We narrowed to 8,526 results for: construct
-
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only
-
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
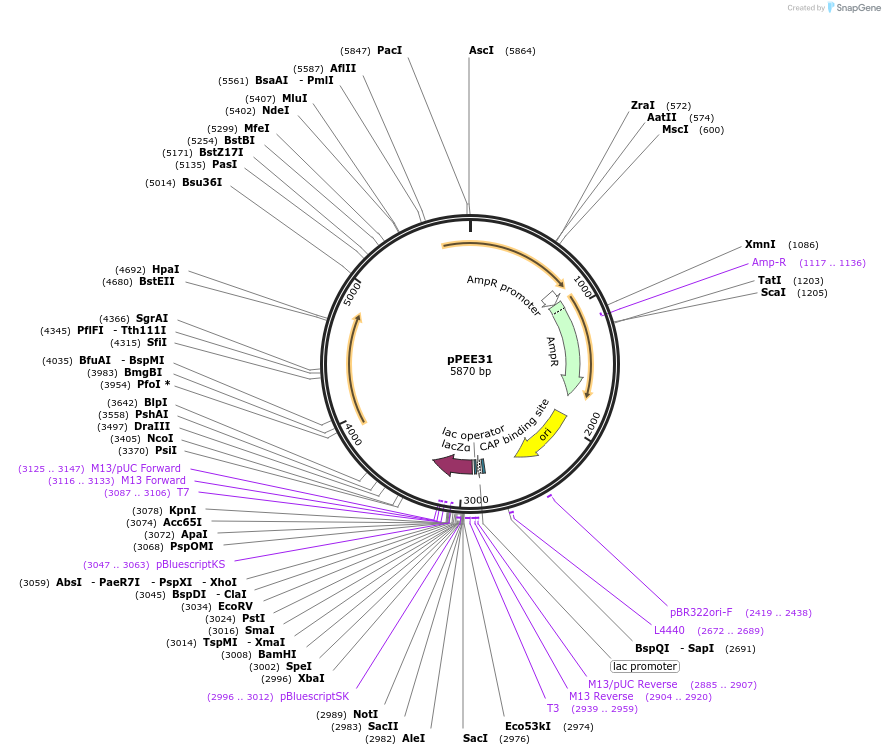
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
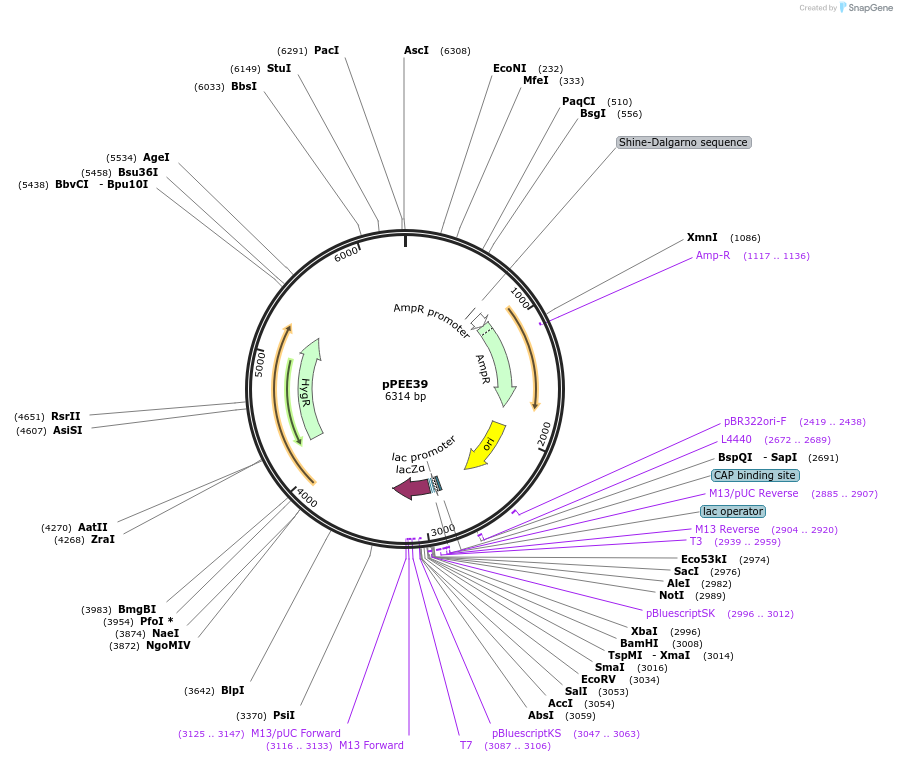
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
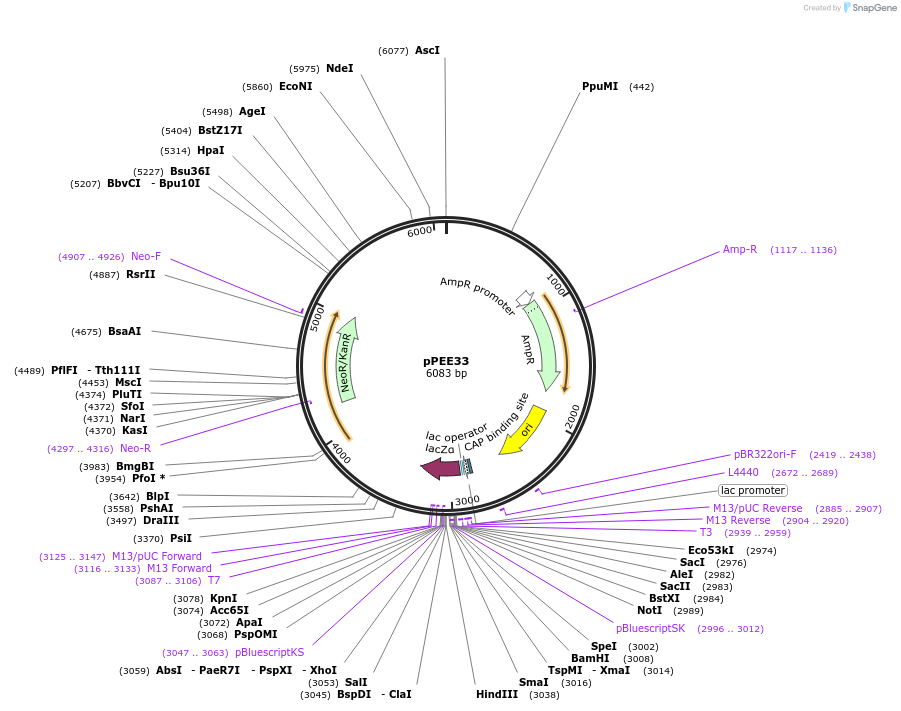
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
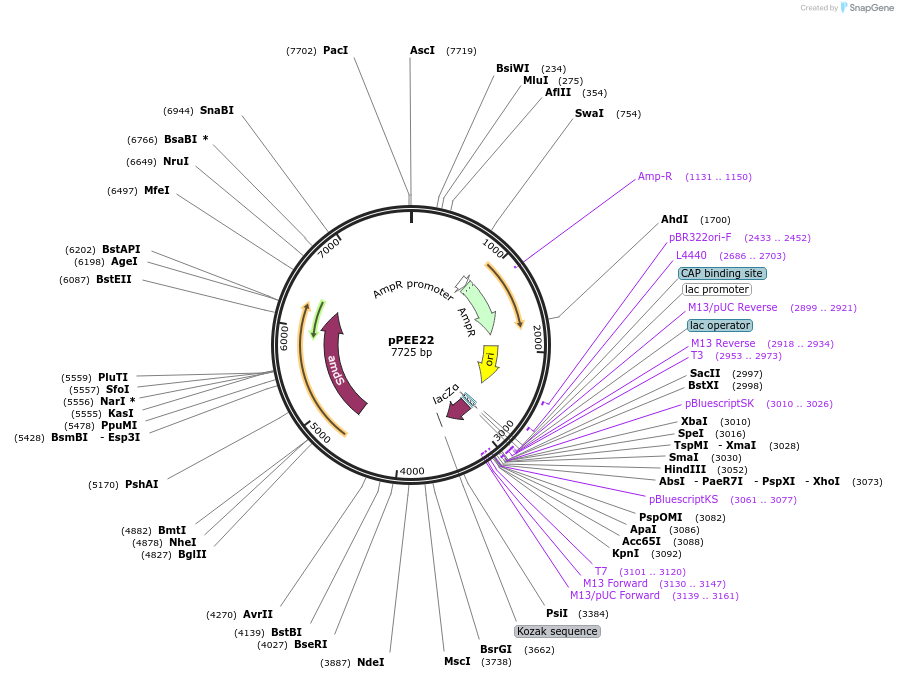
pPEE22
Plasmid#128365PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
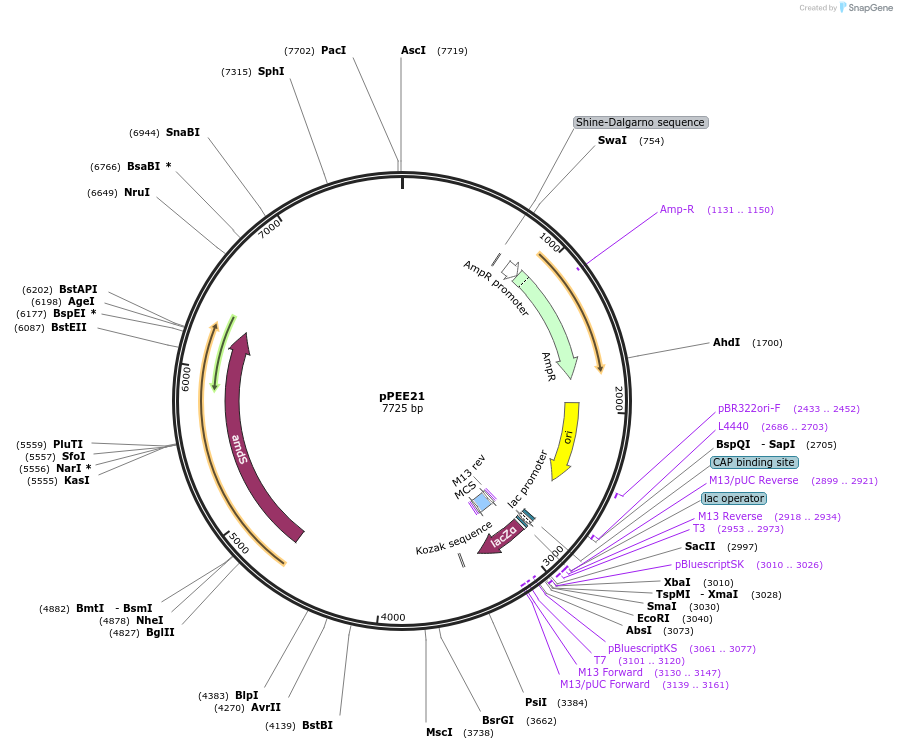
pPEE21
Plasmid#128364PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
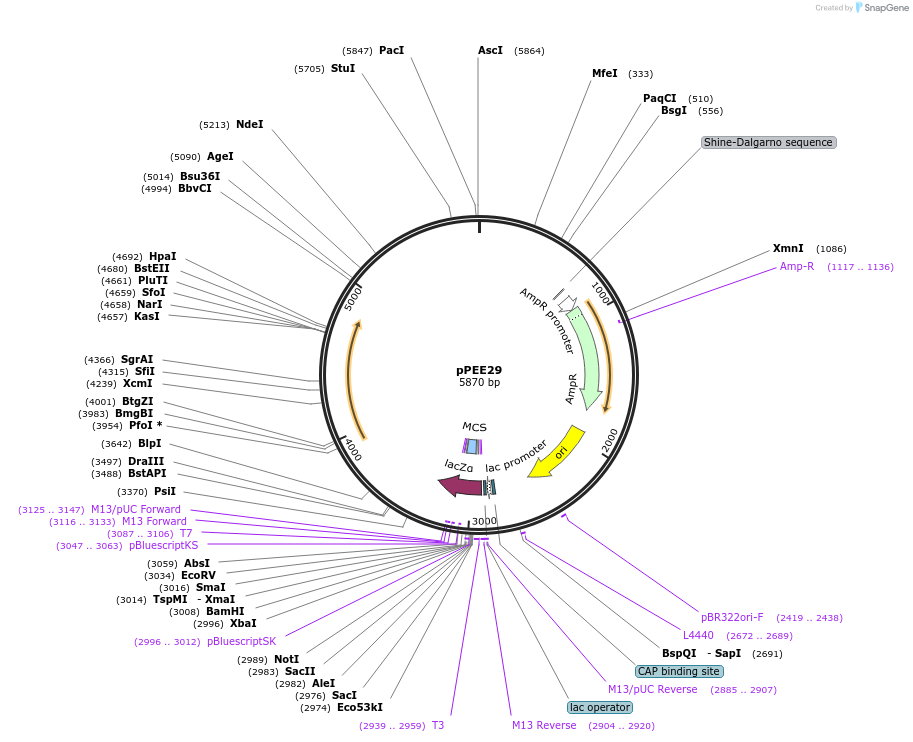
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
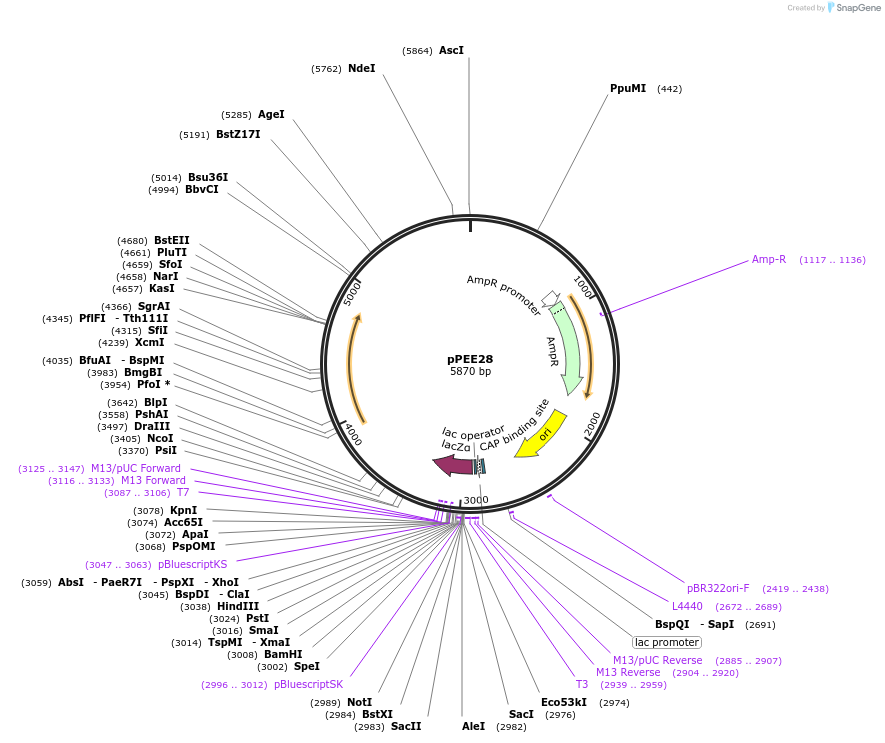
pPEE28
Plasmid#128368PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
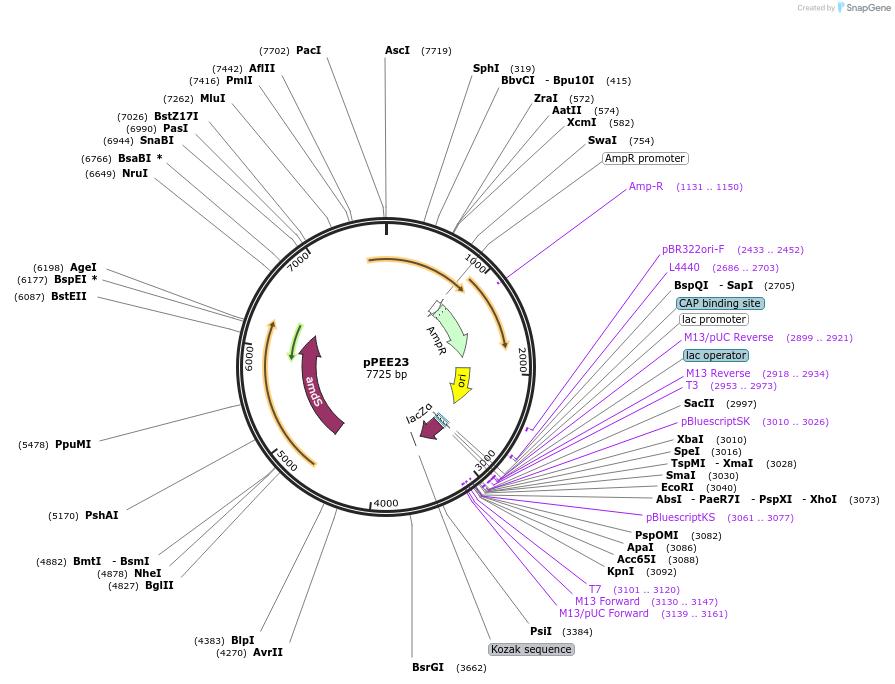
pPEE23
Plasmid#128366PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
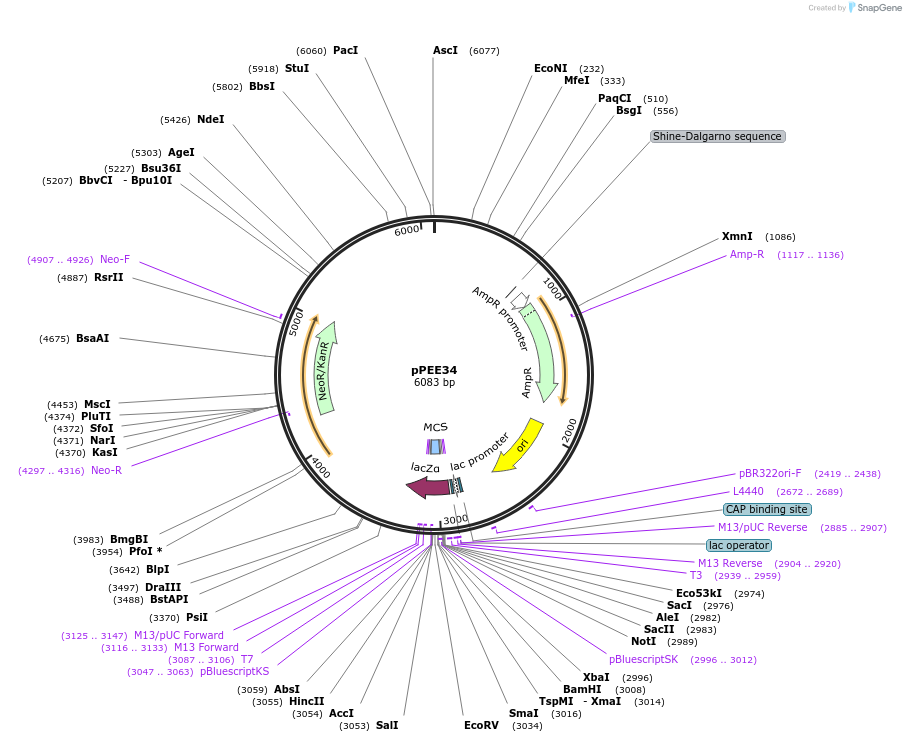
pPEE34
Plasmid#128374PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
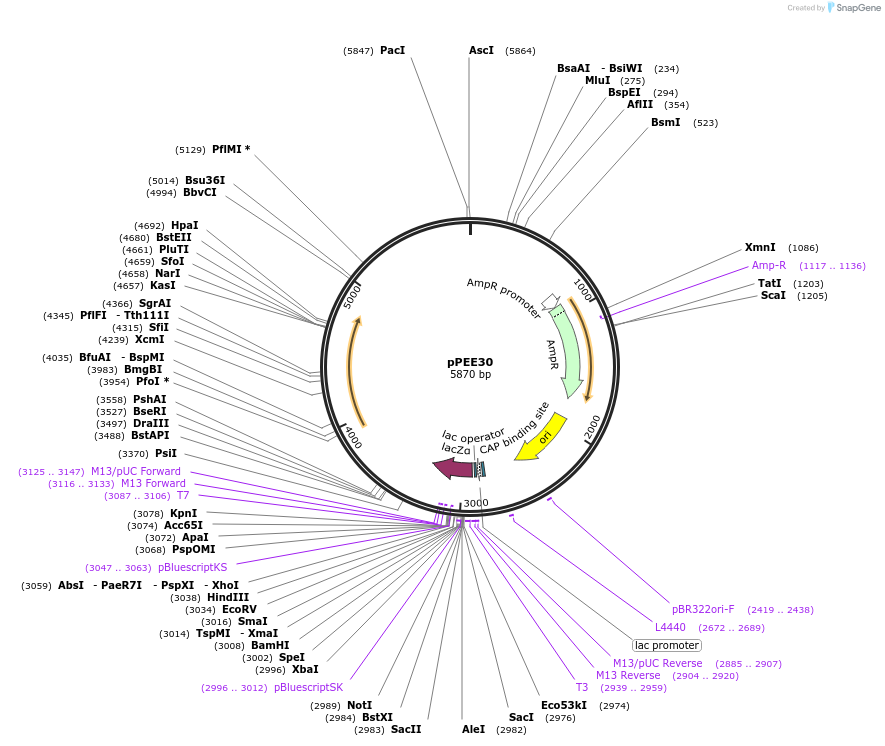
pPEE30
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
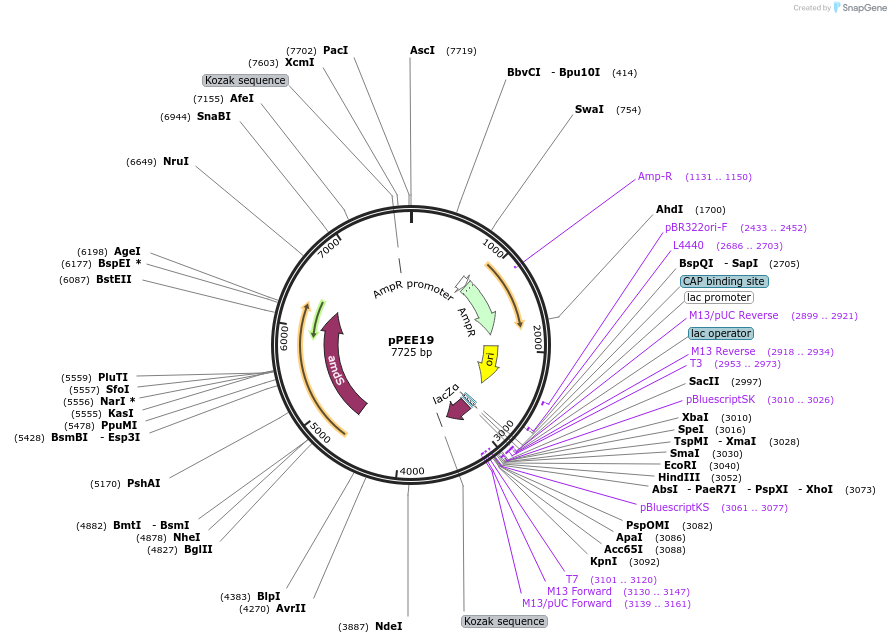
pPEE19
Plasmid#128362PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
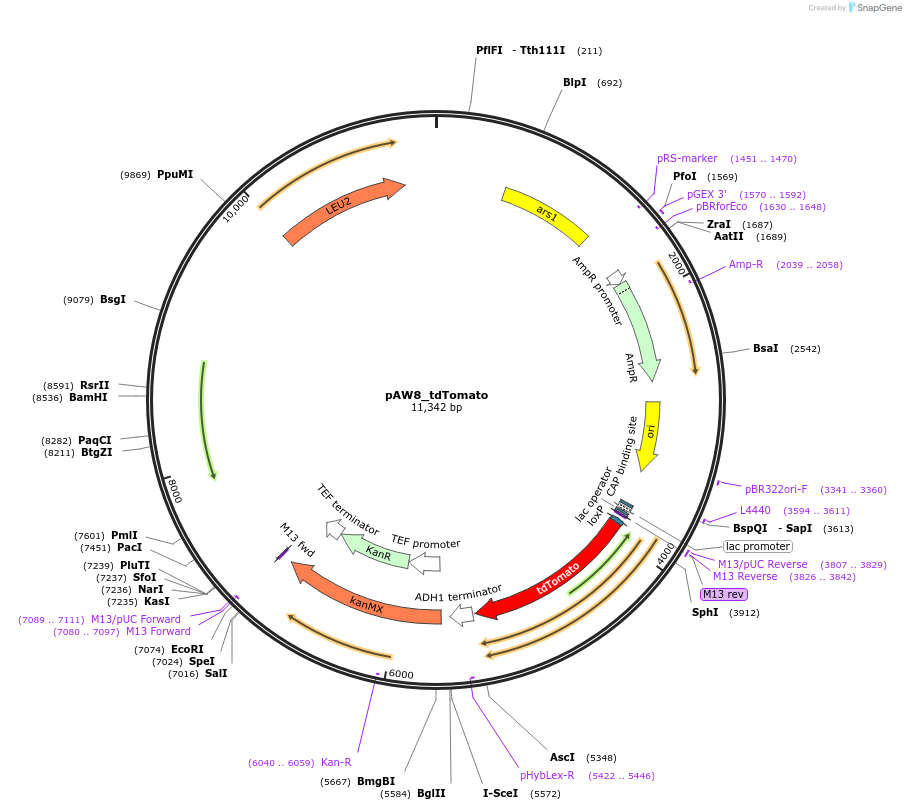
pAW8_tdTomato
Plasmid#110239PurposeCre/Lox C-terminal tagging construct encoding red fluorescent protein tdTomatoDepositorTypeEmpty backboneUseCre/LoxAvailable SinceMay 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
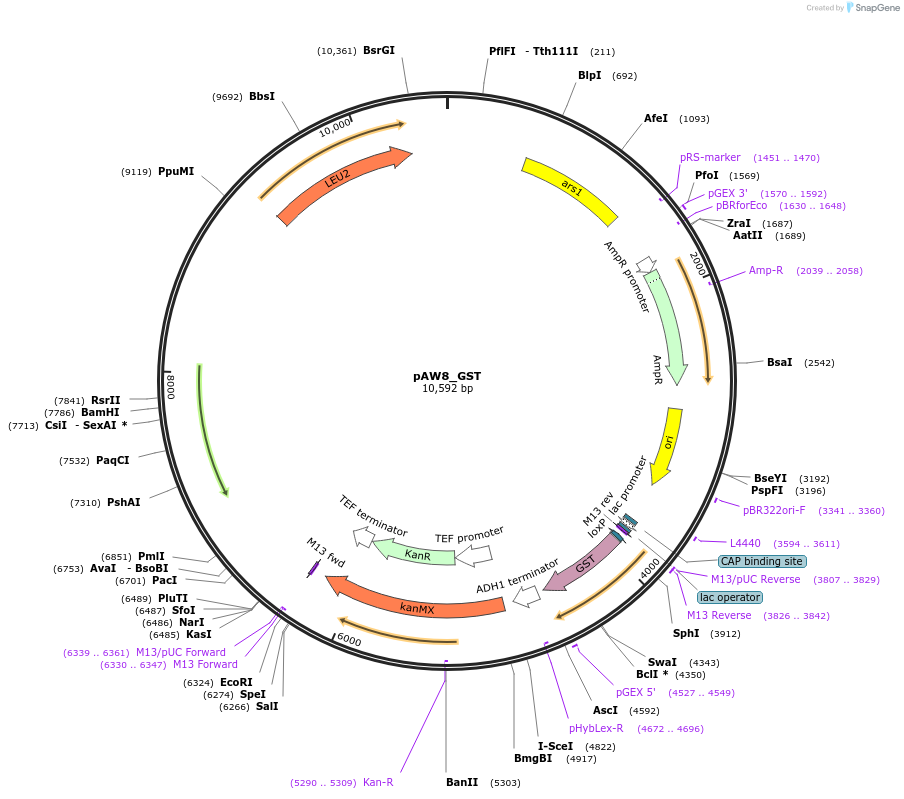
pAW8_GST
Plasmid#110237PurposeCre/Lox C-terminal tagging construct encoding glutathione S-transferaseDepositorTypeEmpty backboneUseCre/LoxAvailable SinceMay 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
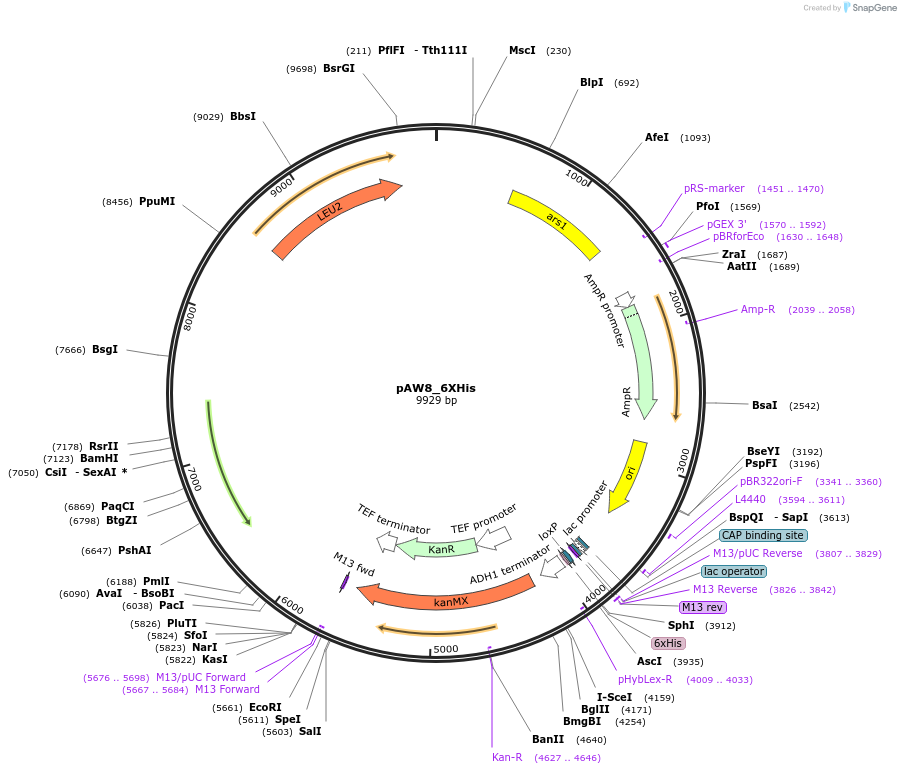
pAW8_6XHis
Plasmid#110236PurposeCre/Lox C-terminal tagging construct encoding six histidine (His) residuesDepositorTypeEmpty backboneUseCre/LoxAvailable SinceApril 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
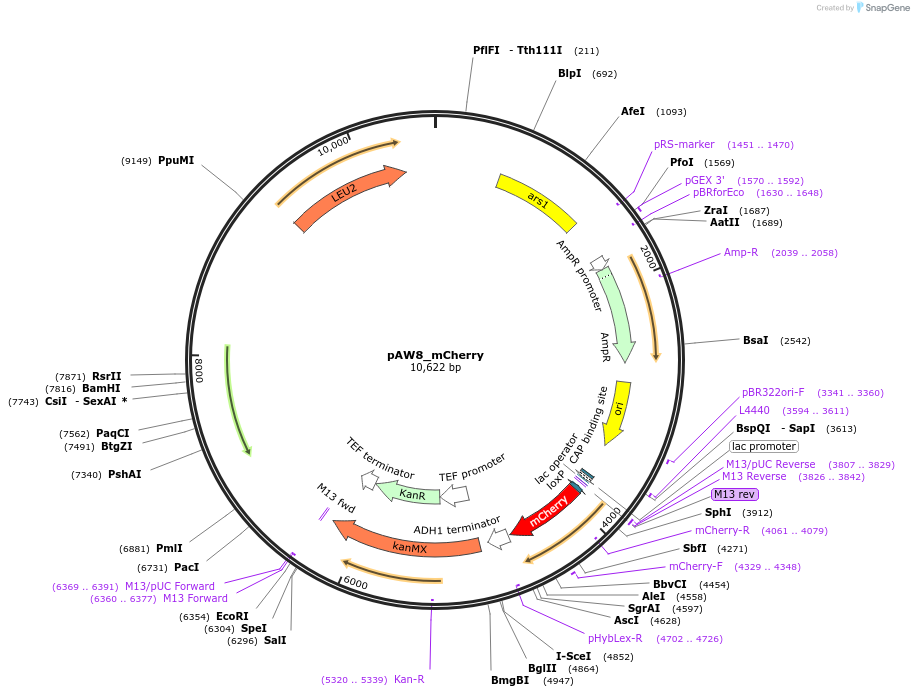
pAW8_mCherry
Plasmid#110238PurposeCre/Lox C-terminal tagging construct encoding red fluorescent protein mCherryDepositorTypeEmpty backboneUseCre/LoxAvailable SinceApril 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
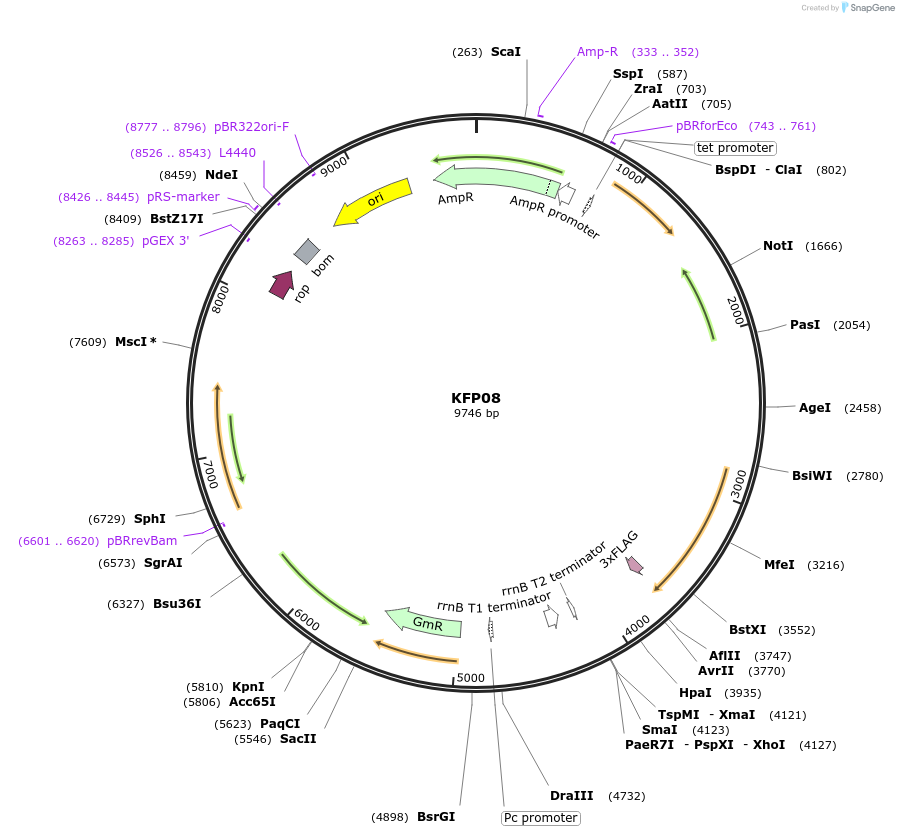
KFP08
Plasmid#119798PurposeSigF2 epitope tagged construct for integration at neutral site 2.2DepositorInsertSynpcc7942_1784
ExpressionBacterialAvailable SinceDec. 21, 2018AvailabilityAcademic Institutions and Nonprofits only