We narrowed to 10,180 results for: yeast
-
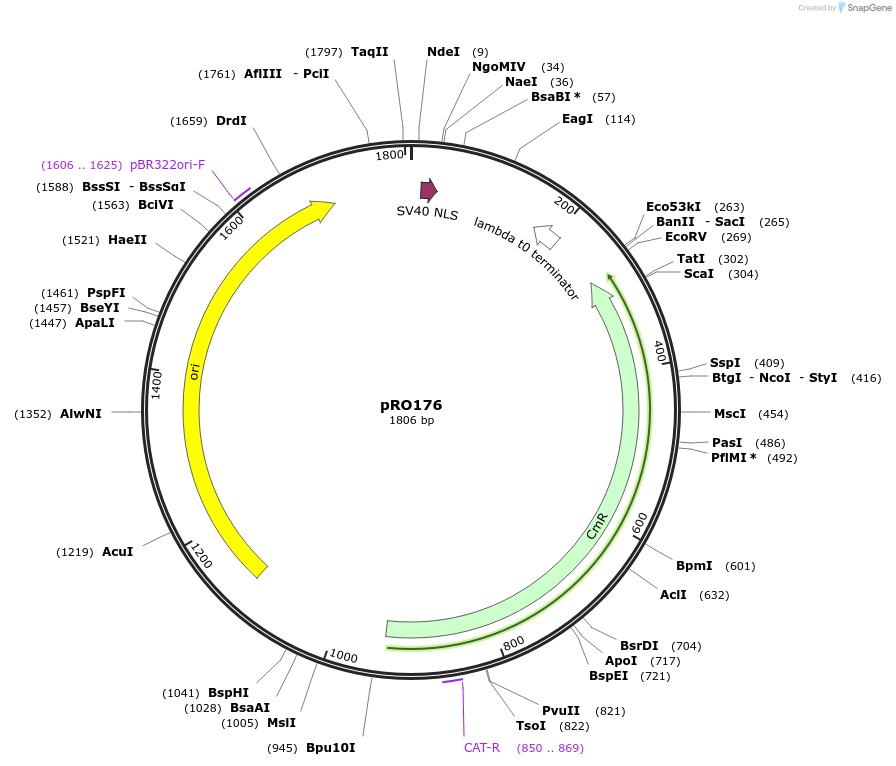
Plasmid#233824PurposeMoClo-YTK; type3A; Activation domains; NLS-VP48DepositorInsertNLS-VP48
ExpressionYeastAvailable SinceMay 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
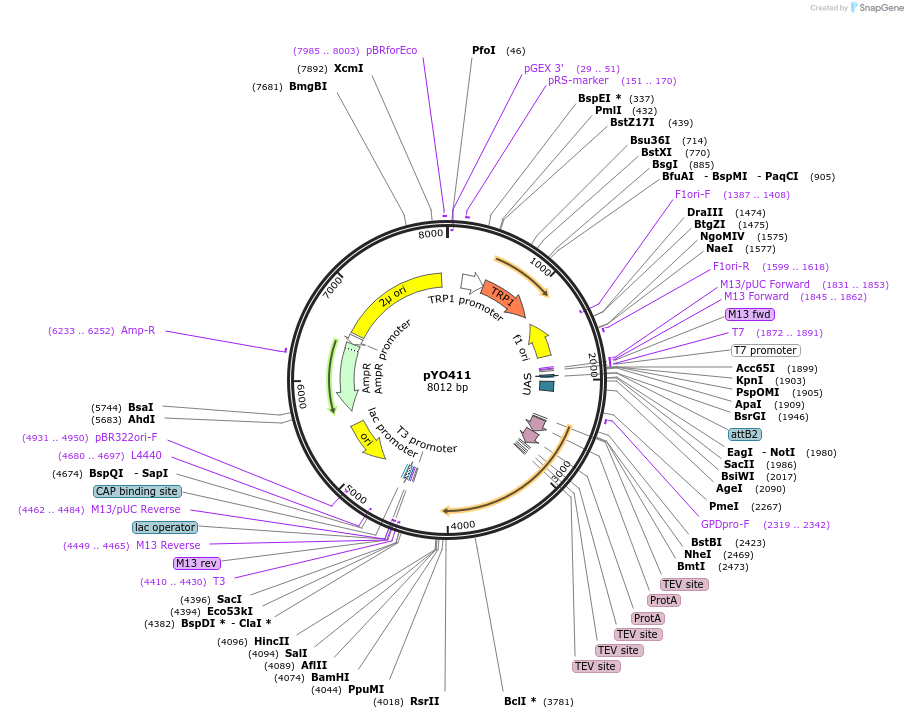
pYO411
Plasmid#235732PurposeProtein expression of ZZ-FL yeast Atg14 in yeastDepositorInsertATG14
Tags3xTEV-ZZExpressionYeastAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
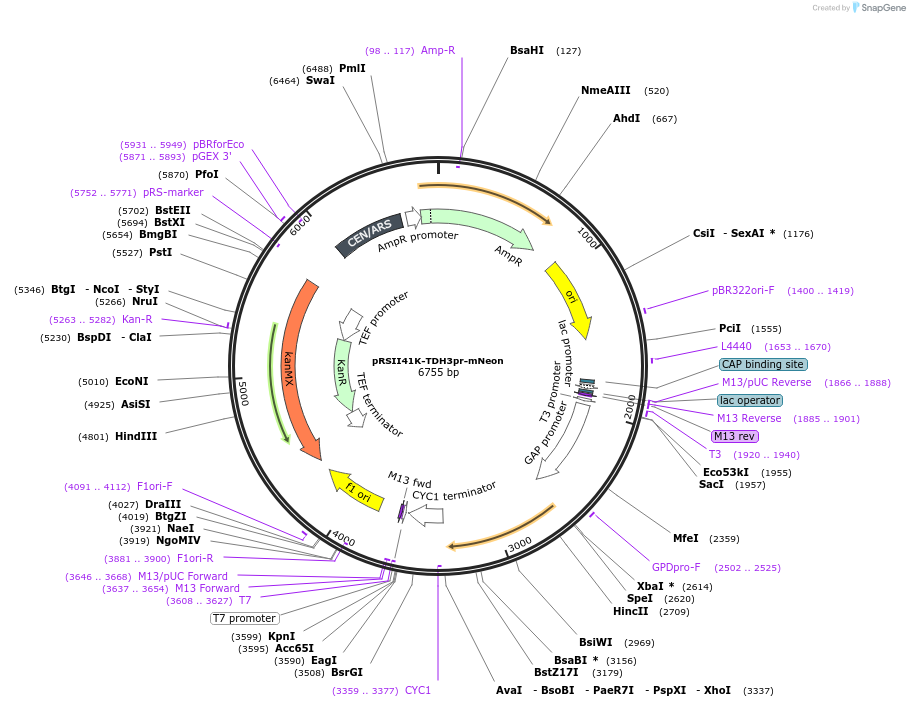
pRSII41K-TDH3pr-mNeon
Plasmid#194534PurposeLow copy vector backbone with G418 selectivity, encodes for TDH3(pr) driven expression of mNeonDepositorInsertkanMX
ExpressionYeastPromoterTDH3Available SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
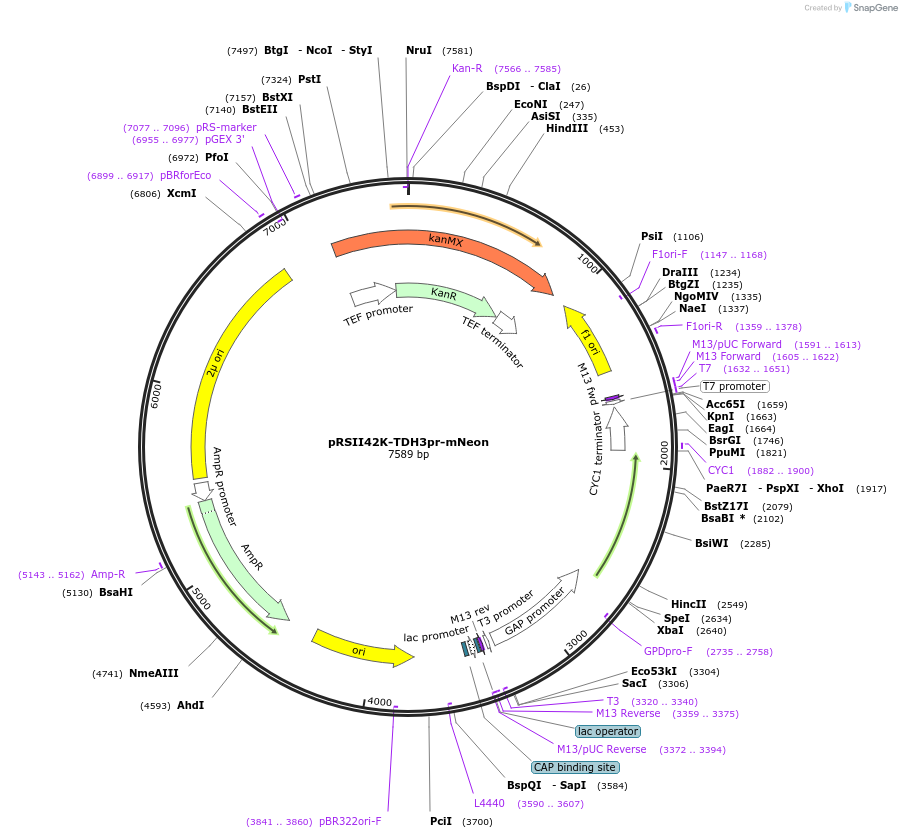
pRSII42K-TDH3pr-mNeon
Plasmid#194535PurposeHigh copy vector backbone with G418 selectivity, encodes for TDH3(pr) driven expression of mNeonDepositorInsertkanMX
ExpressionYeastPromoterTDH3Available SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
pRSII41N-TDH3pr-mNeon
Plasmid#194536PurposeLow copy vector backbone with nourseothricin selectivity, encodes for TDH3(pr) driven expression of mNeonDepositorInsertnatAC
ExpressionYeastPromoterTDH3Available SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
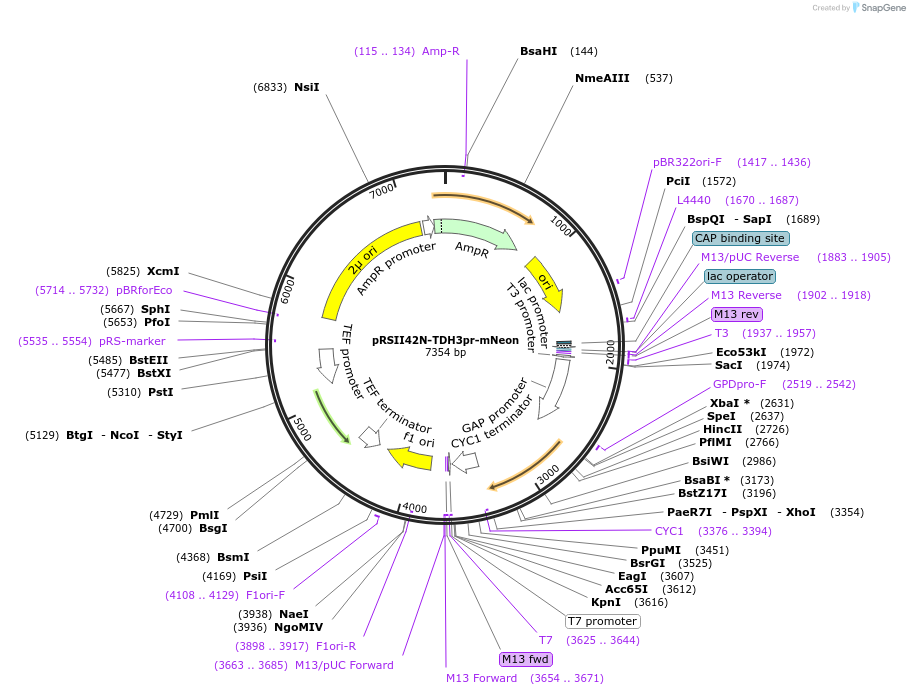
pRSII42N-TDH3pr-mNeon
Plasmid#194537PurposeHigh copy vector backbone with nourseothricin selectivity, encodes for TDH3(pr) driven expression of mNeonDepositorInsertnatAC
ExpressionYeastPromoterTDH3Available SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
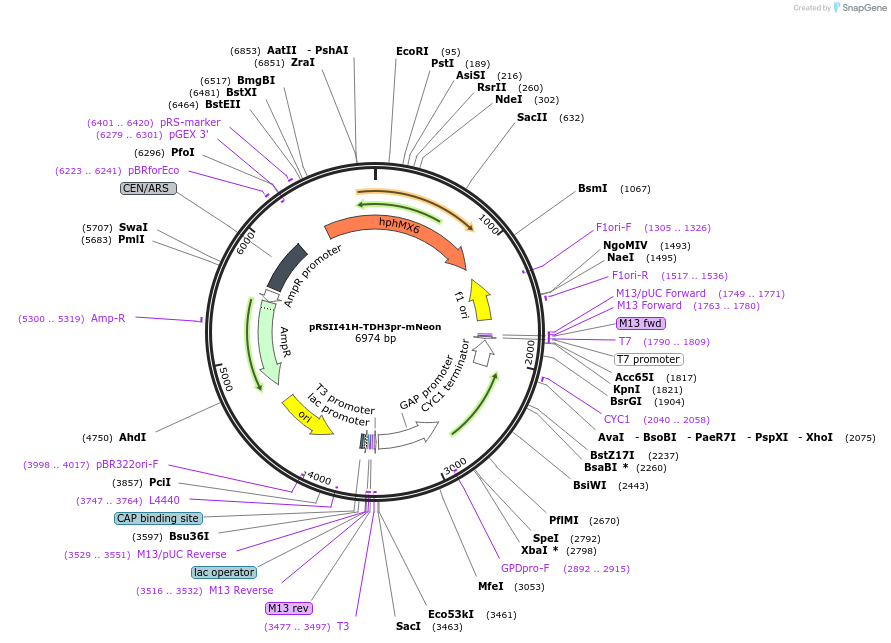
pRSII41H-TDH3pr-mNeon
Plasmid#194538PurposeLow copy vector backbone with hygromycin B selectivity, encodes for TDH3(pr) driven expression of mNeonDepositorInserthphMX
ExpressionYeastPromoterTDH3Available SinceOct. 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
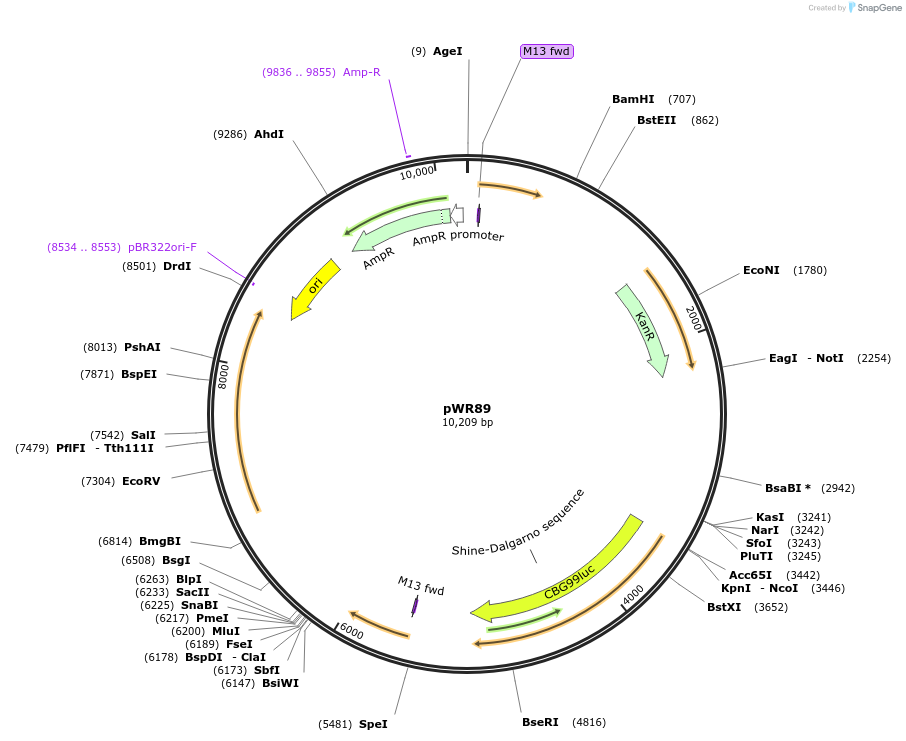
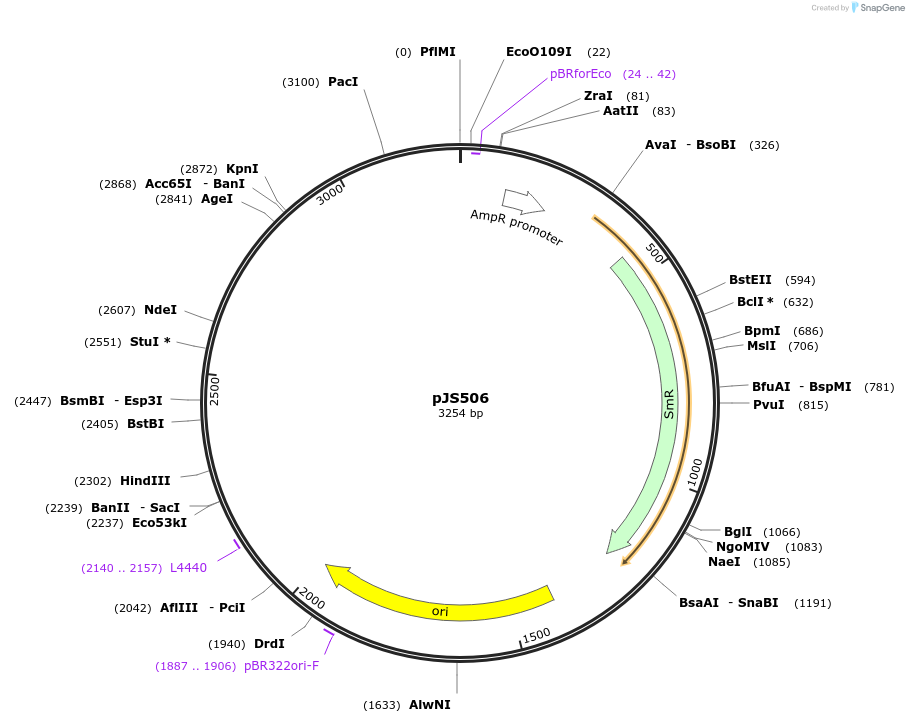
pJS506
Plasmid#200712PurposePromoter region from ATPase PMA1 as Pro5U promoter module for heterologous expression of plasma membrane localized transporters in yeastDepositorInsertPMA1 promoter (PMA1 Budding Yeast)
UseSynthetic Biology; Modular cloningMutationdomesticated for MoClo cloning (removal of intern…Available SinceAug. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAG306-pTet07-synYFP[TTG/AGA]-4xArg[CGA]-nLuc
Plasmid#199759PurposeElongation reporter construct to quantify elongation duration of 4_CGA stallingDepositorInsertsynYFP[TTG/AGA]-4xArg[CGA]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
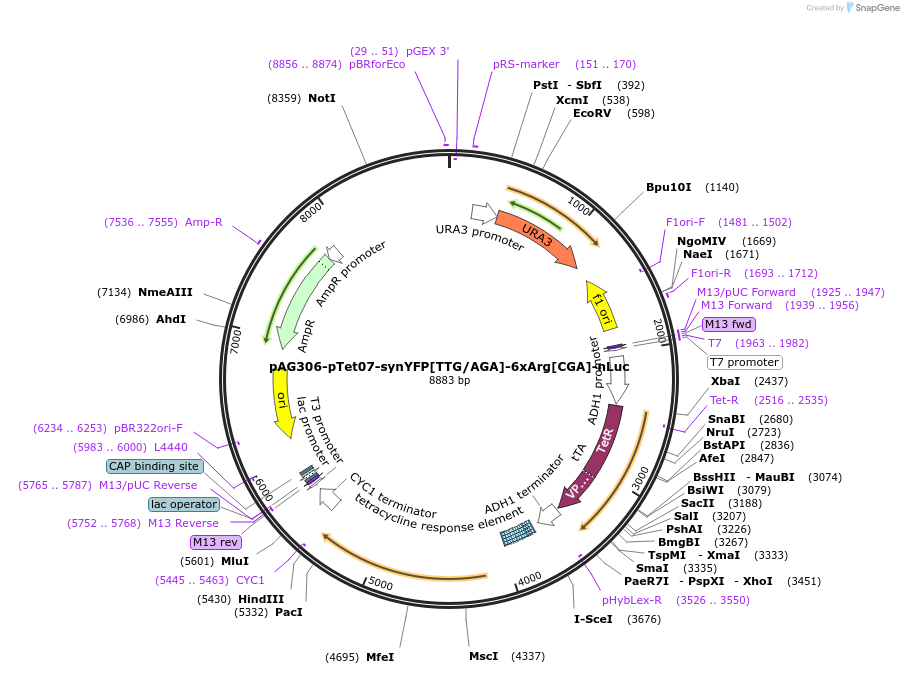
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[CGA]-nLuc
Plasmid#199760PurposeElongation reporter construct to quantify elongation duration of 6_CGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[CGA]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
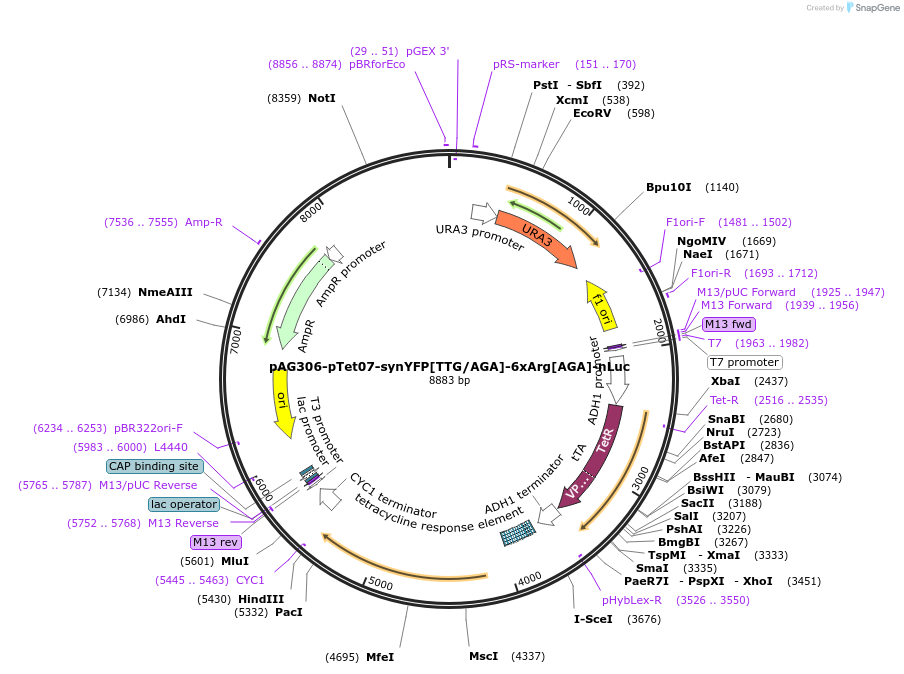
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[AGA]-nLuc
Plasmid#199761PurposeElongation reporter construct to quantify elongation duration of 6_AGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[AGA]-cLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
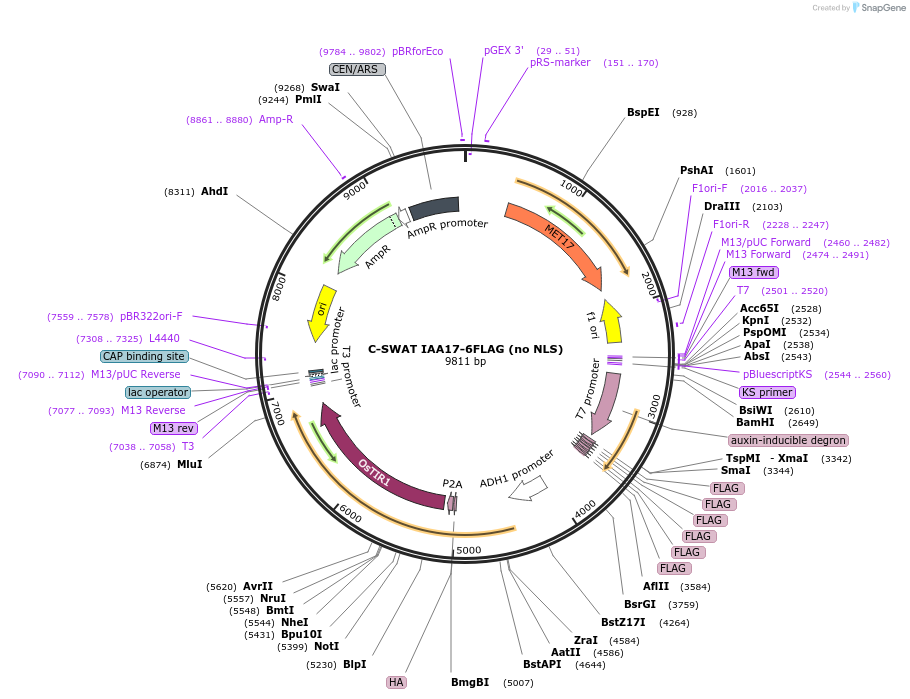
C-SWAT IAA17-6FLAG (no NLS)
Plasmid#193322PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin-Inducible Degron) and 6FLAG. ARF16(PB1)-P2A-osTIR1 has SIR4 homology arms for genome integration and does not have NLS tags..DepositorInsertIAA17
Tags6FLAGExpressionYeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
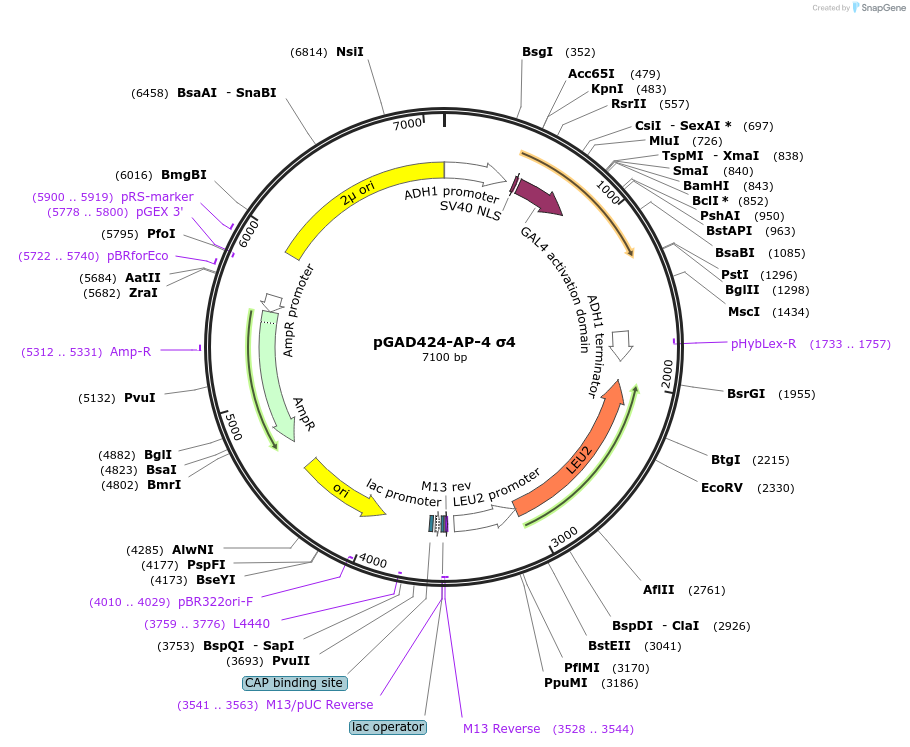
pGAD424-AP-4 σ4
Plasmid#198328PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-4 σ4 fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-4 σ4
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastPromoterADH1Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
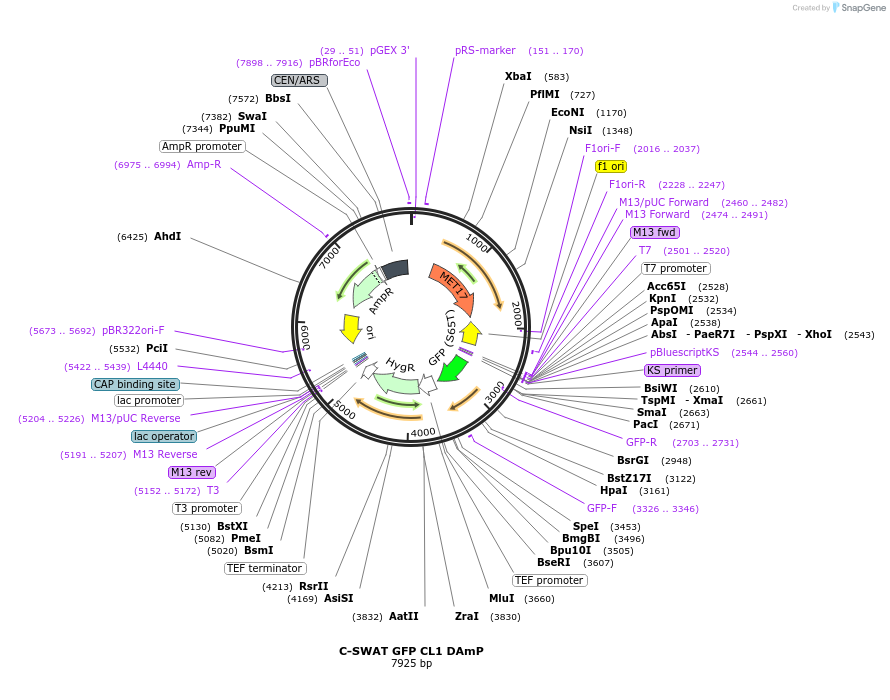
C-SWAT GFP CL1 DAmP
Plasmid#182513PurposeC-SWAT compatible vector for C-terminal tagging with GFP CL1 followed by the DAmP cassette. The stop codon after the CL1 degron is immediately followed by the Hygromycin B (Hph) resistance cassette.DepositorInsertCL1 and DAmP
TagsGFPExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
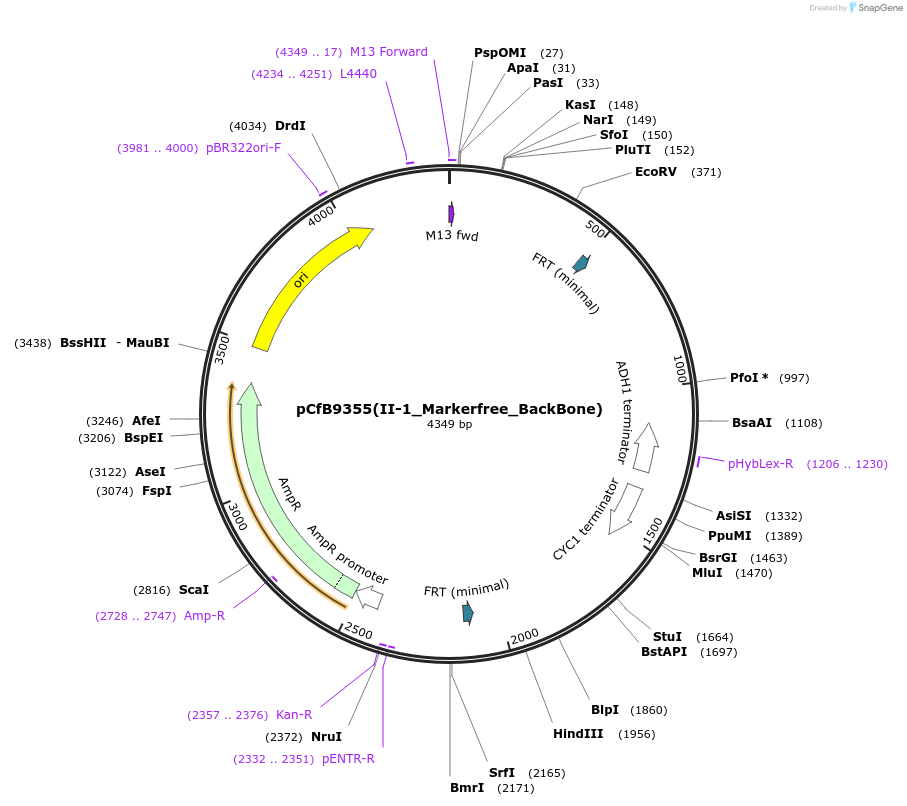
pCfB9355(II-1_Markerfree_BackBone)
Plasmid#161597PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site II-1, (Chr II: 341622..341641)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
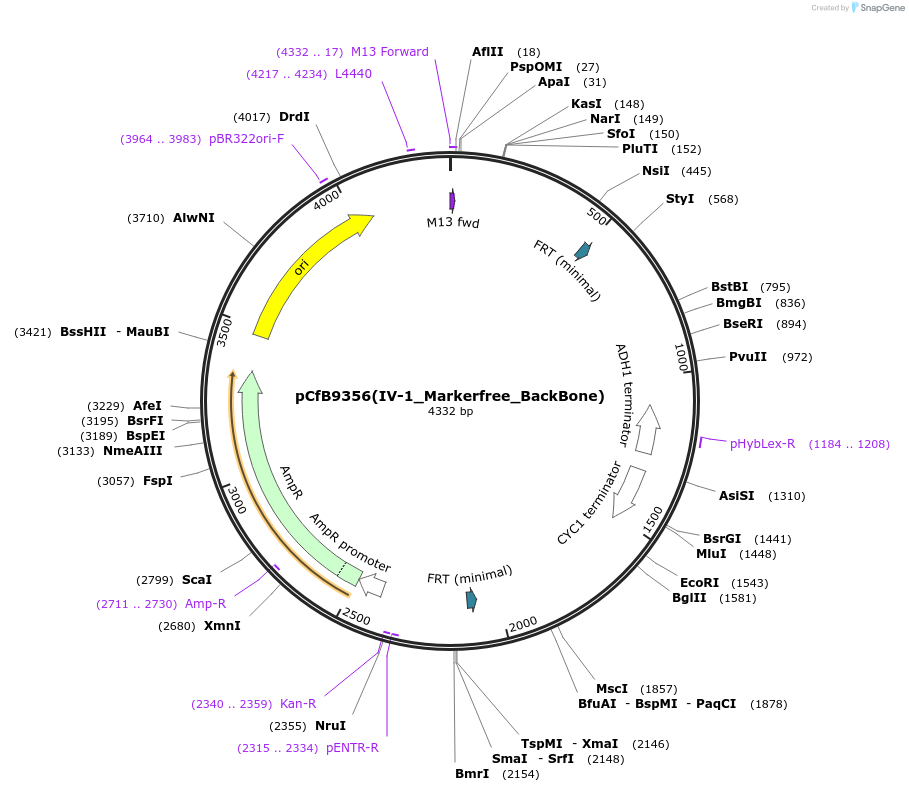
pCfB9356(IV-1_Markerfree_BackBone)
Plasmid#161598PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site IV-1, (Chr IV: 132921..132940)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
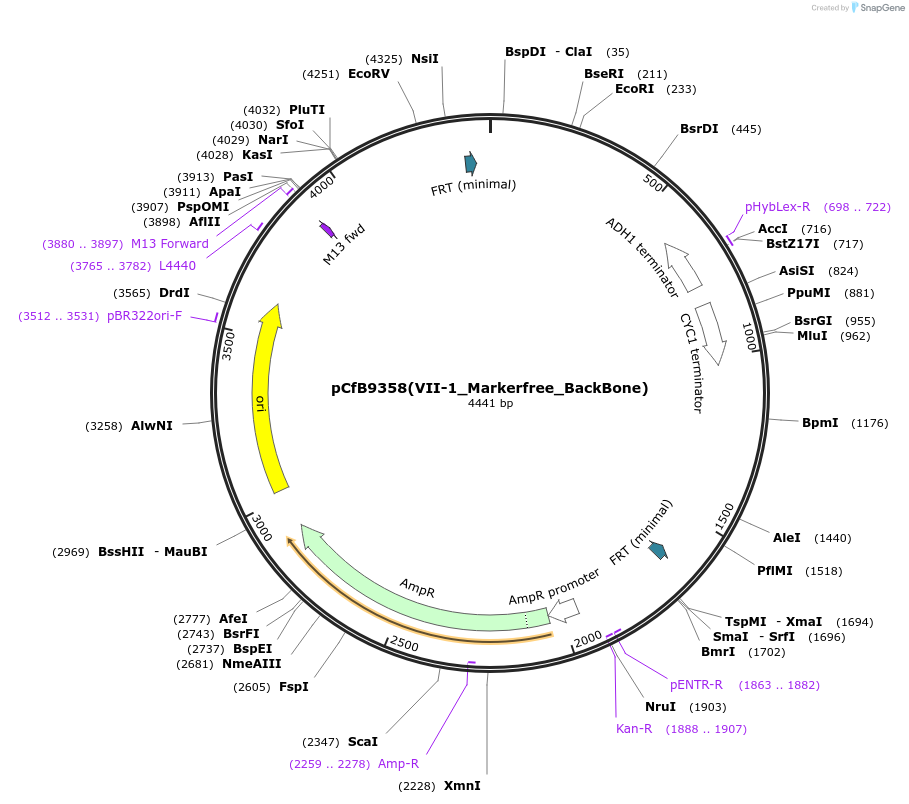
pCfB9358(VII-1_Markerfree_BackBone)
Plasmid#161599PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site VII-1, (Chr VII: 438509..438490)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only