We narrowed to 2,220 results for: SE
-
Plasmid#87663Purposeretroviral vector encoding cre-dependent expression of kikumeDepositorInsertkikume
UseRetroviralTagsExpressionMutationPromoterCAGAvailable SinceMay 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
pBabe-Puro-RalA-S194A
Plasmid#15255DepositorInsertv-ral simian leukemia viral oncogene homolog A (RALA Human)
UseRetroviralTagsExpressionMammalianMutationserine 194 to alaninePromoterAvailable SinceJuly 12, 2007AvailabilityAcademic Institutions and Nonprofits only -
pGL3-p107 2*
Plasmid#32388DepositorInsertp107 promoter (Rbl1 Mouse)
UseLuciferaseTagsLuciferaseExpressionMammalianMutation2nd E2F binding site mutated: GTC-->AAAPromoterAvailable SinceSept. 27, 2011AvailabilityAcademic Institutions and Nonprofits only -
pBabe-Puro-RalA-S11A
Plasmid#15253DepositorInsertv-ral simian leukemia viral oncogene homolog A (RALA Human)
UseRetroviralTagsExpressionMammalianMutationserine 11 to alaninePromoterAvailable SinceJuly 12, 2007AvailabilityAcademic Institutions and Nonprofits only -
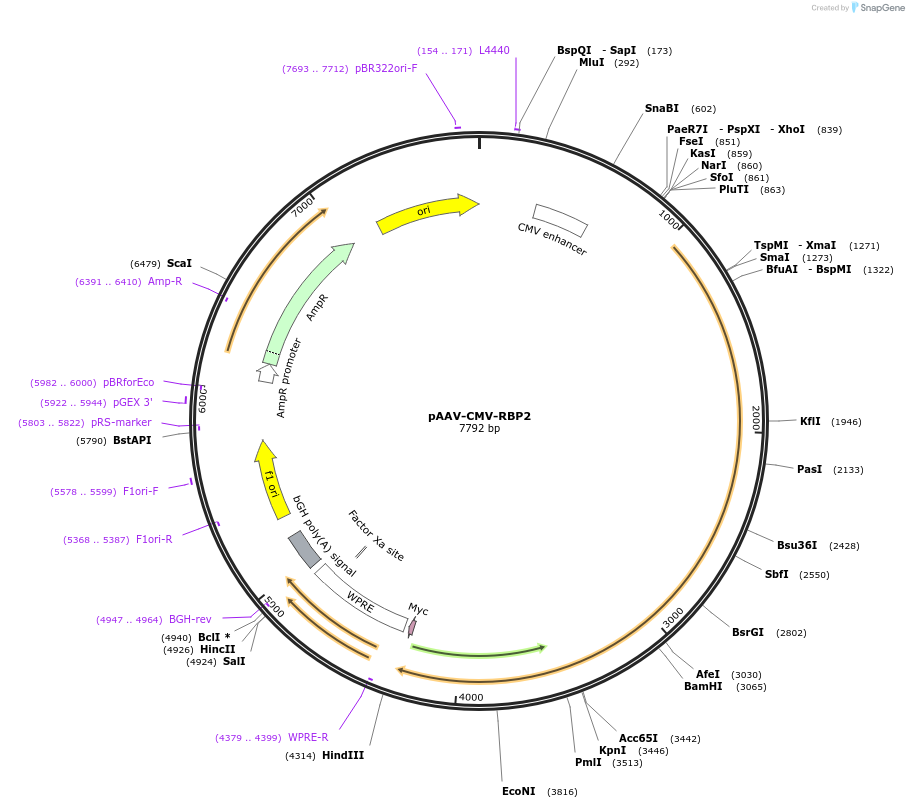
pAAV-CMV-RBP2
Plasmid#241990PurposeMouse RBP2 (accession ID NP_001074857.1), CMV promoter-enhancer, downstream WPRE.DepositorArticleInsertRIM-BP2 (Rimbp2 Mouse)
UseAAVTagsRIM-BP2 (untagged)ExpressionMammalianMutationPromoterCMV enhancerAvailable SinceOct. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
pWR91
Plasmid#203915Purposeempty BLINCAR plasmid for payload delivery, no payload, integrates at CRF1 locusDepositorAvailable SinceJune 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pWR90
Plasmid#203914Purposeempty BLINCAR plasmid for payload delivery, no payload, integrates at GSC2 locusDepositorAvailable SinceJune 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pWR89
Plasmid#203423Purposeempty BLINCAR plasmid for payload delivery, no payload, integrates at EGC3 locusDepositorAvailable SinceJune 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
pRegev_S4.39U>G.51A>C
Plasmid#204173PurposepRegev_S4 with double mutations that restore secondary structure.DepositorInsertExon S4 with 39U>G and 51A>C mutations in beta globin splicing minigene
UseSynthetic BiologyTagsExpressionMammalianMutation39U>G and 51A>C mutationsPromoterTruncated CAGAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
pRegev_S1.34C>A_46G>U
Plasmid#204161PurposepRegev_S1 with double mutations that restore secondary structure.DepositorInsertS1 exon with 34C>A and 46G>U substitutions in beta globin splicing minigene
UseSynthetic BiologyTagsExpressionMammalianMutation34C>A and 46G>U mutationsPromoterTruncated CAGAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
pRegev_S2.39U>G.51A>C
Plasmid#204165PurposepRegev_S2 with double mutations that restore secondary structure.DepositorInsertExon S2 with 39U>G and 51A>C mutations in beta globin splicing minigene
UseSynthetic BiologyTagsExpressionMammalianMutation39U>G and 51A>C mutationsPromoterTruncated CAGAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
pRegev_S5.49C>A.59G>U
Plasmid#204177PurposepRegev_S5 with double mutations that restore secondary structure.DepositorInsertExon S5 with 49C>A and 59G>U mutations in beta globin splicing minigene
UseSynthetic BiologyTagsExpressionMammalianMutation49C>A and 59G>U mutationsPromoterTruncated CAGAvailable SinceAug. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
pJL1_CSL3
Plasmid#200173PurposeAmino acids 1-195 of CSL3 with an N-terminal 6xHis-tag followed by a TEV site in the pJL1 vectorDepositorInsertCSL3
UseTags6x His tag and TEV cleavage siteExpressionBacterialMutationPromoterT7Available SinceMay 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAC-U63-tgRNA-Gal80
Plasmid#169030PurposegRNA-marker vector contain a U6:3 promoter, gRNA scaffold, and a Ubi-Gal80 marker.DepositorTypeEmpty backboneUseCRISPRTagsExpressionInsectMutationPromoterU6:3Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pRS314_U6_A34G_A35G
Plasmid#81232PurposeU6 telestem mutant (A34G/A35G) for in vivo studiesDepositorInsertU6 RNA with A34G/A35G mutation (snR6 Budding Yeast)
UseTagsExpressionYeastMutationU6 telestem mutant (A34G/A35G)PromoterAvailable SinceSept. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
pRS314_U6_G96C_A97C
Plasmid#81235PurposeU6 telestem mutant (G96C/A97C) for in vivo studiesDepositorInsertU6 RNA with G96C/A97C mutation (snR6 Budding Yeast)
UseTagsExpressionYeastMutationU6 telestem mutant (G96C/A97C)PromoterAvailable SinceSept. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
pRS314_U6_U100C_U101C
Plasmid#81236PurposeU6 telestem mutant (U100C/U101C) for in vivo studiesDepositorInsertU6 RNA with U100C_U101C mutation (snR6 Budding Yeast)
UseTagsExpressionYeastMutationU6 telestem mutant (G96C/A97C)PromoterAvailable SinceSept. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
FLAG-MBP1-E322TAG-E309C-W340A-CAAX.pcDNA3-k
Plasmid#127409PurposeExpresses MBP in mammalian cells. Amber codon at AA# 322 as a noncanonical amino acid incorporation siteDepositorInsertMaltose Binding Protein
UseTagsCAAX and FLAGExpressionMammalianMutationAmino acid mutations: E322 to Amber codon, E309C …PromoterCMVAvailable SinceJuly 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
FLAG-MBP1-K295TAG-W340A-S233H-T237C-CAAX.pcDNA3-k
Plasmid#127403PurposeExpresses MBP in mammalian cells. Amber codon at AA# 295 as a noncanonical amino acid incorporation siteDepositorInsertMaltose Binding Protein
UseTagsCAAX and FLAGExpressionMammalianMutationAmino acid mutations: K295 to Amber codon, W340A,…PromoterCMVAvailable SinceJuly 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
FLAG-MBP1-E322TAG-K305H-E309H-W340A-CAAX.pcDNA3-k
Plasmid#127407PurposeExpresses MBP in mammalian cells. Amber codon at AA# 322 as a noncanonical amino acid incorporation siteDepositorInsertMaltose Binding Protein
UseTagsCAAX and FLAGExpressionMammalianMutationAmino acid mutations: E322 to Amber codon, K305H,…PromoterCMVAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only