We narrowed to 1,336 results for: HRAS
-
Plasmid#214043PurposeBacterial expression plasmid for SAVED-CHAT and PCaspase N-terminal fragment (aa 1-153) from Haliangium ochraceumDepositorInsertSAVED-CHAT
ExpressionBacterialMutationWTPromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
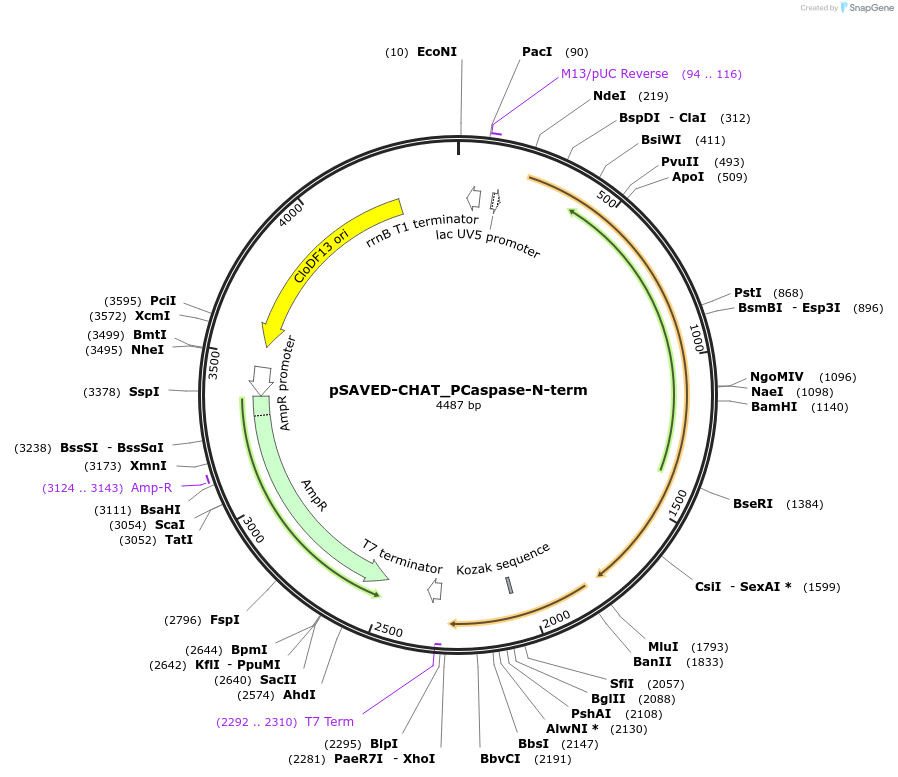
pSAVED-CHAT_PCaspase-N-term
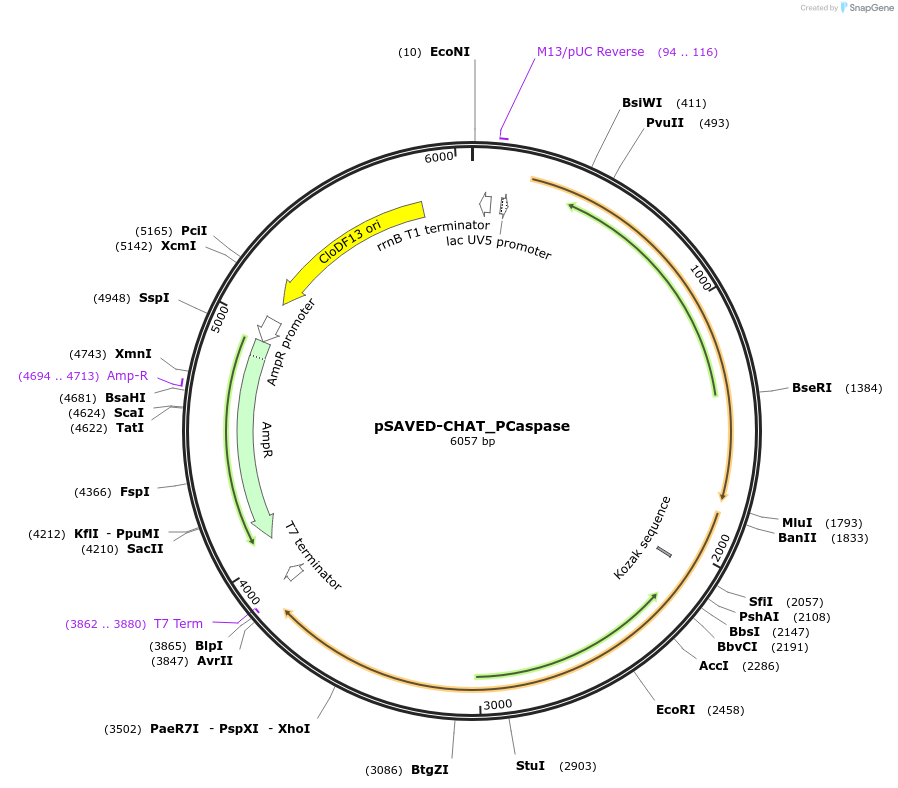
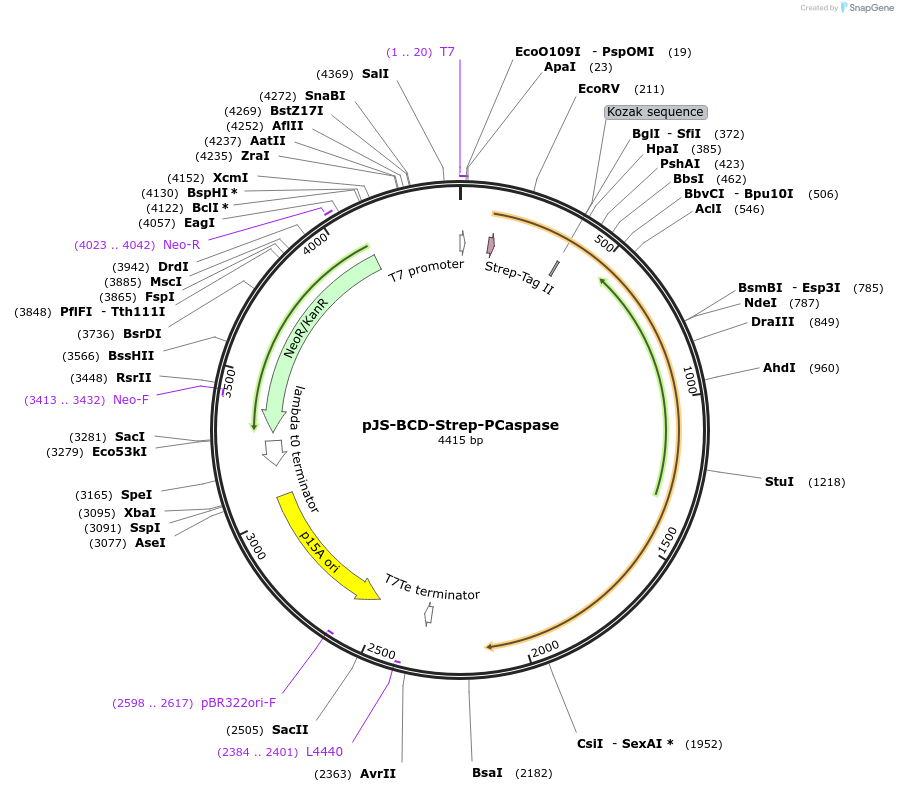
Plasmid#214044PurposeBacterial expression plasmid for SAVED-CHAT and PCaspase N-terminal fragment (aa 1-153) from Haliangium ochraceumDepositorInsertSAVED-CHAT-PCaspase
ExpressionBacterialMutationaa 1-153PromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
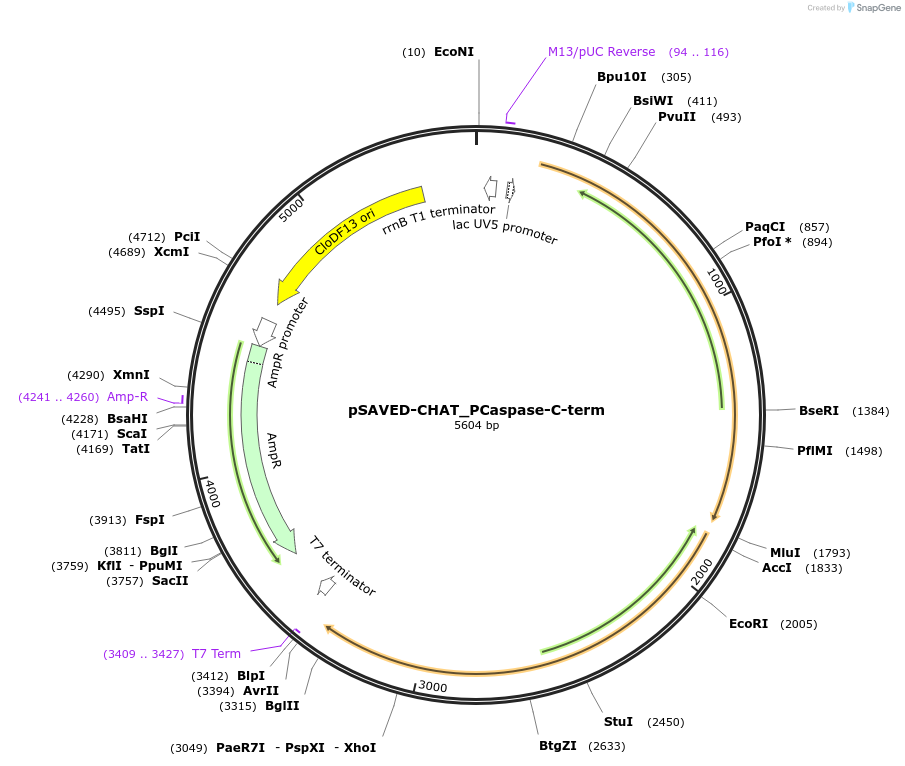
pSAVED-CHAT_PCaspase-C-term
Plasmid#214045PurposeBacterial expression plasmid for SAVED-CHAT and PCaspase C-terminal fragment (aa 154-666) from Haliangium ochraceumDepositorInsertSAVED-CHAT-PCaspase
ExpressionBacterialMutationaa 154-666PromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
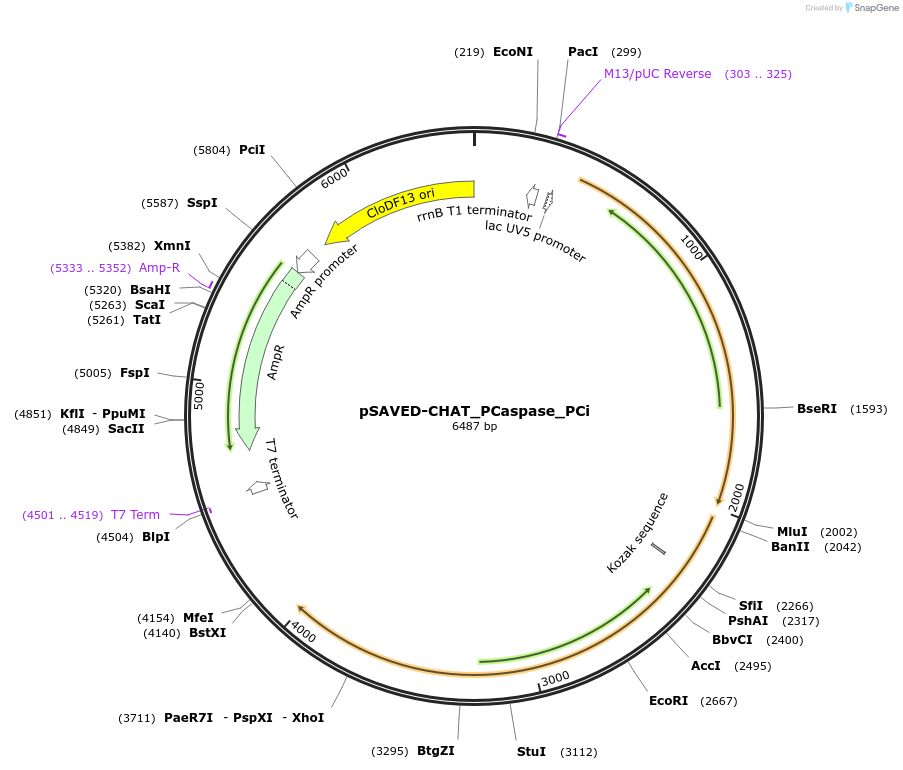
pSAVED-CHAT_PCaspase_PCi
Plasmid#214049PurposeBacterial expression plasmid for SAVED-CHAT, PCaspase, and PCi from Haliangium ochraceumDepositorInsertSAVED-CHAT-Pcaspase-PCi
ExpressionBacterialMutationWTPromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
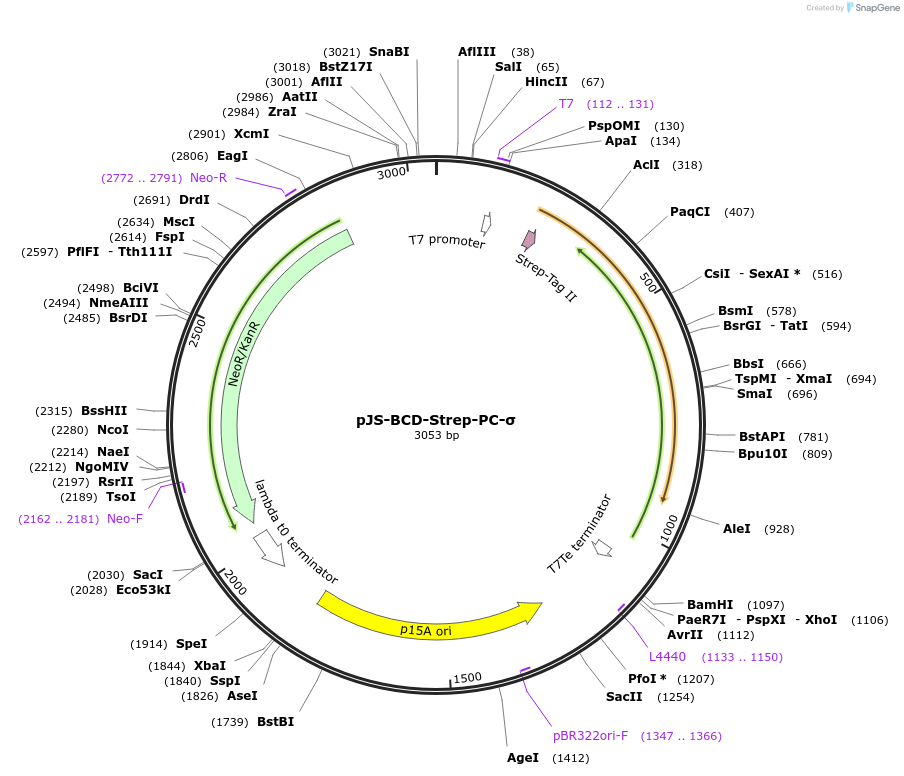
pJS-BCD-Strep-PC-σ
Plasmid#214037PurposeBacterial expression plasmid for strep-tagged PC-σ from Haliangium ochraceumDepositorInsertPC-σ
TagsStrep-tag IIExpressionBacterialMutationWTPromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
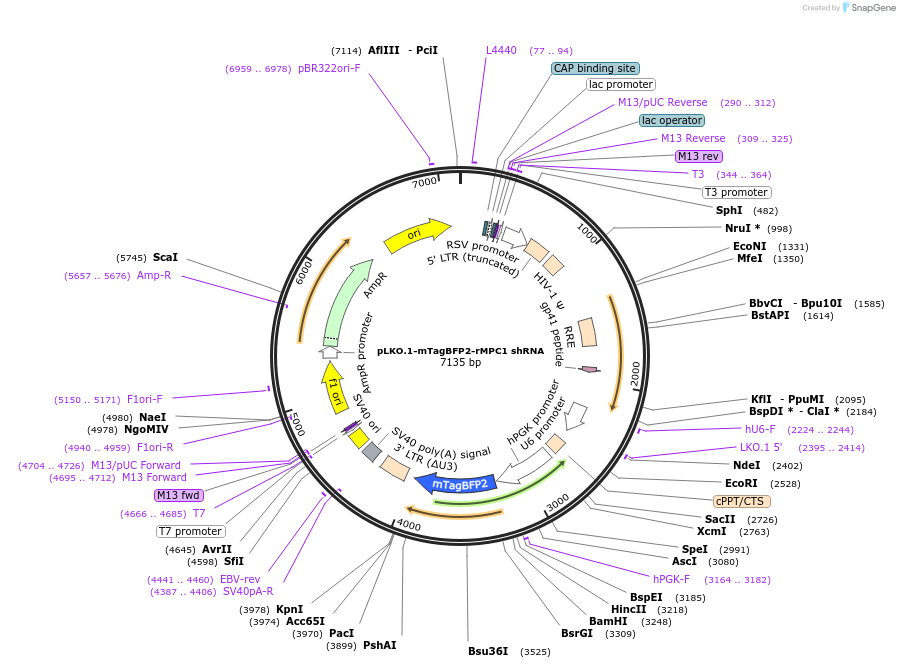
pLKO.1-mTagBFP2-rMPC1 shRNA
Plasmid#229016PurposeExpression of an shRNA construct that knocks down rat MPC1. This plasmid also encodes a blue fluorescent protein tag to verify transfectionDepositorAvailable SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
pclbw-opa1(isoform 7DeltaS1)-myc
Plasmid#62846PurposeMammalian Expression of myc-tagged Opa1 isoform 7 Delta S1DepositorInsertmmOPA1isoform7 DeltaS1 - myc (Opa1 Mouse)
UseRetroviralTags9xMycExpressionMammalianPromotercmv-bactinAvailable SinceJan. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
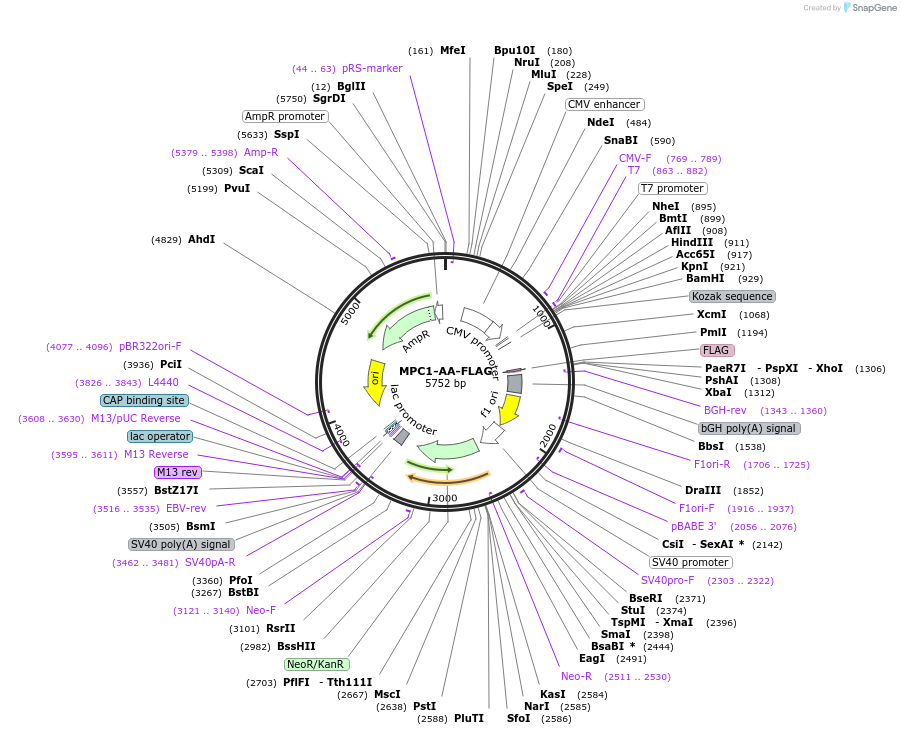
MPC1-AA-FLAG
Plasmid#228904PurposeExpression of mouse MPC1 in which K45 and K46 motif sites were replaced with alanine blocking acetylation with a C terminal FLAG tagDepositorInsertMPC1 (Mpc1 Mouse)
TagsFLAGExpressionMammalianMutationK45 and K46 motif sites were both replaced with a…PromoterCMVAvailable SinceFeb. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
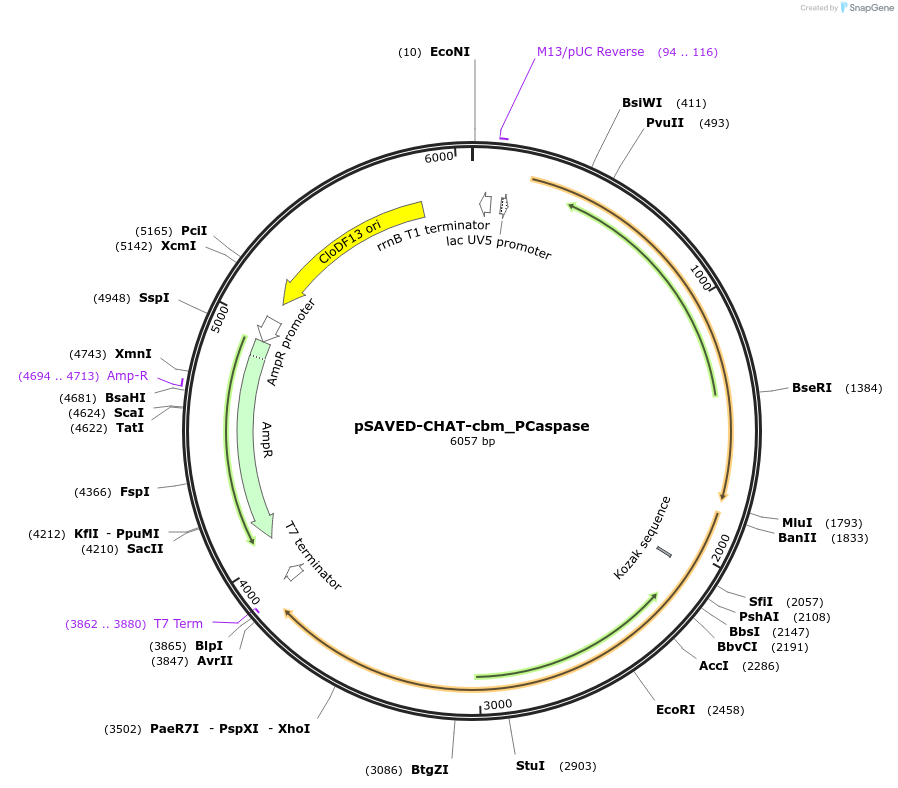
pSAVED-CHAT-cbm_PCaspase
Plasmid#214047PurposeBacterial expression plasmid for SAVED-CHAT cA3 binding pocket mutant and PCaspase from Haliangium ochraceumDepositorInsertSAVED-CHAT-PCaspase
ExpressionBacterialMutationK121E, Q122A, N274A, R276E, Y285A, F286APromoterlacUV5 promoterAvailable SinceAug. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
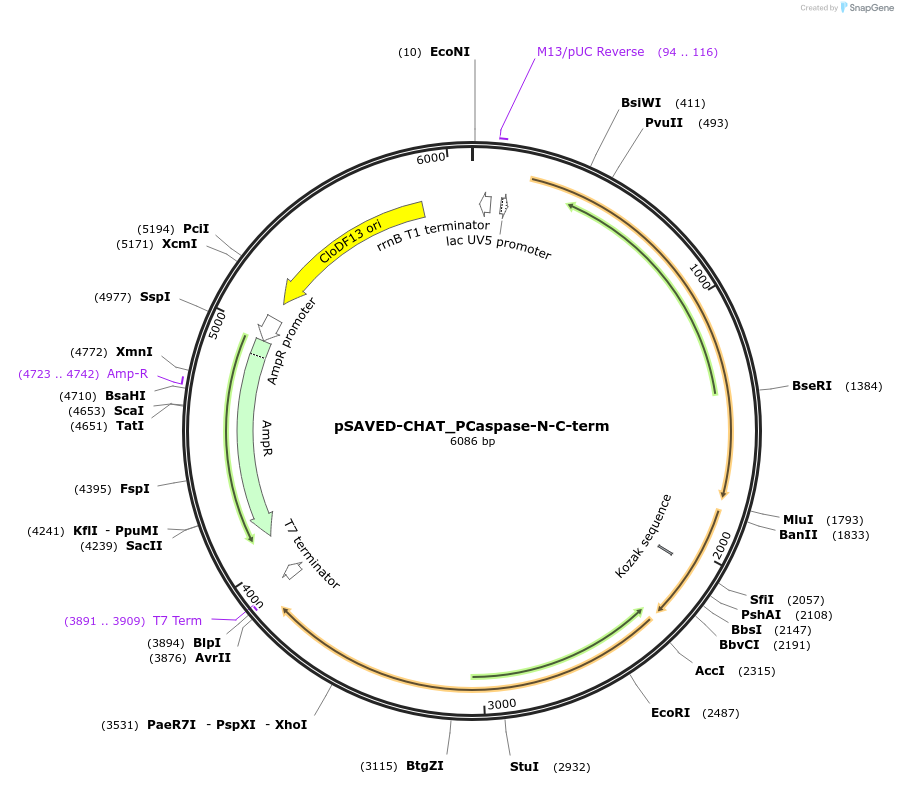
pSAVED-CHAT_PCaspase-N-C-term
Plasmid#214046PurposeBacterial expression plasmid for SAVED-CHAT, PCaspase N-terminal fragment (aa 1-153), and C-terminal fragment (154-666) from Haliangium ochraceumDepositorInsertSAVED-CHAT-PCaspase
ExpressionBacterialMutationaa 1-153, 154-666PromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
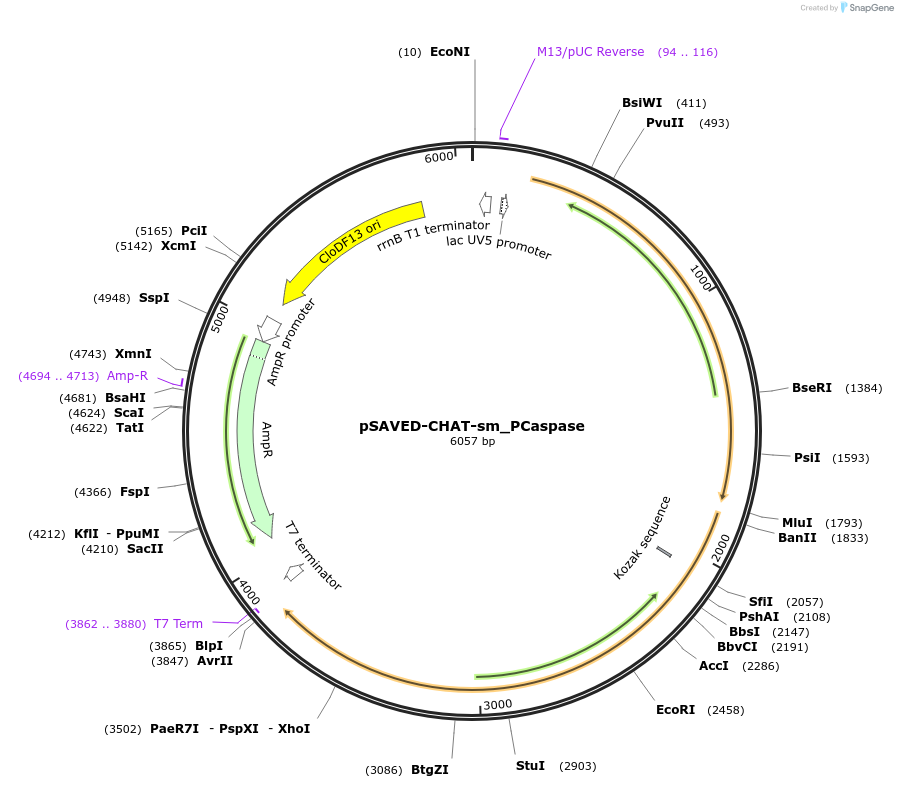
pSAVED-CHAT-sm_PCaspase
Plasmid#214048PurposeBacterial expression plasmid for SAVED-CHAT CHAT-CHAT singlet interface mutant and PCaspase from Haliangium ochraceumDepositorInsertSAVED-CHAT-PCaspase
ExpressionBacterialMutationR456E, D459K, D476K, R479EPromoterlacUV5 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
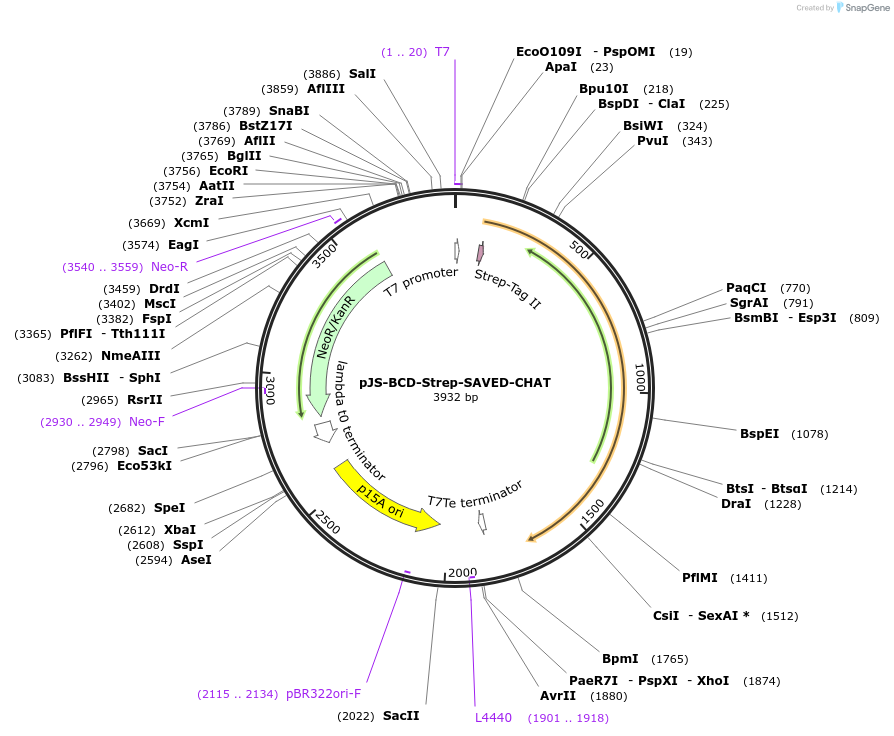
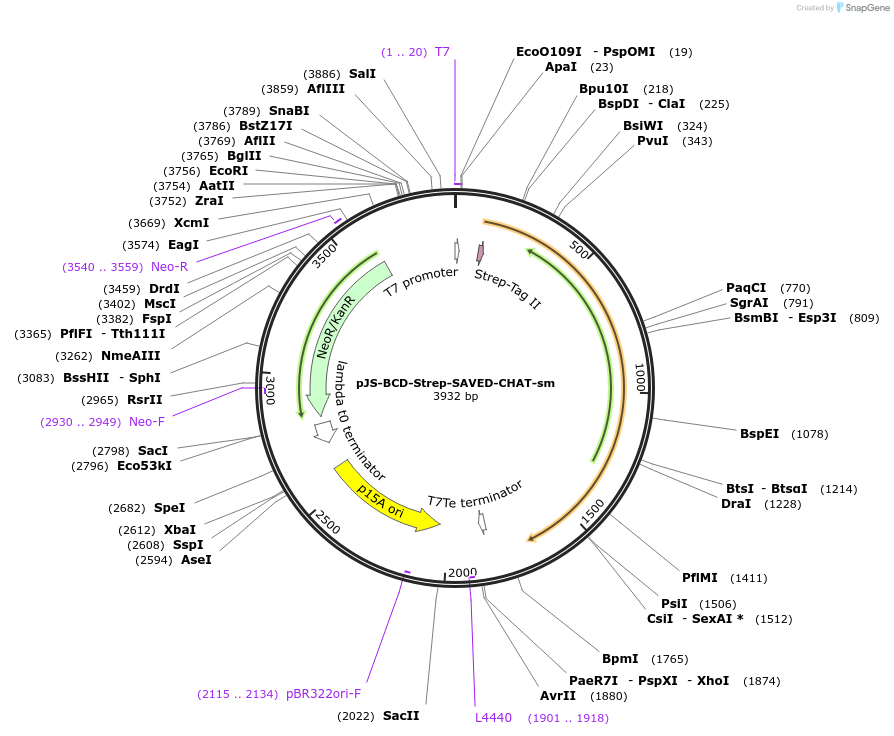
pJS-BCD-Strep-SAVED-CHAT
Plasmid#214030PurposeBacterial expression plasmid for strep-tagged wild-type SAVED-CHAT from Haliangium ochraceumDepositorInsertSAVED-CHAT
TagsStrep-tag IIExpressionBacterialMutationWTPromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
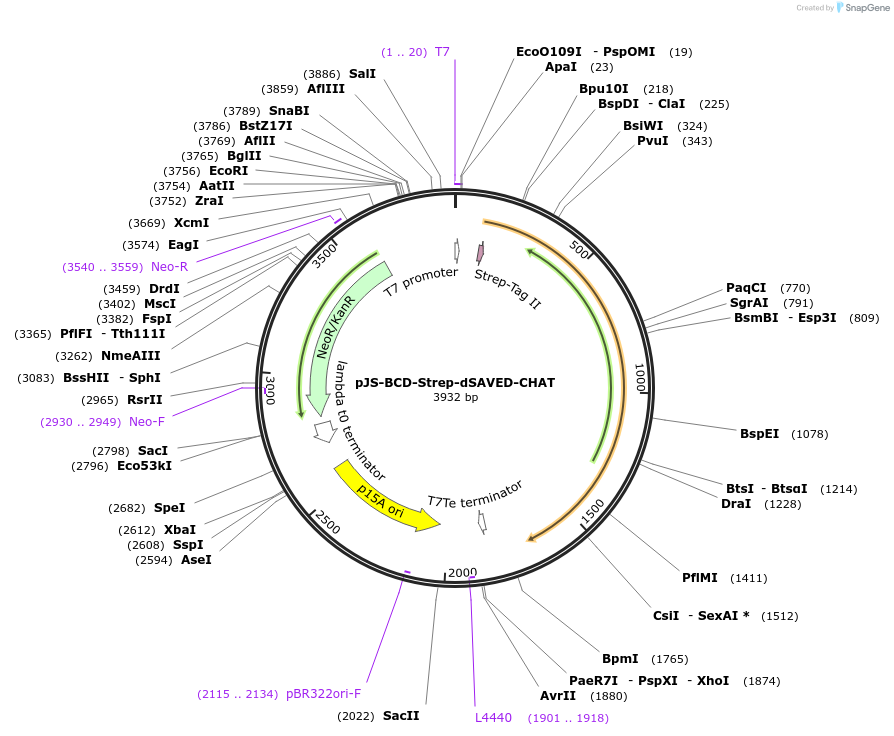
pJS-BCD-Strep-dSAVED-CHAT
Plasmid#214031PurposeBacterial expression plasmid for strep-tagged catalytically dead SAVED-CHAT mutant from Haliangium ochraceumDepositorInsertSAVED-CHAT
TagsStrep-tag IIExpressionBacterialMutationH375A, C422APromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
pJS-BCD-Strep-SAVED-CHAT-sm
Plasmid#214033PurposeBacterial expression plasmid for strep-tagged SAVED-CHAT CHAT-CHAT singlet interface mutant from Haliangium ochraceumDepositorInsertSAVED-CHAT
TagsStrep-tag IIExpressionBacterialMutationR456E, D459K, D476K, R479EPromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
pJS-BCD-Strep-PCaspase
Plasmid#214034PurposeBacterial expression plasmid for strep-tagged PCaspase (Prokaryotic Caspase) from Haliangium ochraceumDepositorInsertPCaspase
TagsStrep-tag IIExpressionBacterialMutationWTPromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
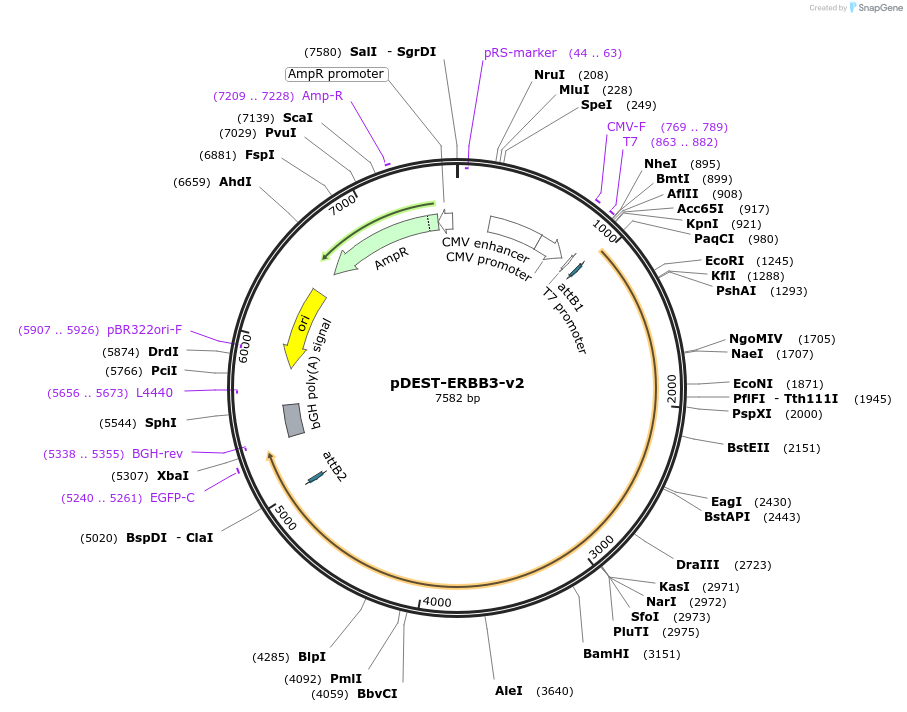
pDEST-ERBB3-v2
Plasmid#154894PurposeExpresses ERBB3 (a.k.a HER3) fused to Venus fragment 2 (v2) for use in bimolecular fluorescence complementation (BiFC) assaysDepositorAvailable SinceJuly 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
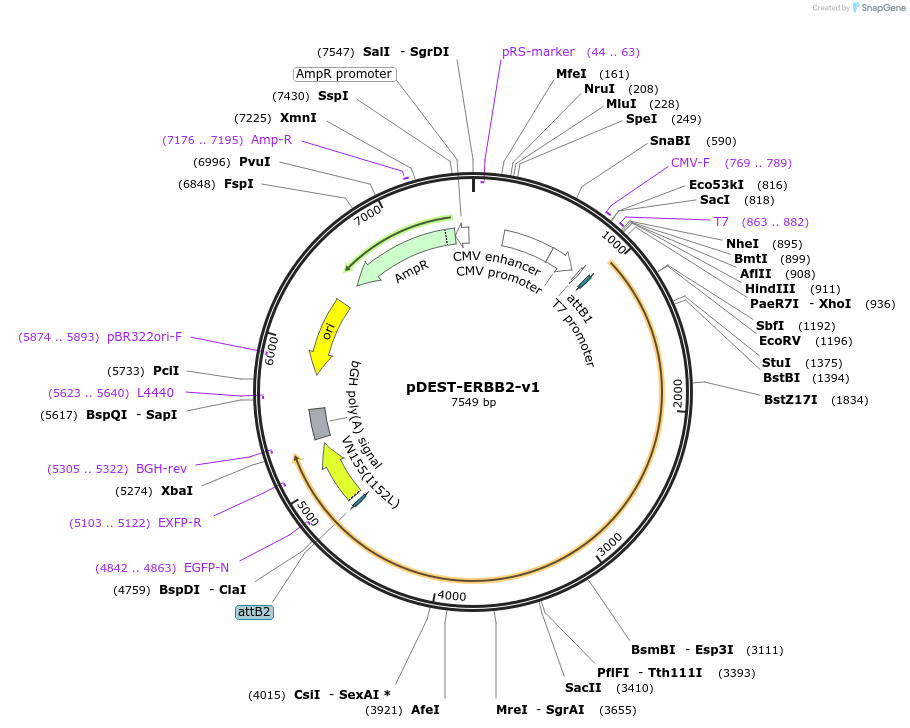
pDEST-ERBB2-v1
Plasmid#154901PurposeExpresses ERBB2 fused to Venus fragment 1 (v1) for use in bimolecular fluorescence complementation (BiFC) assaysDepositorAvailable SinceJuly 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
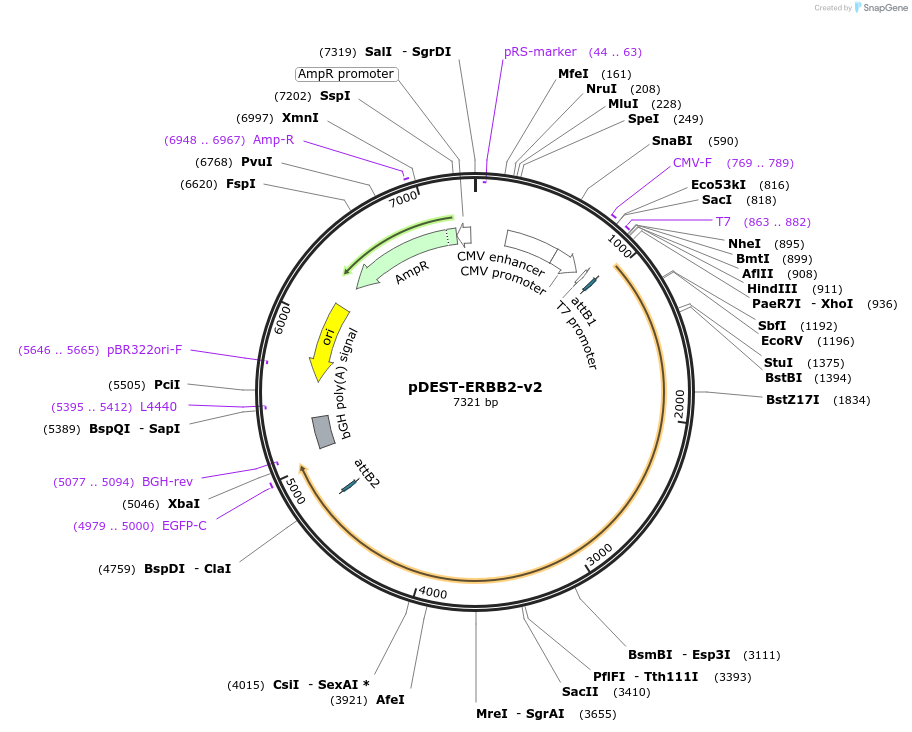
pDEST-ERBB2-v2
Plasmid#154895PurposeExpresses ERBB2 (a.k.a HER2) fused to Venus fragment 2 (v2) for use in bimolecular fluorescence complementation (BiFC) assaysDepositorAvailable SinceJuly 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
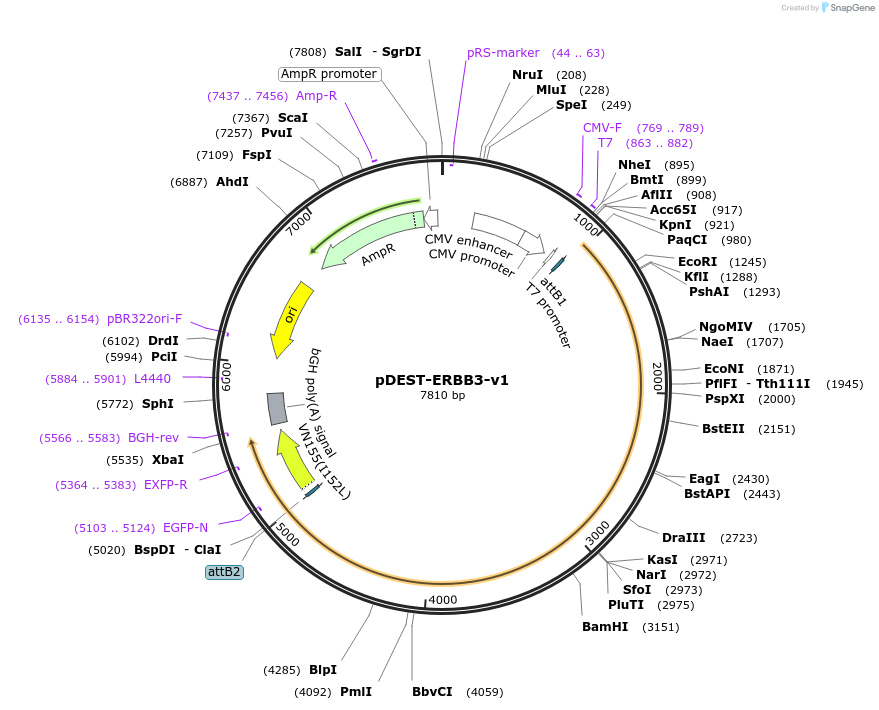
pDEST-ERBB3-v1
Plasmid#154893PurposeExpresses ERBB3 (a.k.a HER3) fused to Venus fragment 1 (v1) for use in bimolecular fluorescence complementation (BiFC) assaysDepositorAvailable SinceJuly 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
MLM3636-Prnp-CDS-Pos
Plasmid#61857PurposeExpresses a gRNA plasmid targeting the prion gene's exon 3, positive strandDepositorAvailable SinceFeb. 3, 2015AvailabilityAcademic Institutions and Nonprofits only