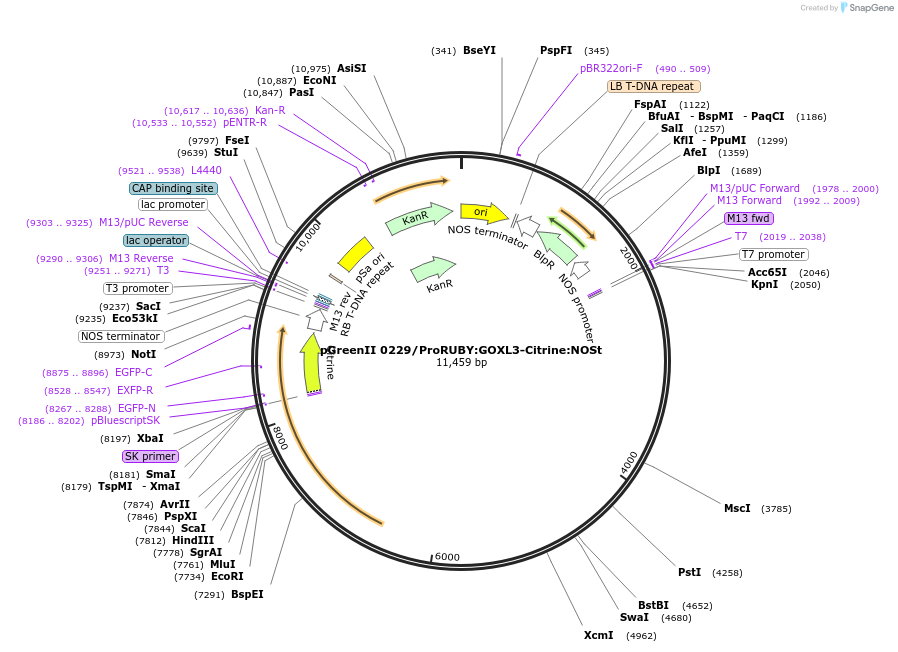
pGreenII 0229/ProRUBY:GOXL3-Citrine:NOSt
Plasmid#175565PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-3DepositorInsertGOXL3
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
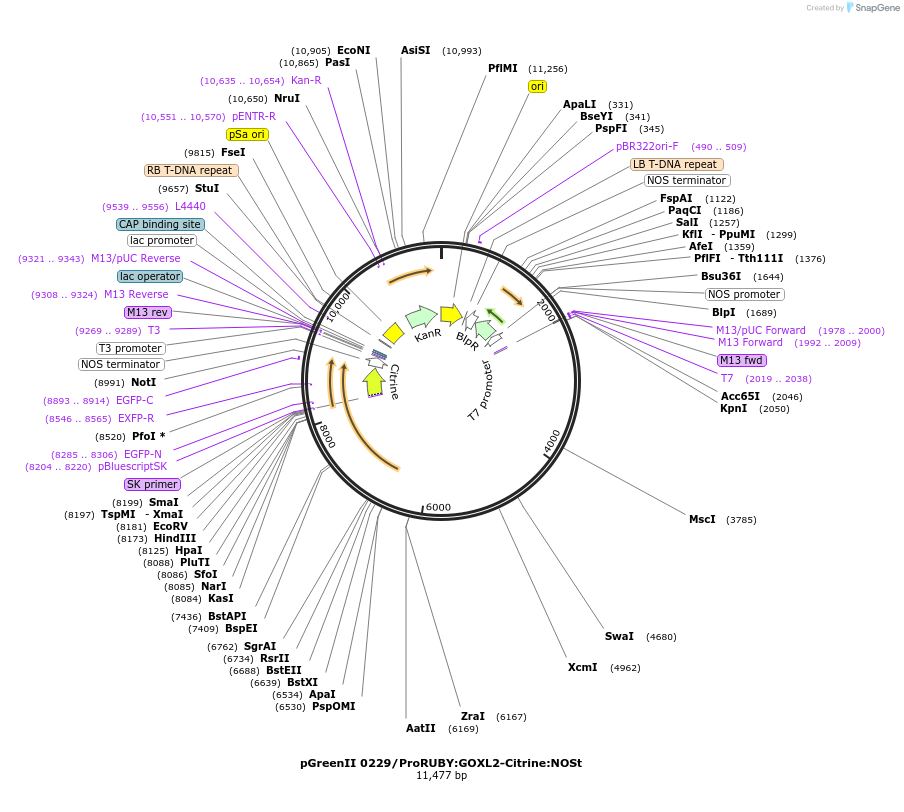
pGreenII 0229/ProRUBY:GOXL2-Citrine:NOSt
Plasmid#175564PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-2DepositorInsertGOXL2
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 19, 2021AvailabilityAcademic Institutions and Nonprofits only