We narrowed to 14,004 results for: Ung;
-
Plasmid#48652PurposeBacterial NM crRNA expression: targets NM to protospacer B (ACTTTAAAAGTATTCGCCAT), p15A/chloramphenicolDepositorInsertBacterial NM crRNA to prototspacer B
UseCRISPRTagsExpressionMutationPromoterJ23100 promoterAvailable SinceOct. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
PM-SP!TA
Plasmid#48649PurposeBacterial SP crRNA expression: targets SP to protospacer A (TACCATCTCAAGCTTGTTGA), p15A/chloramphenicolDepositorInsertBacterial SP crRNA to prototspacer A
UseCRISPRTagsExpressionMutationPromoterJ23100 promoterAvailable SinceOct. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP1
Plasmid#83326PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP1
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceAug. 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP12
Plasmid#83337PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP12
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP11
Plasmid#83336PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP11
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP10
Plasmid#83335PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP10
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP9
Plasmid#83334PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP9
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP8
Plasmid#83333PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP8
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP7
Plasmid#83332PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP7
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP6
Plasmid#83331PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP6
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP4
Plasmid#83329PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP4
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP3
Plasmid#83328PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP3
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
Pentaprobe PP2
Plasmid#83327PurposePentaprobe for assesing the RNA binding potential of a proteinDepositorInsertPP2
UseRna transcriptionTagsExpressionMammalianMutationPromoterCMVAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
FB376
Plasmid#209129PurposeMultipartite assembly of the TU for YFP expression with the TU for the auxotrophy marker pyr4 from T. reseeiDepositorInsertPgpdA:yfp:TtrpC::Ppyr4:pyr4:Tpyr4
UseSynthetic BiologyTagsExpressionMutationPromoterAvailable SinceJan. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
pED14xW-3
Plasmid#134806PurposeIPTG inducible expression of W-TAGA-Evo1 qtRNADepositorInsertW-TAGA-Evo1 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a6
Plasmid#134787PurposeLuciferase reporter for quadruplet codon TAGADepositorInsertluxAB-357-TAGA
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-TAGAPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a20
Plasmid#134788PurposeLuciferase reporter for quadruplet codon CAGGDepositorInsertluxAB-357-CAGG
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-CAGGPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a34
Plasmid#134789PurposeLuciferase reporter for quadruplet codon AGGGDepositorInsertluxAB-357-AGGG
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-AGGGPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a36
Plasmid#134790PurposeLuciferase reporter for quadruplet codon CGGTDepositorInsertluxAB-357-CGGT
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-CGGTPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a39
Plasmid#134791PurposeLuciferase reporter for quadruplet codon TACADepositorInsertluxAB-357-TACA
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-TACAPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a71
Plasmid#134792PurposeLuciferase reporter for quadruplet codon AGGADepositorInsertluxAB-357-AGGA
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-AGGAPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED1a89
Plasmid#134793PurposeLuciferase reporter for quadruplet codon CGGCDepositorInsertluxAB-357-CGGC
UseLuciferase and Synthetic BiologyTagsExpressionBacterialMutation357-CGGCPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED14xQ-3
Plasmid#134795PurposeIPTG inducible expression of Q-TAGA-Evo1 qtRNADepositorInsertQ-TAGA-Evo1 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED14xQ-4
Plasmid#134796PurposeIPTG inducible expression of Q-TAGA-Evo2 qtRNADepositorInsertQ-TAGA-Evo2 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED14xR-3
Plasmid#134798PurposeIPTG inducible expression of R-TAGA-Evo1 qtRNADepositorInsertR-TAGA-Evo1 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED14xR-4
Plasmid#134799PurposeIPTG inducible expression of R-TAGA-Evo2 qtRNADepositorInsertR-TAGA-Evo2 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pED14xS-3
Plasmid#134801PurposeIPTG inducible expression of S-TAGA-Evo2 qtRNADepositorInsertS-TAGA-Evo2 qtRNA
UseSynthetic BiologyTagsExpressionBacterialMutationPromoterAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
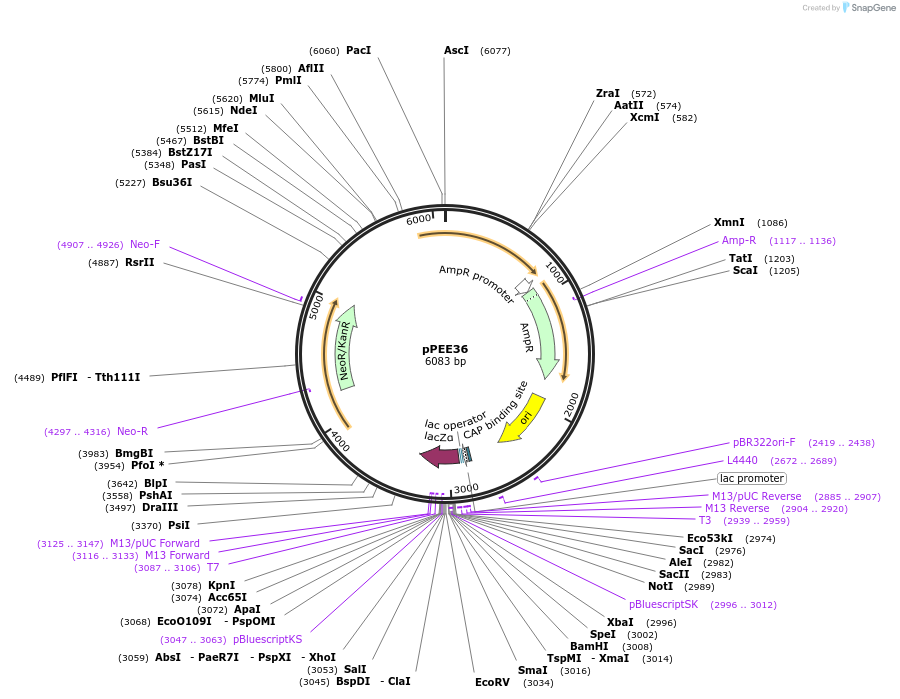
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedTagsExpressionMutationPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only