We narrowed to 27,048 results for: STI;
-
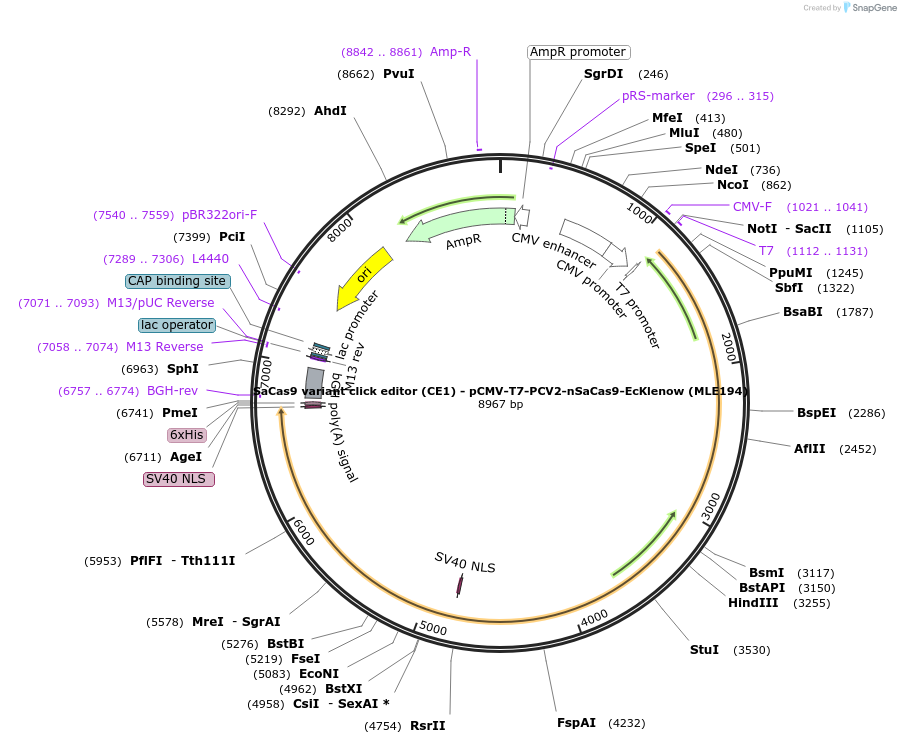
Plasmid#217801PurposeVariant CE1 construct with SaCas9, expressed from CMV or T7 promoters.DepositorInsertPCV2-XTEN-nSaCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSaCas9(N580A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
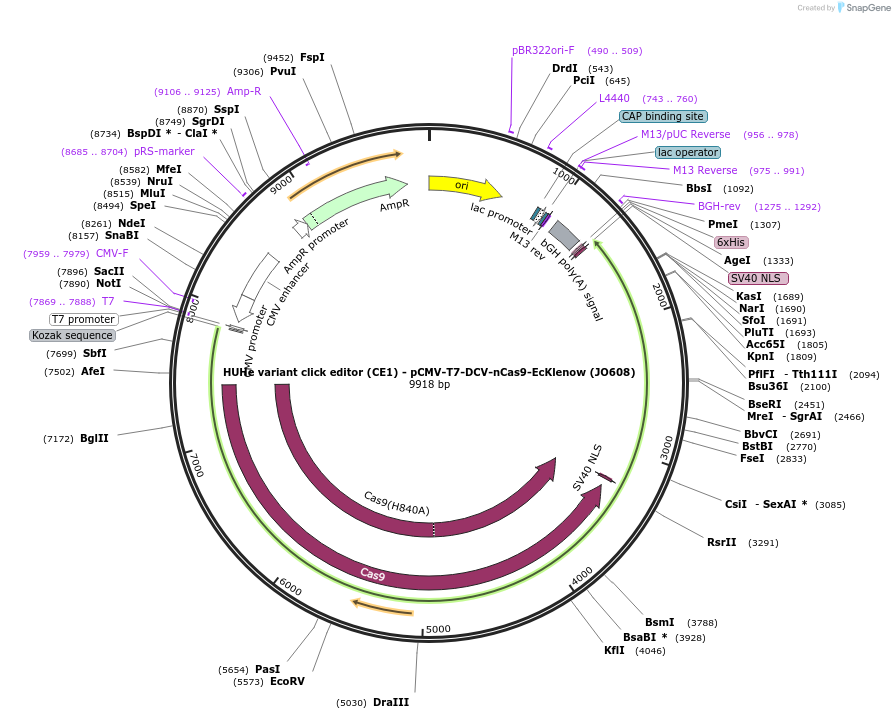
HUHe variant click editor (CE1) - pCMV-T7-DCV-nCas9-EcKlenow (JO608)
Plasmid#208946PurposeA variant CE1 construct with DCV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertDCV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
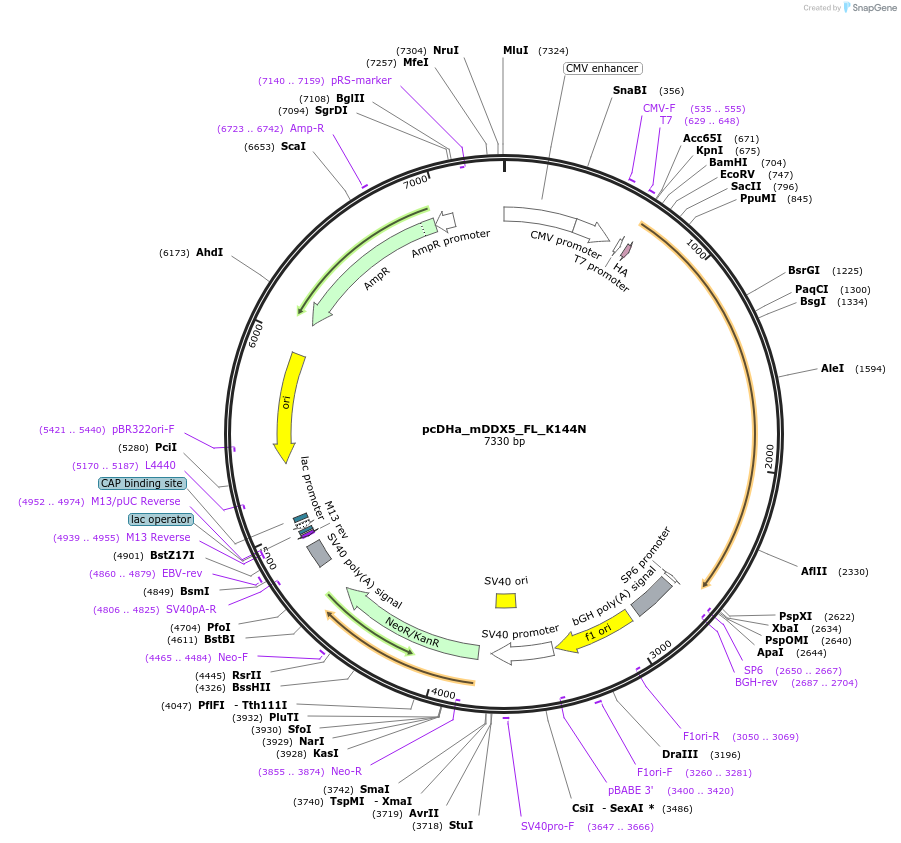
pcDHa_mDDX5_FL_K144N
Plasmid#88870Purposeconstitutive expression of Ha-DDX5 K144N in mammalian cellsDepositorAvailable SinceJune 12, 2017AvailabilityAcademic Institutions and Nonprofits only -
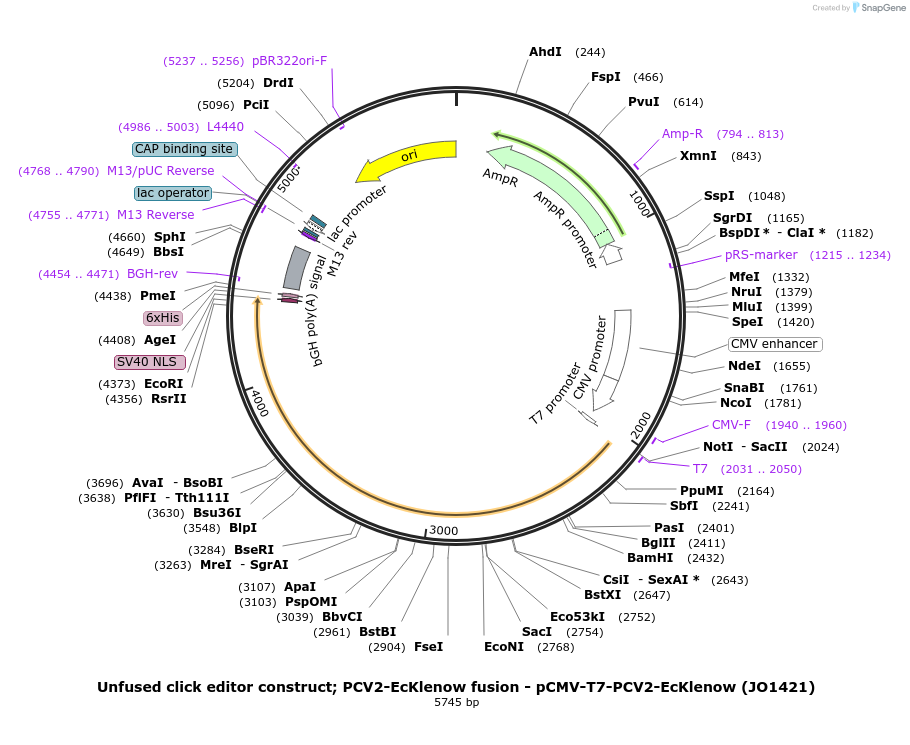
Unfused click editor construct; PCV2-EcKlenow fusion - pCMV-T7-PCV2-EcKlenow (JO1421)
Plasmid#217804PurposeUnfused click editor construct expressing PCV2-EcKlenow, expressed from CMV or T7 promoters.DepositorInsertPCV2-linker-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationEcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
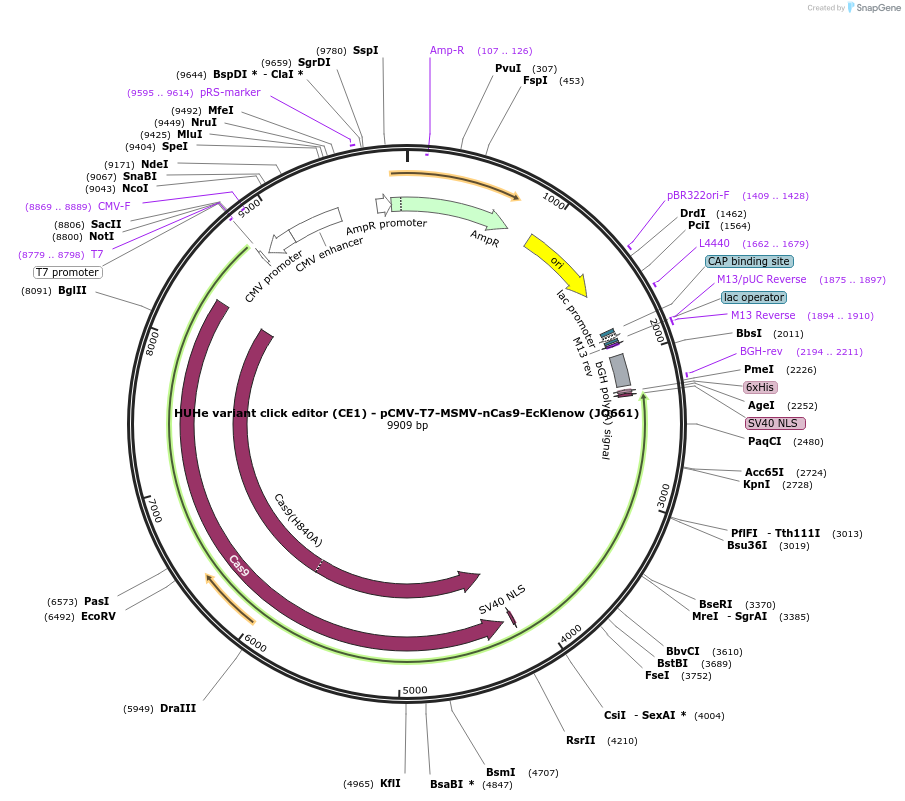
HUHe variant click editor (CE1) - pCMV-T7-MSMV-nCas9-EcKlenow (JO661)
Plasmid#208947PurposeA variant CE1 construct with MSMV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertMSMV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
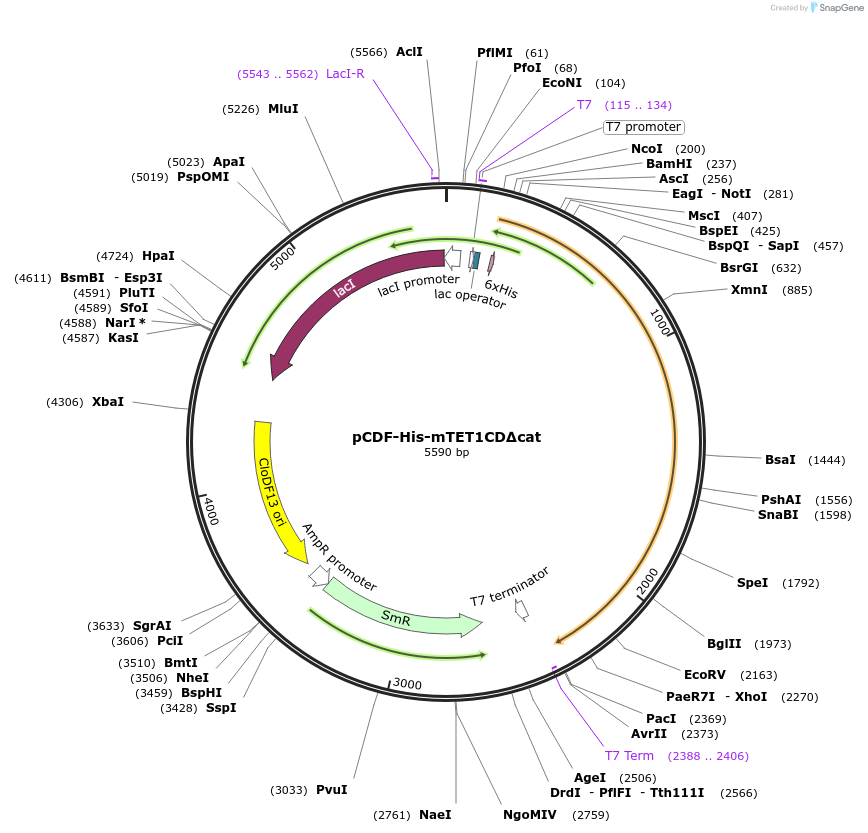
pCDF-His-mTET1CD∆cat
Plasmid#81054Purposebacterial expression of catalytic dead catalytic domain of murine TET1DepositorInsertTET1 (Tet1 Mouse)
Tags6HISExpressionBacterialMutationcatalytic domain amino acid 1367-2057 with histid…PromoterT7Available SinceAug. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
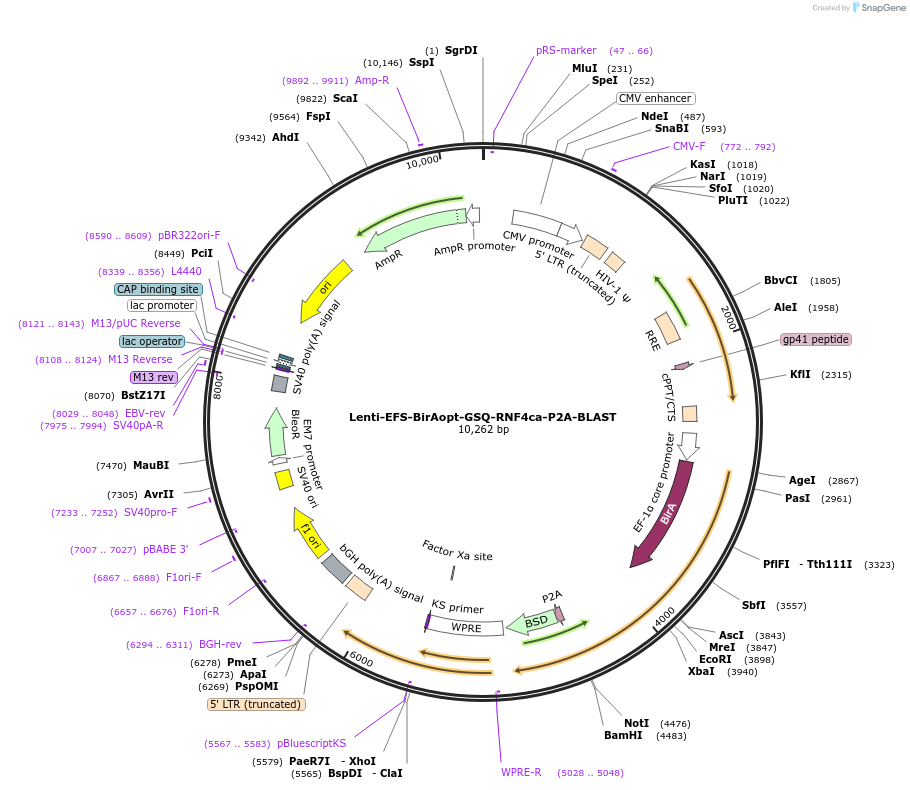
Lenti-EFS-BirAopt-GSQ-RNF4ca-P2A-BLAST
Plasmid#208047PurposeEnables constitutive expression of N-terminal BirAopt-fused RNF4ca (catalytically inactive RING mutant); to perform bioE3; selection with blasticidinDepositorInsertRNF4 (RNF4 Human)
UseLentiviralTagsBirAoptMutationCys>Ala mutations in conserved RING domainPromoterEFSAvailable SinceNov. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
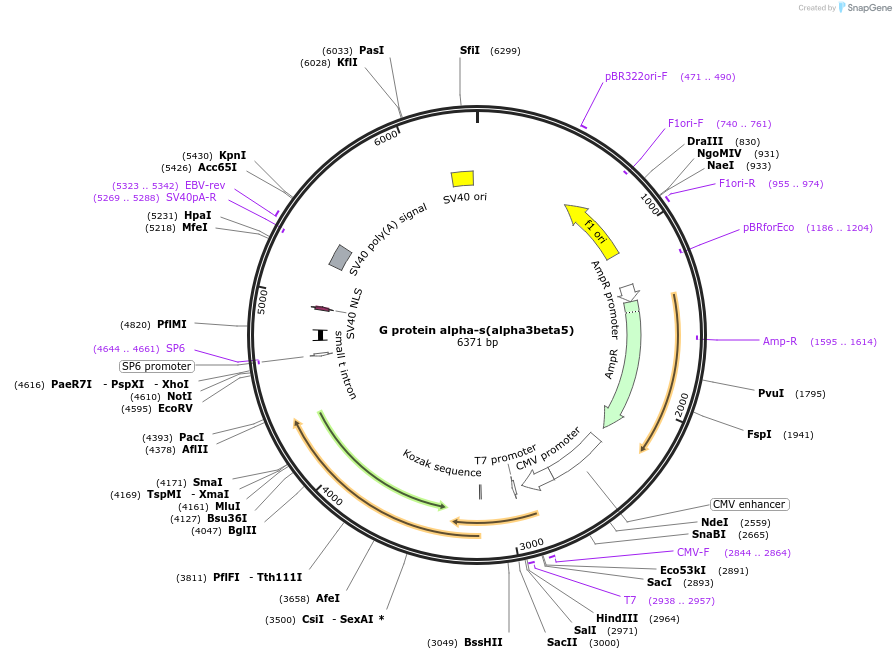
G protein alpha-s(alpha3beta5)
Plasmid#55799PurposeThis G protein alpha-s mutant exhibits increased affinity for and decreased ability to be activated by Gs-protein-coupled receptors as well as decreased ability to activate adenylyl cyclase.DepositorInsertG protein alpha-s with alpha-i2 substitutions in alpha3/beta5 loop (Gnas Rat)
Tagsinternal EE epitope (residues 189-194 in alpha-s …ExpressionMammalianMutationN271K, K274D, R280K, T284D, I285T in alpha-s sequ…PromoterCMVAvailable SinceJuly 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
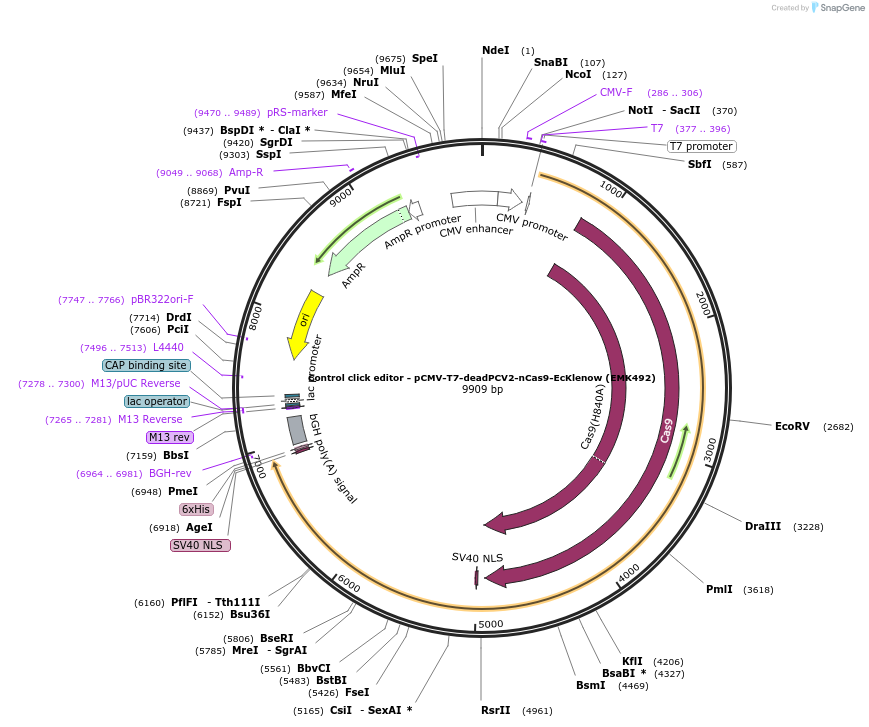
Control click editor - pCMV-T7-deadPCV2-nCas9-EcKlenow (EMK492)
Plasmid#208943PurposeA control CE1 construct with catalytically attenuated PCV2 HUH, expressed from CMV or T7 promoters.DepositorInsertdPCV2-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationPCV2(Y96F); nSpCas9(H840A); EcKlenow(-exo;D355A/E…PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
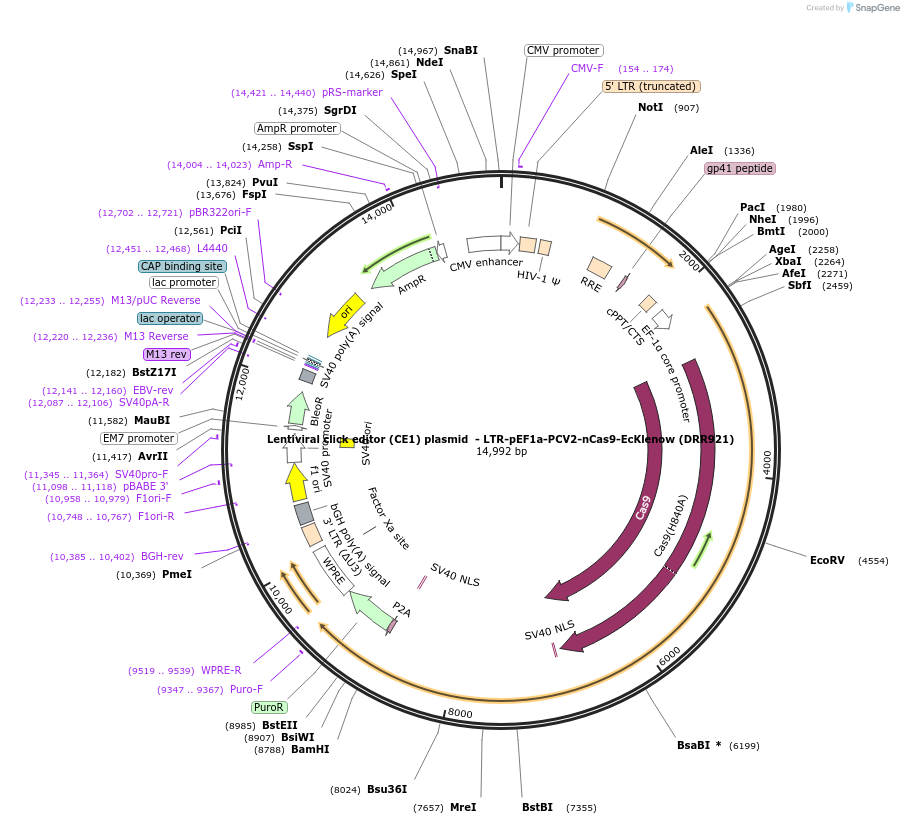
Lentiviral click editor (CE1) plasmid - LTR-pEF1a-PCV2-nCas9-EcKlenow (DRR921)
Plasmid#217798PurposeLentiviral plasmid to produce the prototypical CE1 construct (PCV2-nCas9-EcKlenow), expressed from the EF1a promoterDepositorInsertXTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR and LentiviralTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterEF1aAvailable SinceSept. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
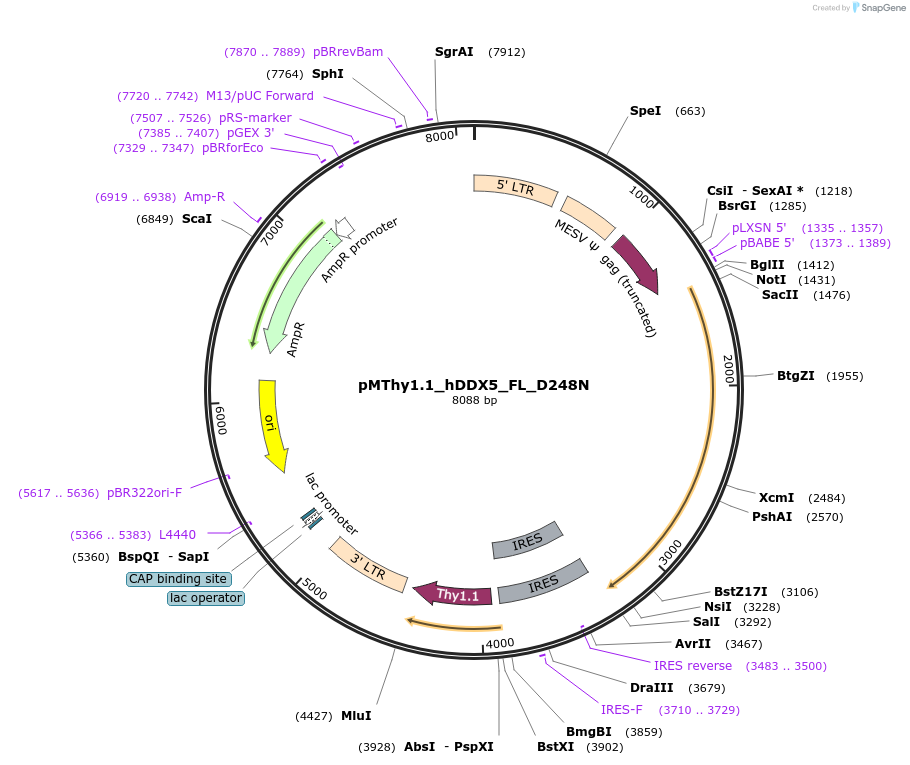
pMThy1.1_hDDX5_FL_D248N
Plasmid#88874Purposeconstitutive expression of DDX5 D248N in mammalian cellsDepositorAvailable SinceJune 12, 2017AvailabilityAcademic Institutions and Nonprofits only -
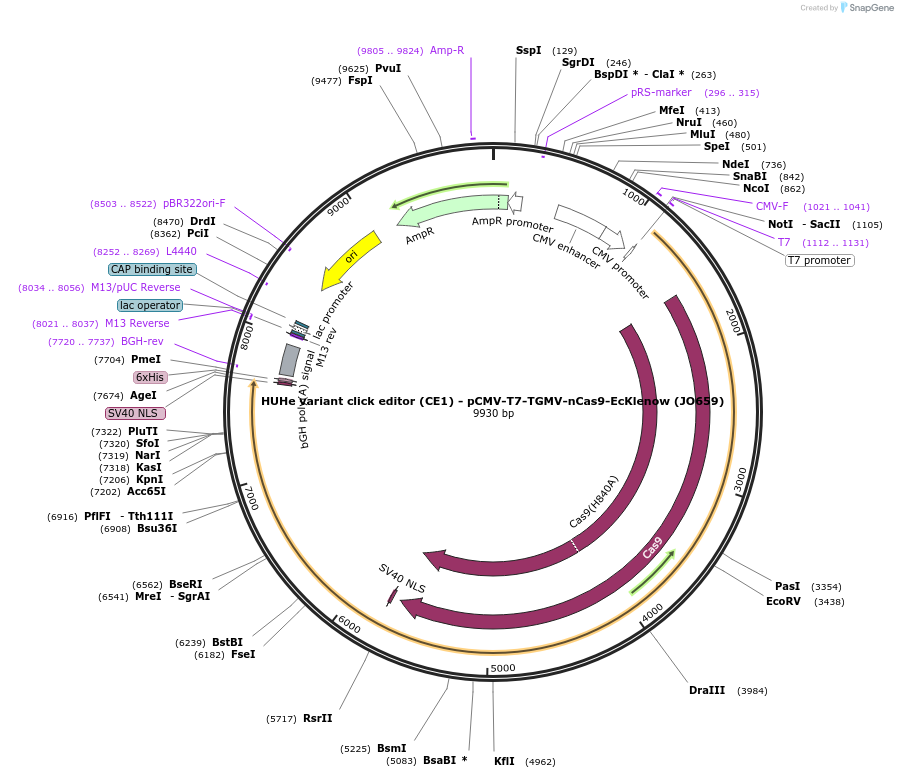
HUHe variant click editor (CE1) - pCMV-T7-TGMV-nCas9-EcKlenow (JO659)
Plasmid#208951PurposeA variant CE1 construct with TGMV HUH enconuclease, expressed from CMV or T7 promoters.DepositorInsertTGMV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
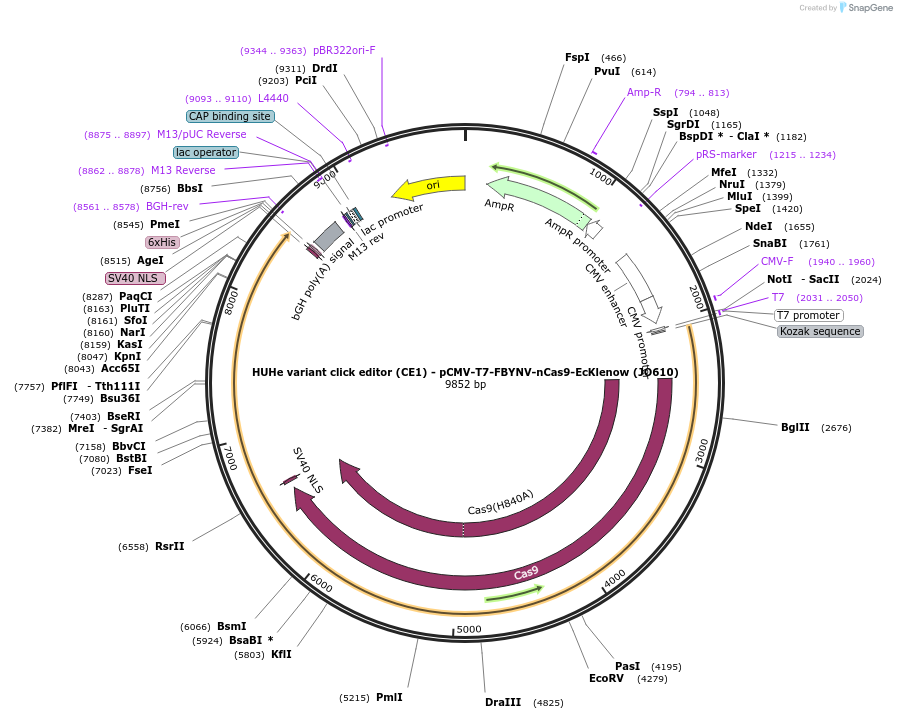
HUHe variant click editor (CE1) - pCMV-T7-FBYNV-nCas9-EcKlenow (JO610)
Plasmid#208950PurposeA variant CE1 construct with FBYNV HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertFBYNV-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
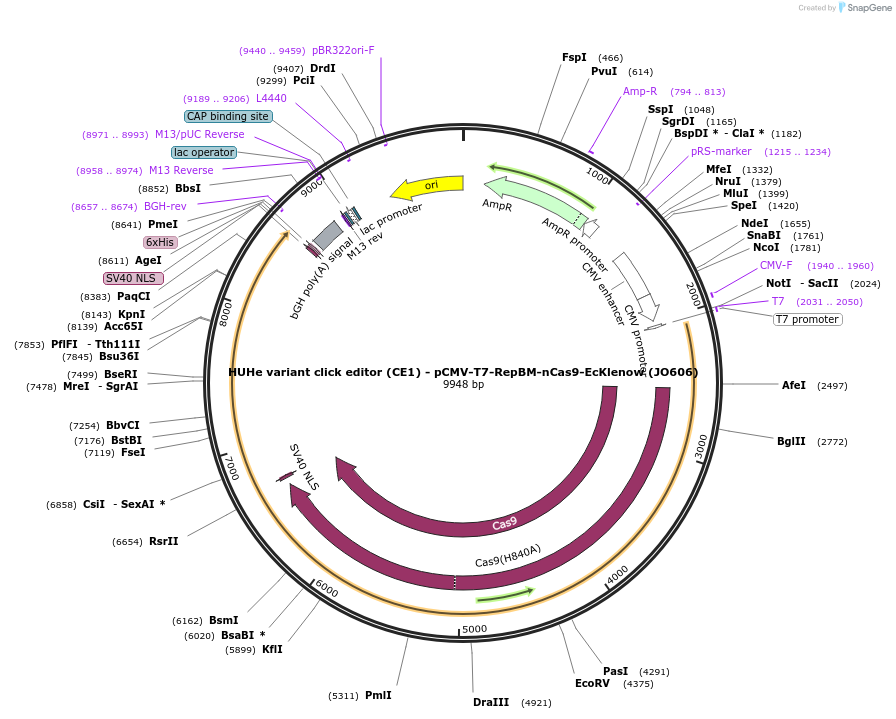
HUHe variant click editor (CE1) - pCMV-T7-RepBM-nCas9-EcKlenow (JO606)
Plasmid#208949PurposeA variant CE1 construct with RepBM HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertRepBm-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
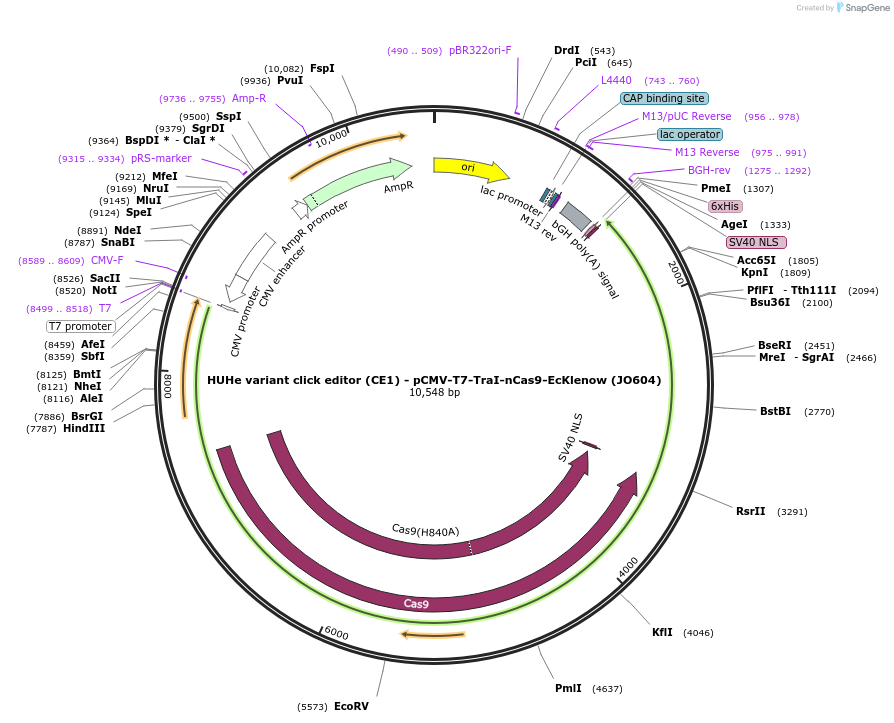
HUHe variant click editor (CE1) - pCMV-T7-TraI-nCas9-EcKlenow (JO604)
Plasmid#208948PurposeA variant CE1 construct with TraI HUH endonuclease, expressed from CMV or T7 promoters.DepositorInsertTraI-XTEN-nSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A); EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
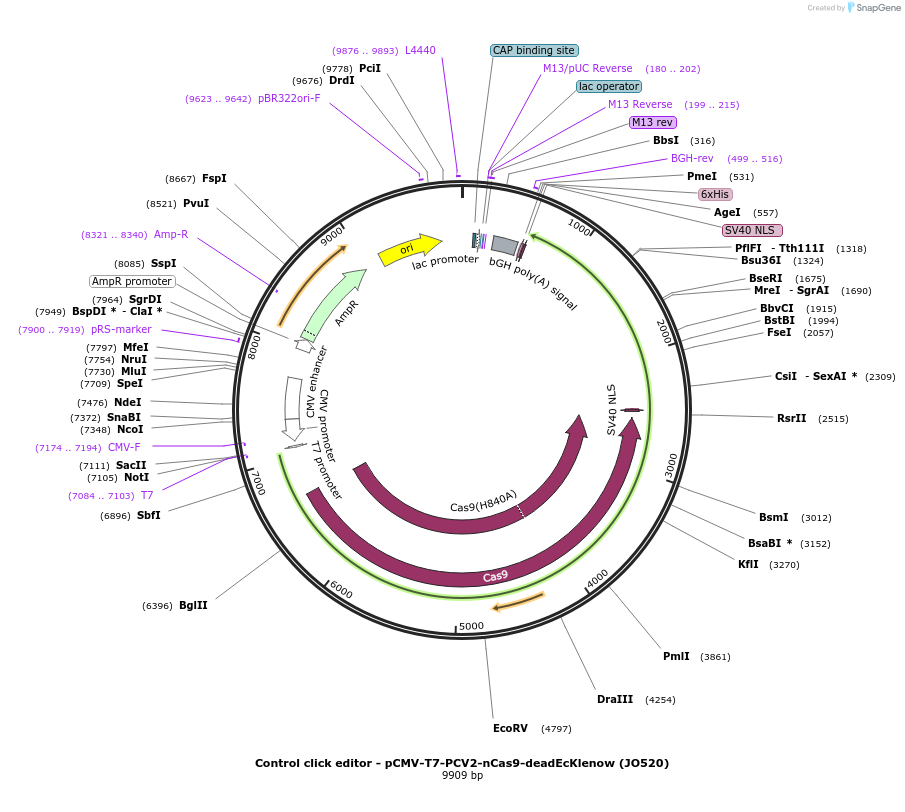
Control click editor - pCMV-T7-PCV2-nCas9-deadEcKlenow (JO520)
Plasmid#208945PurposeA control CE1 construct with catalytically attenuated EcKlenow DNA polymerase, expressed from CMV or T7 promoters.DepositorInsertPCV2-XTEN-nSpCas9-BPNLS-dEcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationnSpCas9(H840A);deadEcKlenow(-exo;D355A,D357A,D705…PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
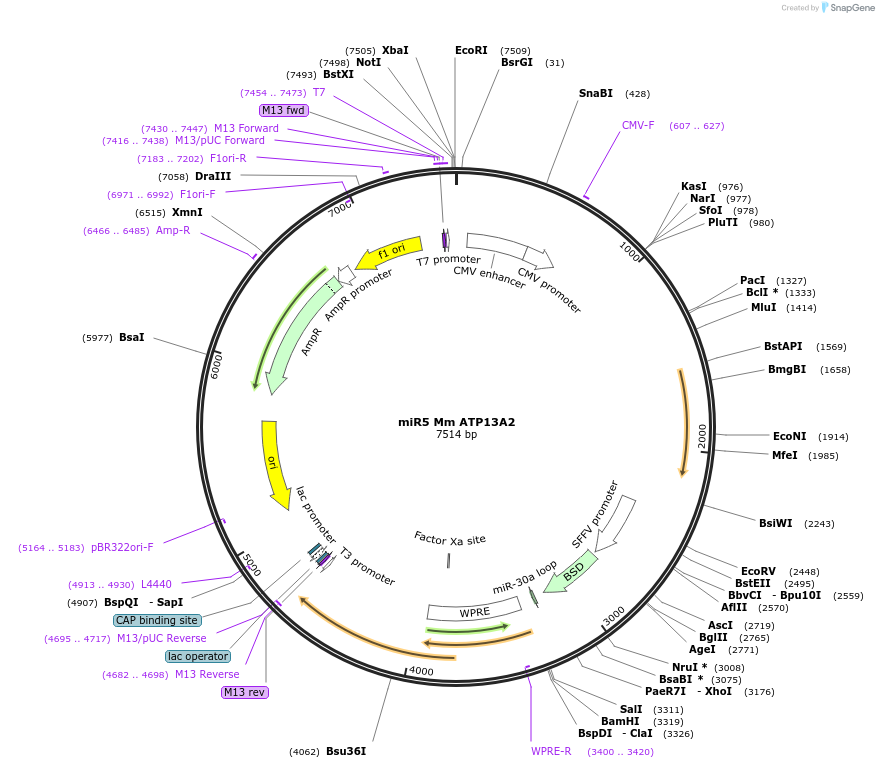
miR5 Mm ATP13A2
Plasmid#171822Purposetransfer plasmid for lentiviral vector production with miR for Mm ATP13A2DepositorAvailable SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
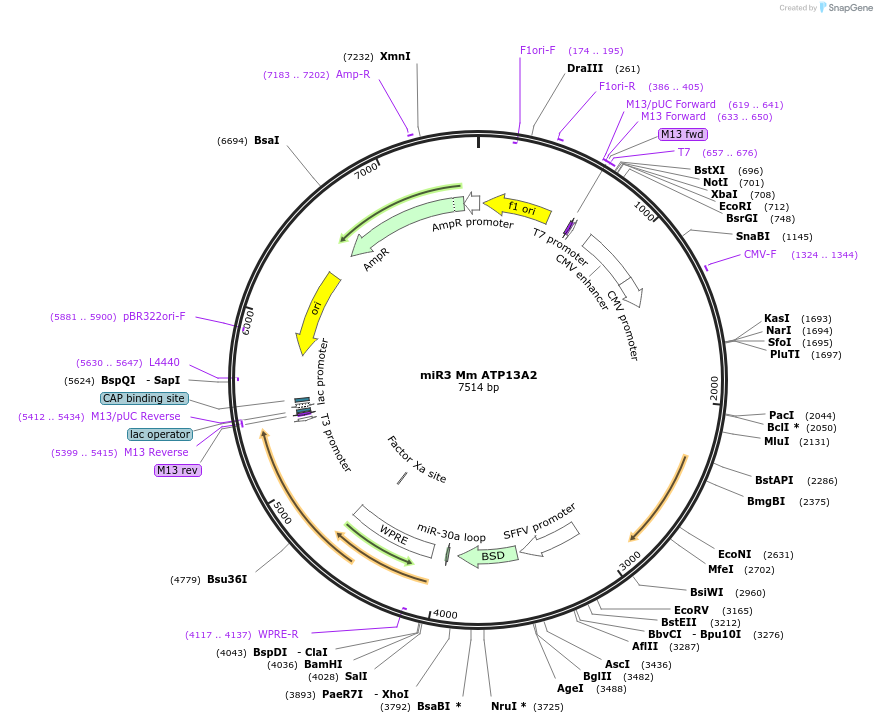
miR3 Mm ATP13A2
Plasmid#171771Purposetransfer plasmid for lentiviral vector production with miR for Mm ATP13A2DepositorAvailable SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
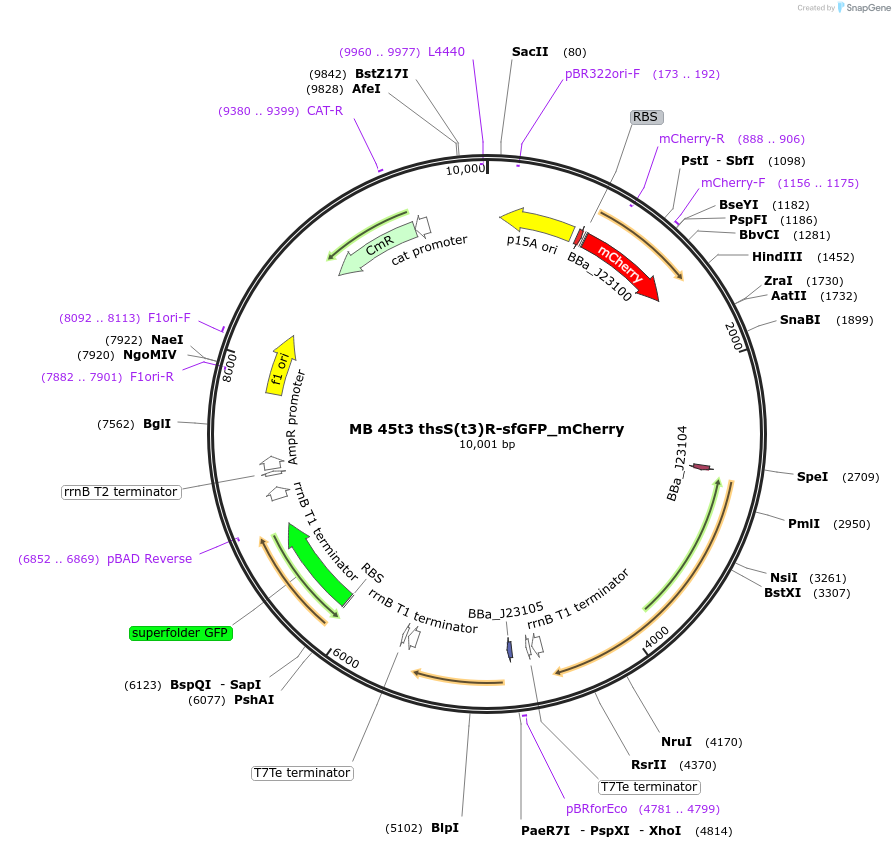
MB 45t3 thsS(t3)R-sfGFP_mCherry
Plasmid#232470PurposeOptimized thiosulfate sensor with fluorescent reporter (GFP), constitutive mCherryDepositorInsertsthsS(t3)
thsR
PphsA342
sfGFP
AxeTxe
mCherry
ExpressionBacterialMutationContains the following mutations K286Q, Q350H, I…PromoterJ23100 (TTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGC), J23…Available SinceSept. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
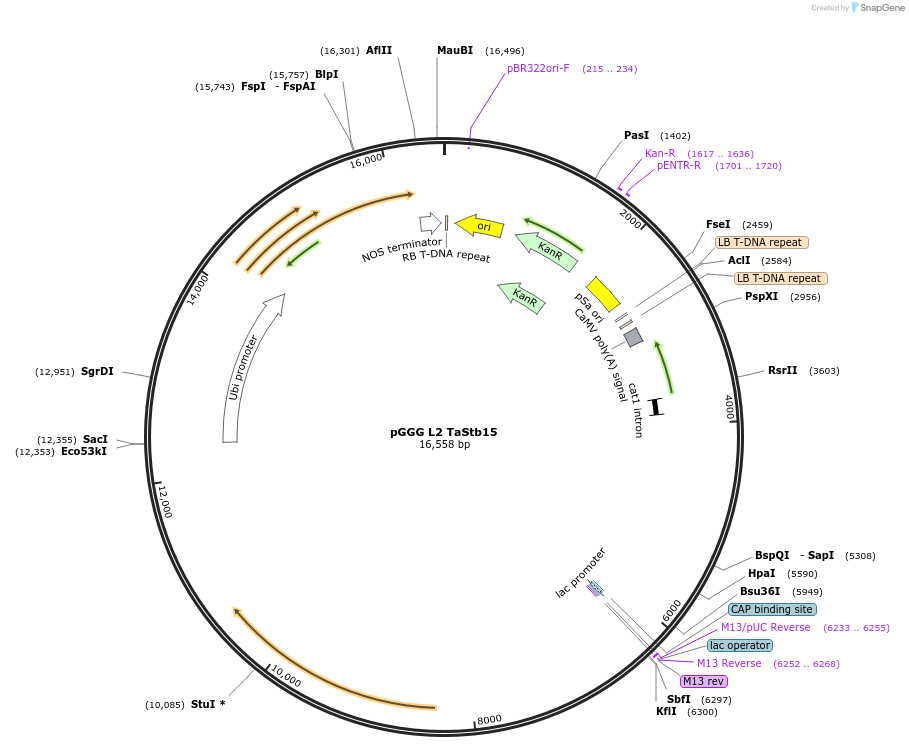
pGGG L2 TaStb15
Plasmid#226630PurposeWheat transformation vector used to express wheat Septoria tritici blotch (Stb) resistance gene 15DepositorInsertTriticum aestivum (wheat) Septoria tritici blotch 15 (TaStb15) disease resistance gene
UseSynthetic BiologyExpressionPlantPromoterNative TaStb15Available SinceAug. 15, 2025AvailabilityAcademic Institutions and Nonprofits only