We narrowed to 11,440 results for: Kars
-
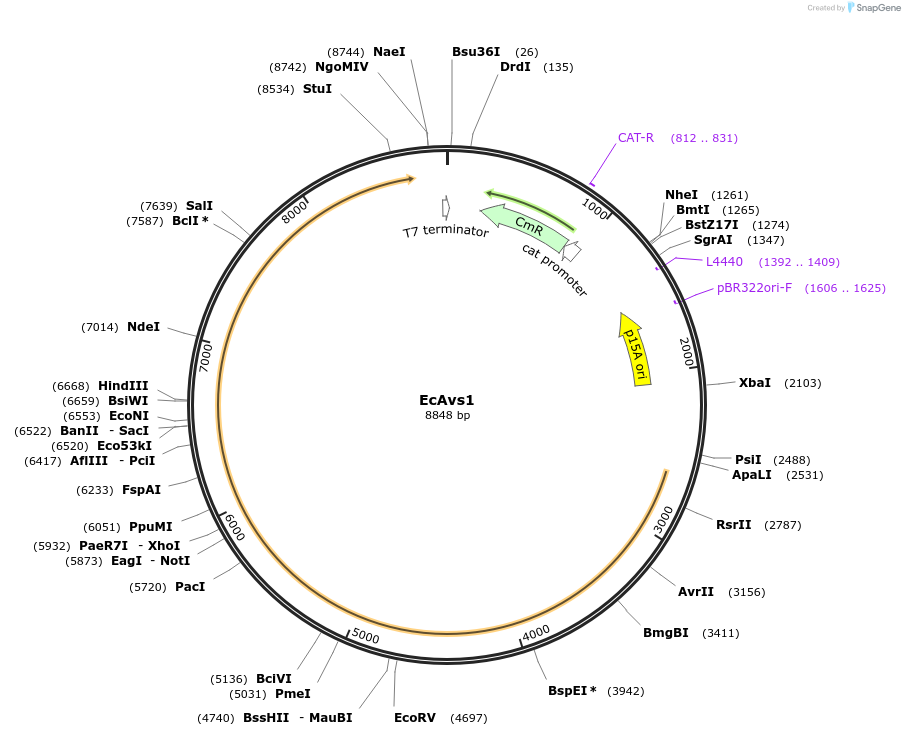
Plasmid#188787PurposeAvs1 from Escherichia coli NCTC9036DepositorInsertEcAvs1
ExpressionBacterialAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
SeAvs1
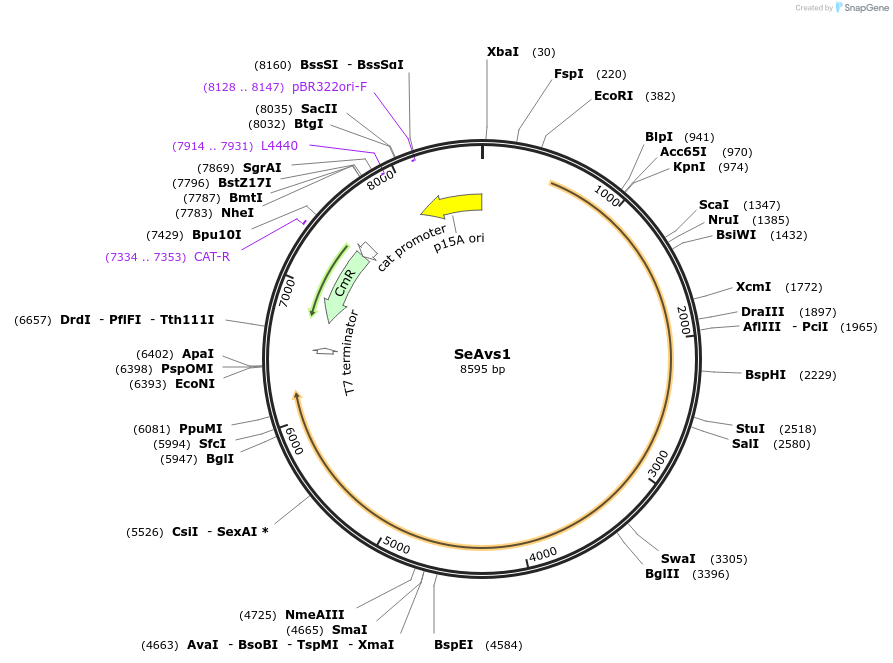
Plasmid#188786PurposeAvs1 from Salmonella enterica NCTC10718DepositorInsertSeAvs1
ExpressionBacterialAvailable SinceSept. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
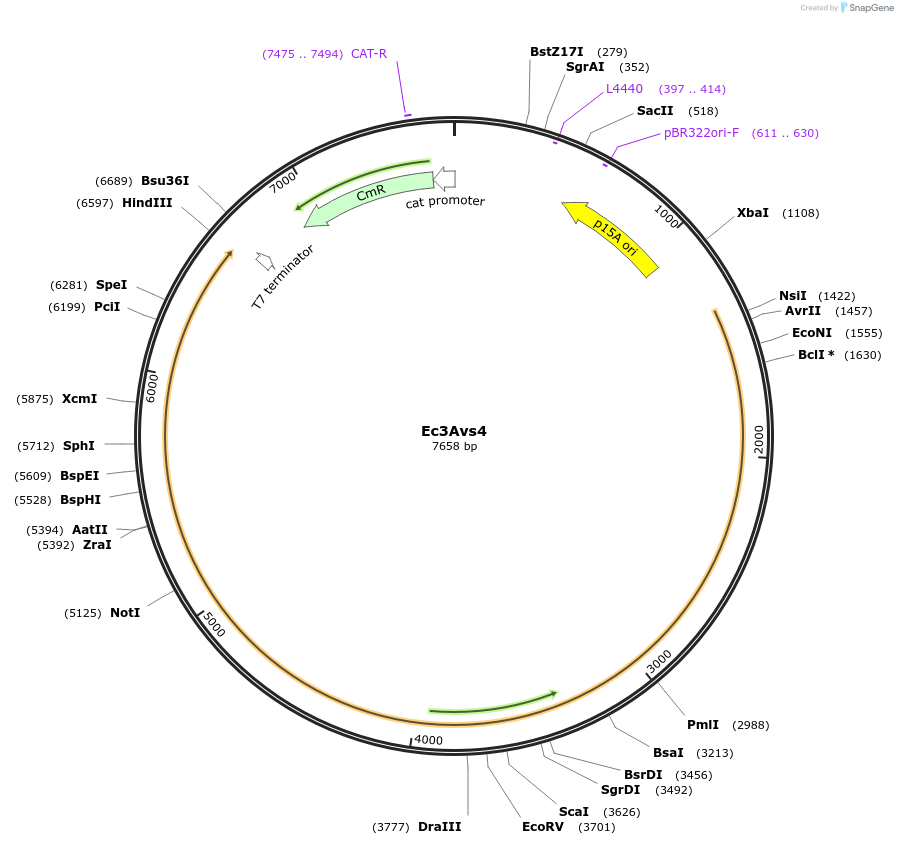
Ec3Avs4
Plasmid#188798PurposeAvs4 from Escherichia coli NCTC11132DepositorInsertEc3Avs4
ExpressionBacterialAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
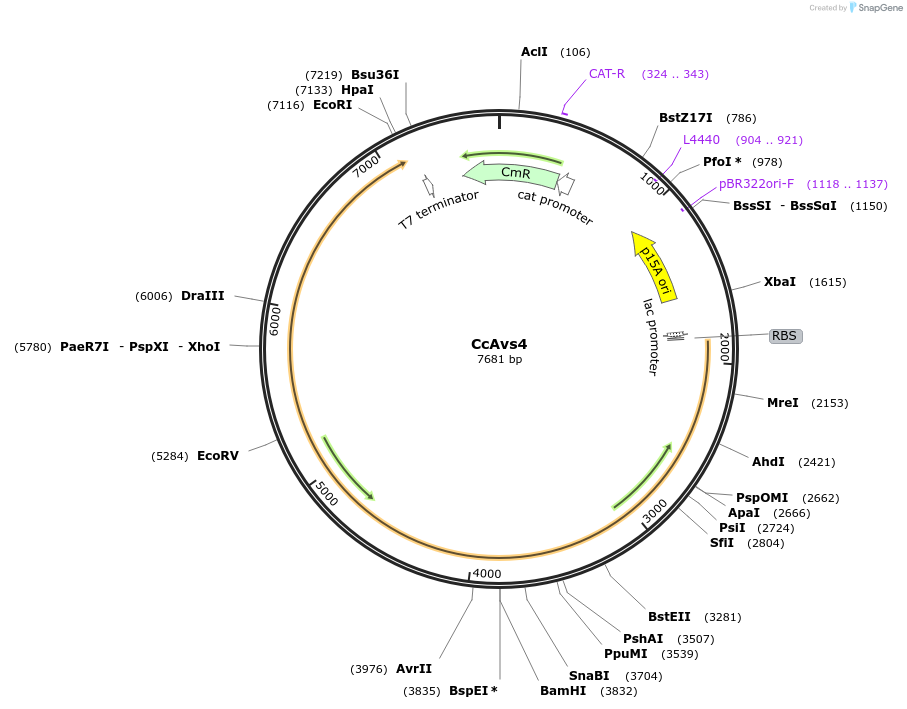
CcAvs4
Plasmid#188797PurposeAvs4 from Corallococcus coralloides DSM2259DepositorInsertCcAvs4
ExpressionBacterialPromoterlacAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
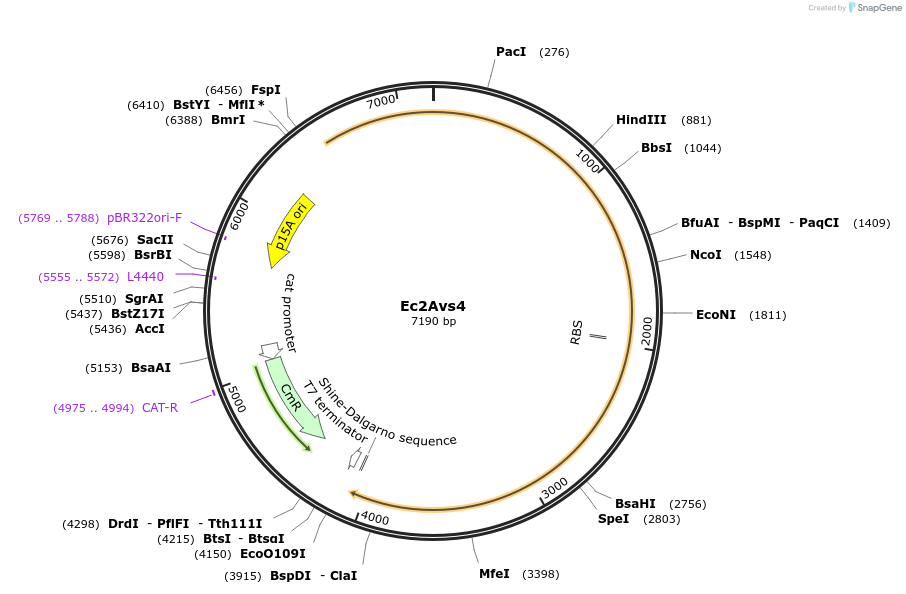
Ec2Avs4
Plasmid#188795PurposeAvs4 from Escherichia coli 382_hDepositorInsertEc2Avs4
ExpressionBacterialAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
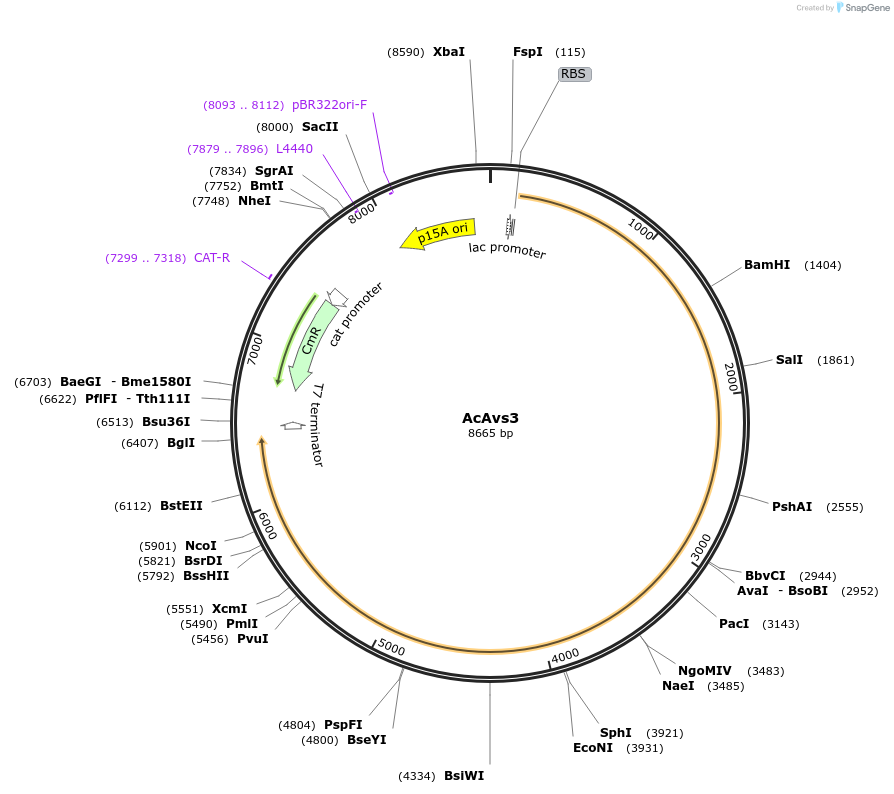
AcAvs3
Plasmid#188793PurposeAvs3 from Arcobacter cryaerophilus M830ADepositorInsertAcAvs3
ExpressionBacterialPromoterlacAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
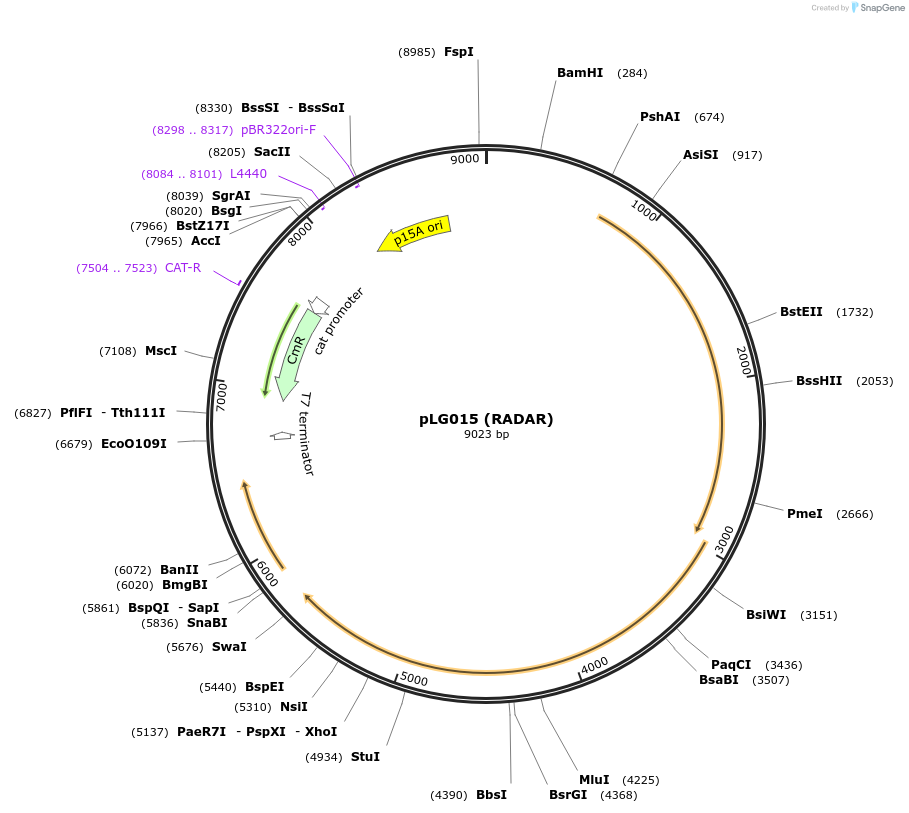
pLG015 (RADAR)
Plasmid#157893PurposeExpresses the RADAR defense systemDepositorInsertRADAR subtype containing auxillary gene rdrD
ExpressionBacterialAvailable SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
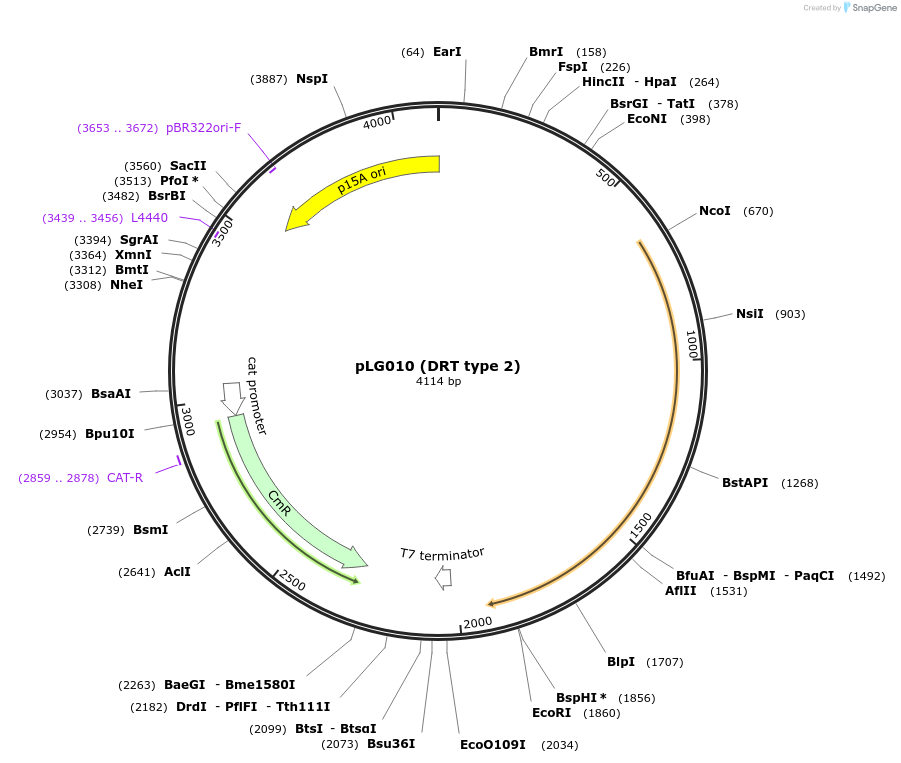
pLG010 (DRT type 2)
Plasmid#157888PurposeExpresses the DRT type 2 defense systemDepositorInsertDRT type 2
ExpressionBacterialAvailable SinceJan. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
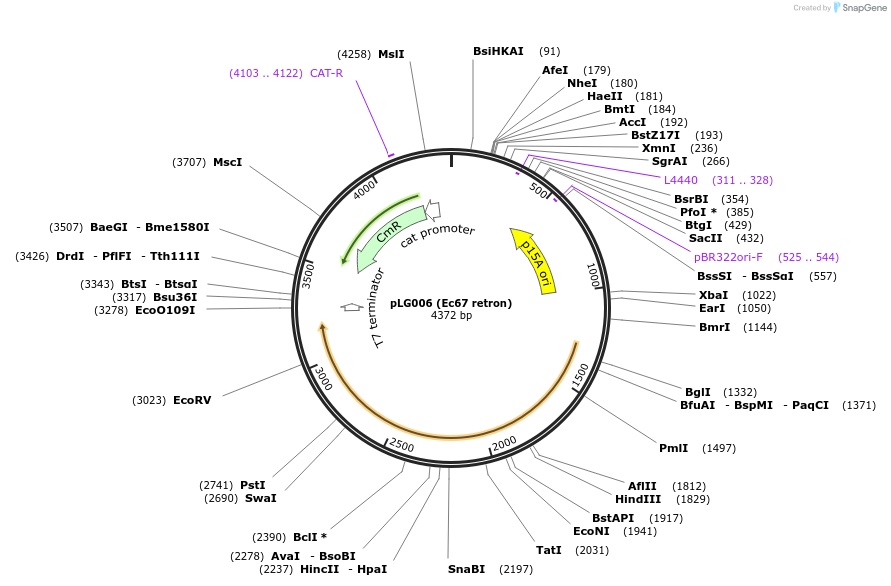
pLG006 (Ec67 retron)
Plasmid#157884PurposeExpresses the Ec67 retron defense systemDepositorInsertEc67 retron
ExpressionBacterialAvailable SinceDec. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
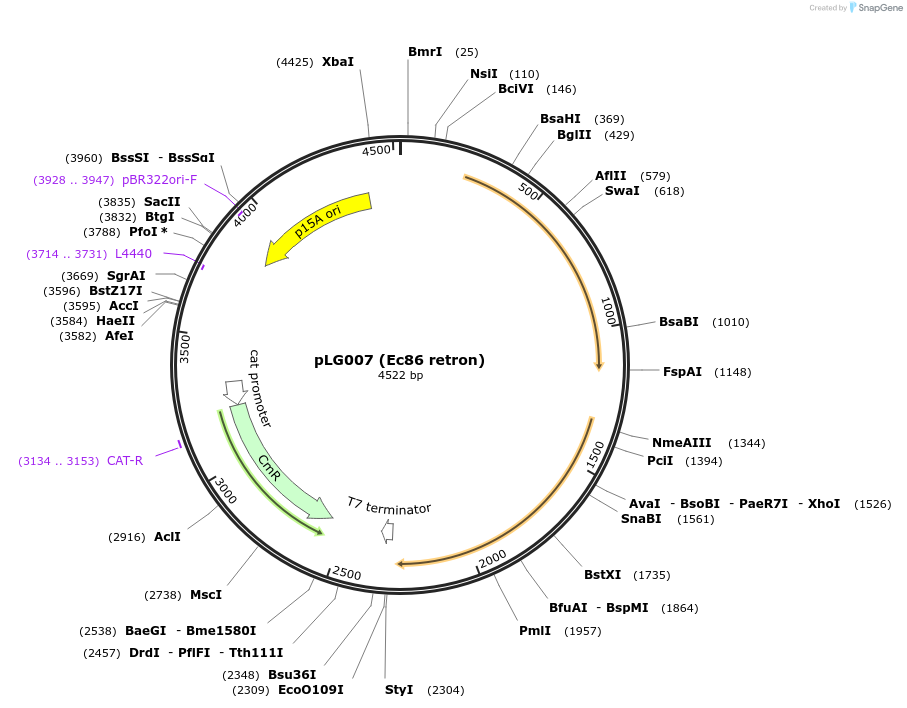
pLG007 (Ec86 retron)
Plasmid#157885PurposeExpresses the Ec86 retron defense systemDepositorInsertEc86 retron
ExpressionBacterialAvailable SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
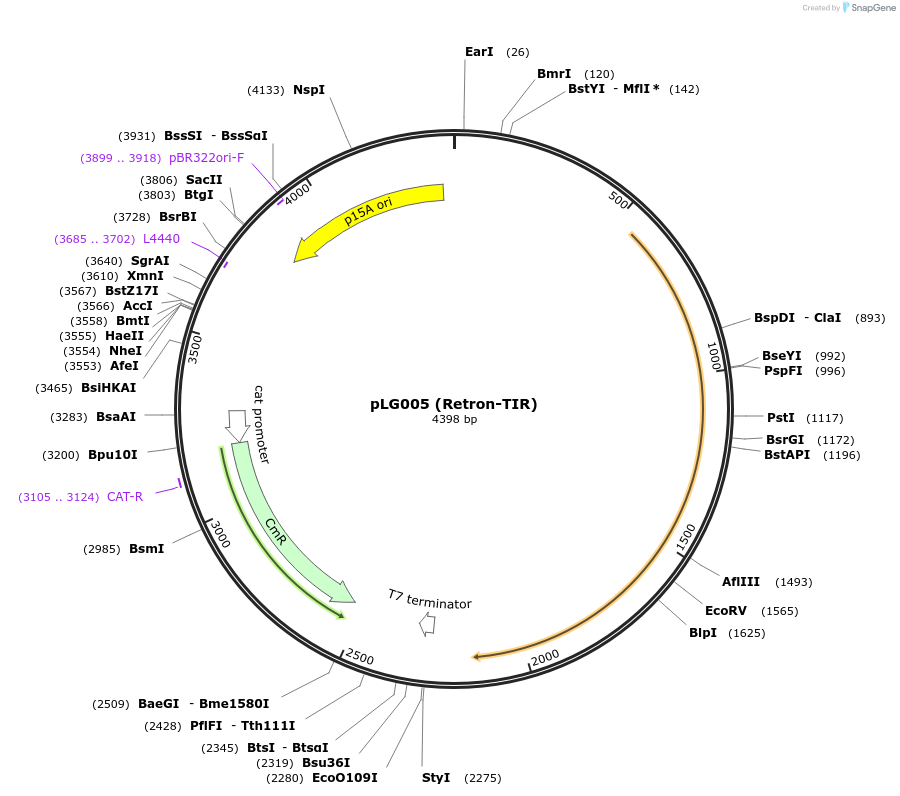
pLG005 (Retron-TIR)
Plasmid#157883PurposeExpresses the retron-TIR defense systemDepositorInsertRetron-TIR
ExpressionBacterialAvailable SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
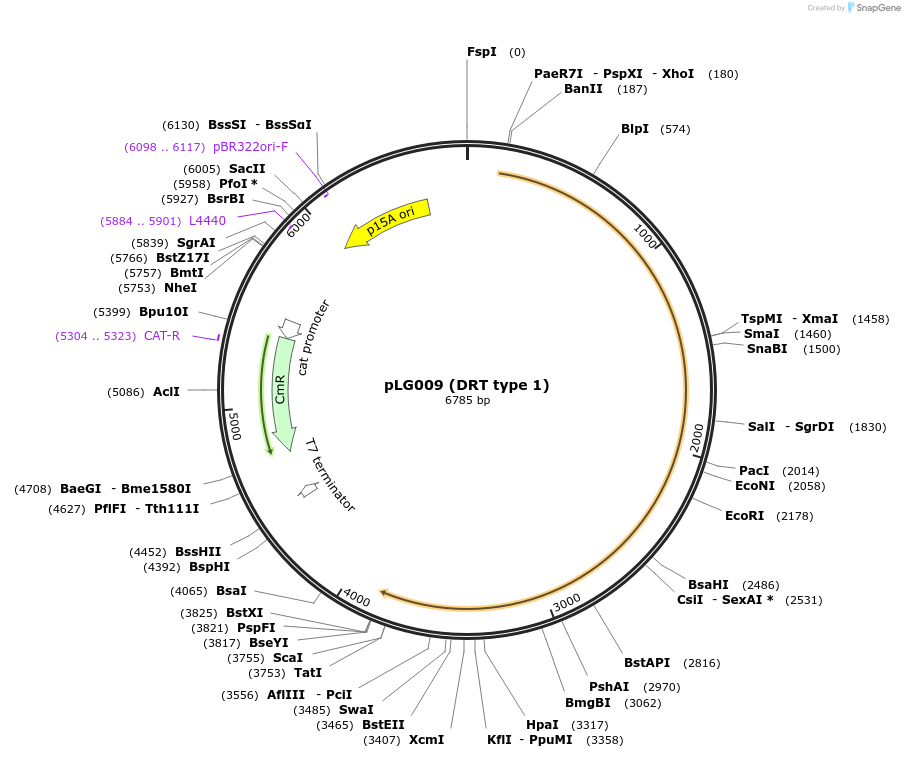
pLG009 (DRT type 1)
Plasmid#157887PurposeExpresses the DRT type 1 defense systemDepositorInsertDRT type 1 (RT-nitrilase)
ExpressionBacterialAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
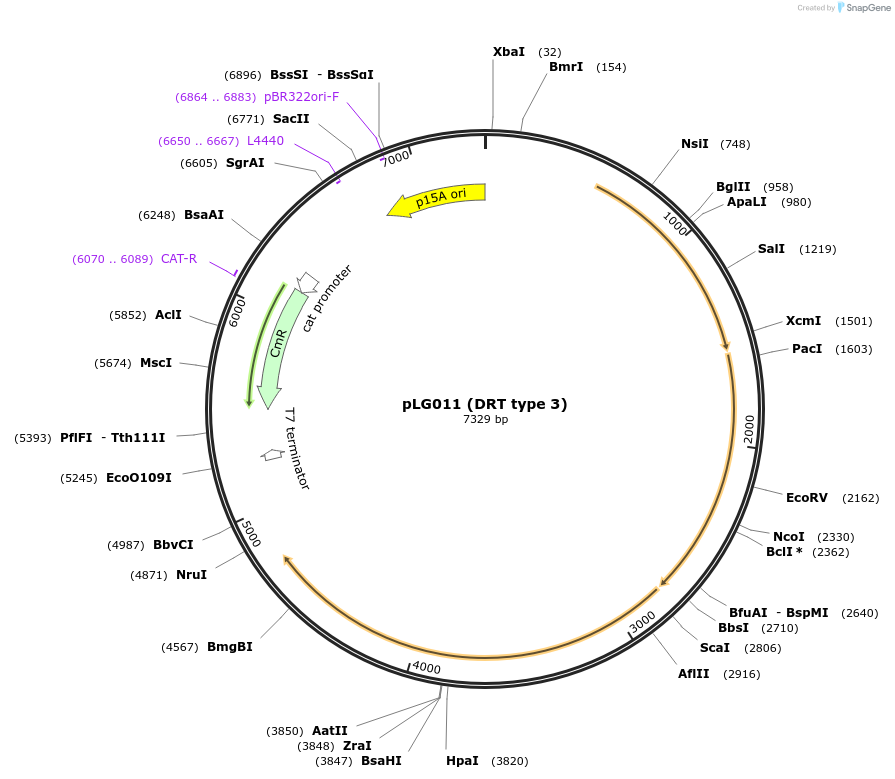
pLG011 (DRT type 3)
Plasmid#157889PurposeExpresses the DRT type 3 defense systemDepositorInsertDRT type 3 (RT + RT)
ExpressionBacterialAvailable SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
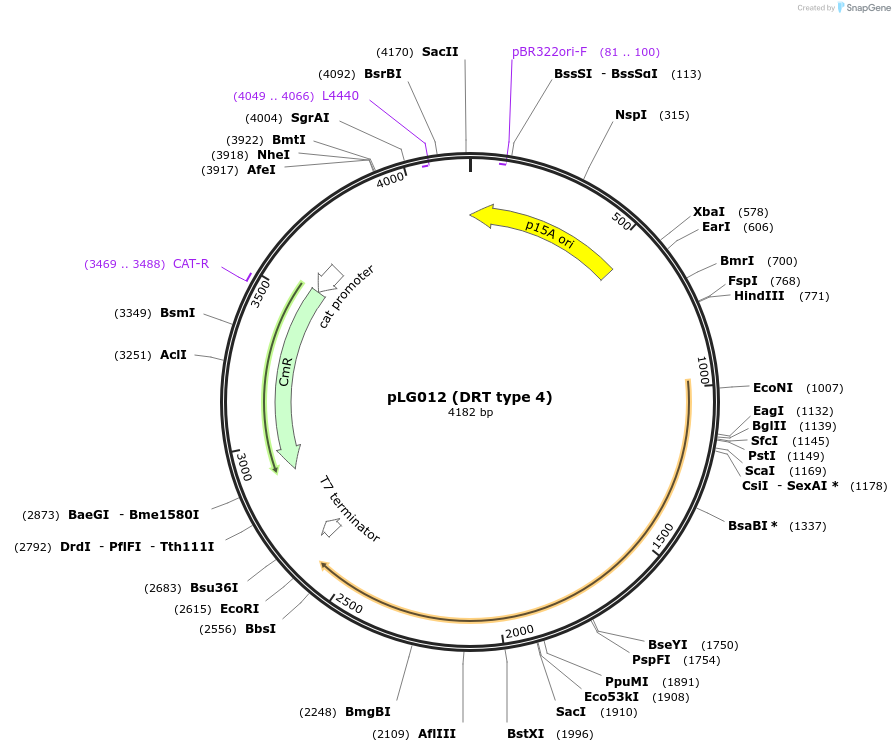
pLG012 (DRT type 4)
Plasmid#157890PurposeExpresses the DRT type 4 defense systemDepositorInsertDRT type 4
ExpressionBacterialAvailable SinceJan. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
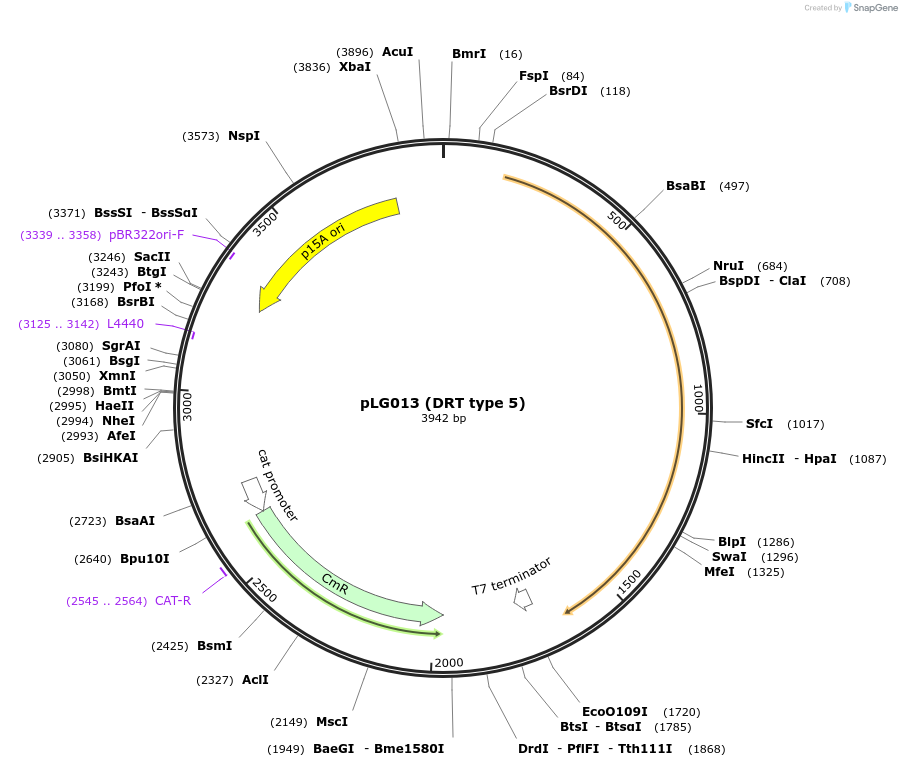
pLG013 (DRT type 5)
Plasmid#157891PurposeExpresses the DRT type 5 defense systemDepositorInsertDRT type 5
ExpressionBacterialAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
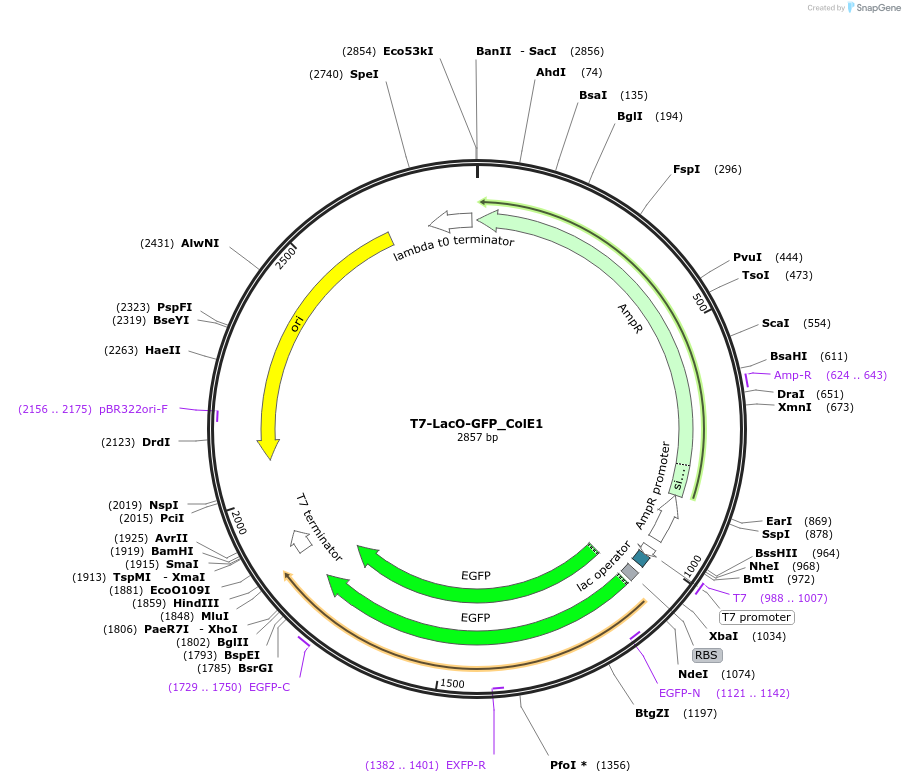
T7-LacO-GFP_ColE1
Plasmid#171648PurposeExpresses GFP in BL21(DE3) E. coli. Positive control for expression experiments. Contains ColE1 origin of replication.DepositorInsertT7-LacO-GFPssrA-T7 Stop
TagsssrA degradation tag (DAS+4)ExpressionBacterialPromoterT7Available SinceOct. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
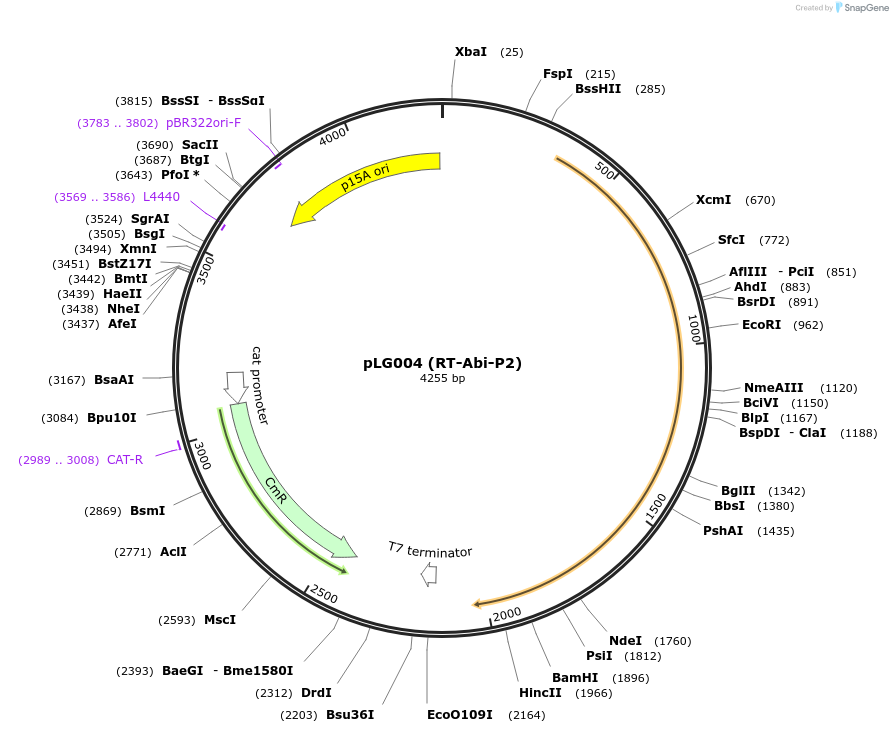
pLG004 (RT-Abi-P2)
Plasmid#157882PurposeExpresses the RT-Abi-P2 defense systemDepositorInsertRT-Abi-P2
ExpressionBacterialAvailable SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
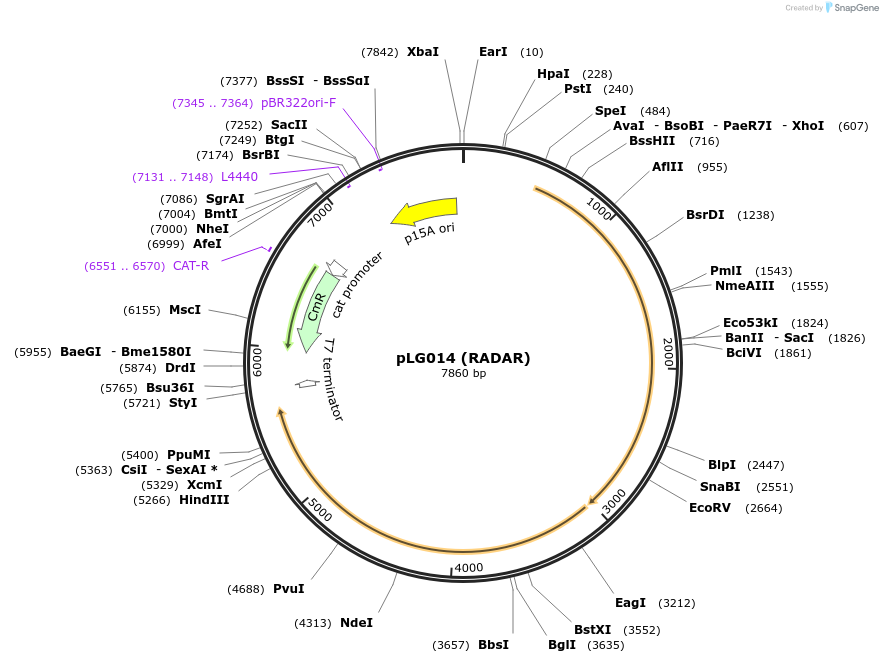
pLG014 (RADAR)
Plasmid#157892PurposeExpresses the RADAR defense systemDepositorInsertRADAR
ExpressionBacterialAvailable SinceJan. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
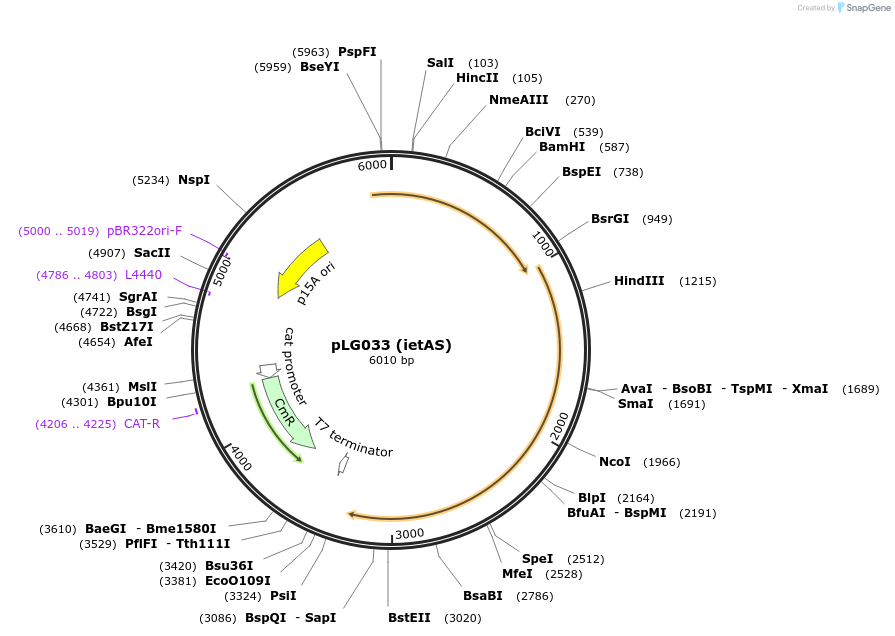
pLG033 (ietAS)
Plasmid#157911PurposeExpresses the ietAS defense systemDepositorInsertietAS (ATPase + serine protease)
ExpressionBacterialAvailable SinceDec. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
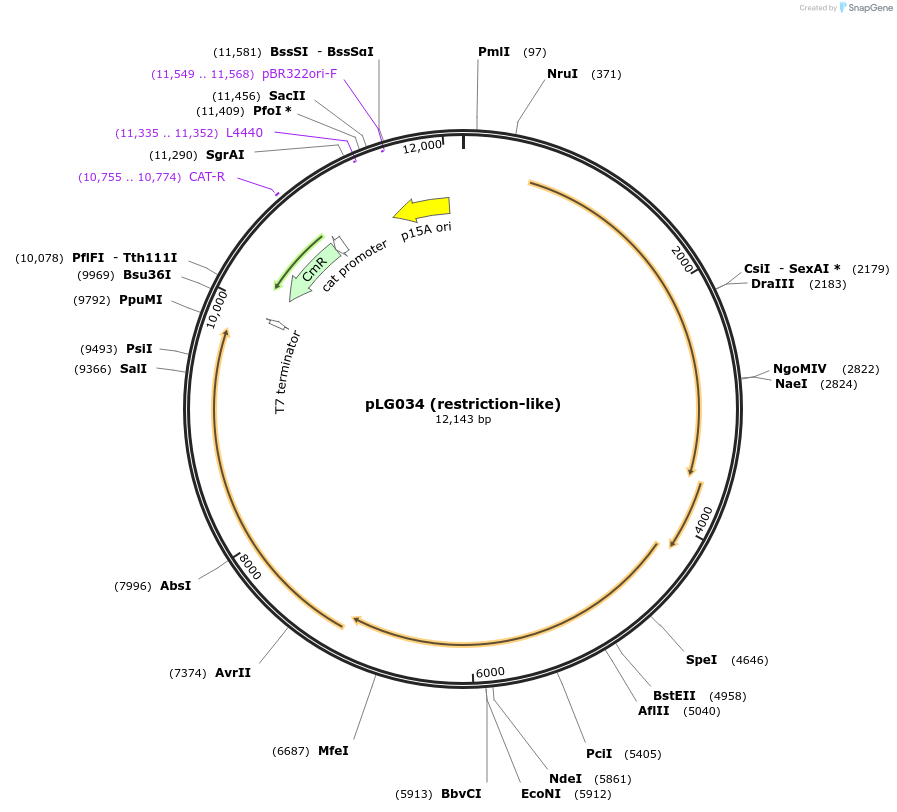
pLG034 (restriction-like)
Plasmid#157912PurposeExpresses a restriction-like defense systemDepositorInsert4-gene restriction-like system
ExpressionBacterialAvailable SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only