We narrowed to 14,201 results for: Ung;
-
Plasmid#40841PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm17-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-5 (mm18-21)
Plasmid#40840PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm18-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-4 (mm19-21)
Plasmid#40839PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm19-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-3 (mm20-21)
Plasmid#40838PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm20-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-2 (mm21)
Plasmid#40837PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-16 (mm14-21)
Plasmid#40851PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm14-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-15 (mm15-21)
Plasmid#40850PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm15-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS14 (mm16-21)
Plasmid#40849PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm16-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-13
Plasmid#40848PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–Bartel
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-12 (mm1-8)
Plasmid#40847PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-11 (bulge 9-15)
Plasmid#40846PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge9-15
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-10 (bulge 9-13)
Plasmid#40845PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-13
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-9 (bulge 9-11)
Plasmid#40844PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-11
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
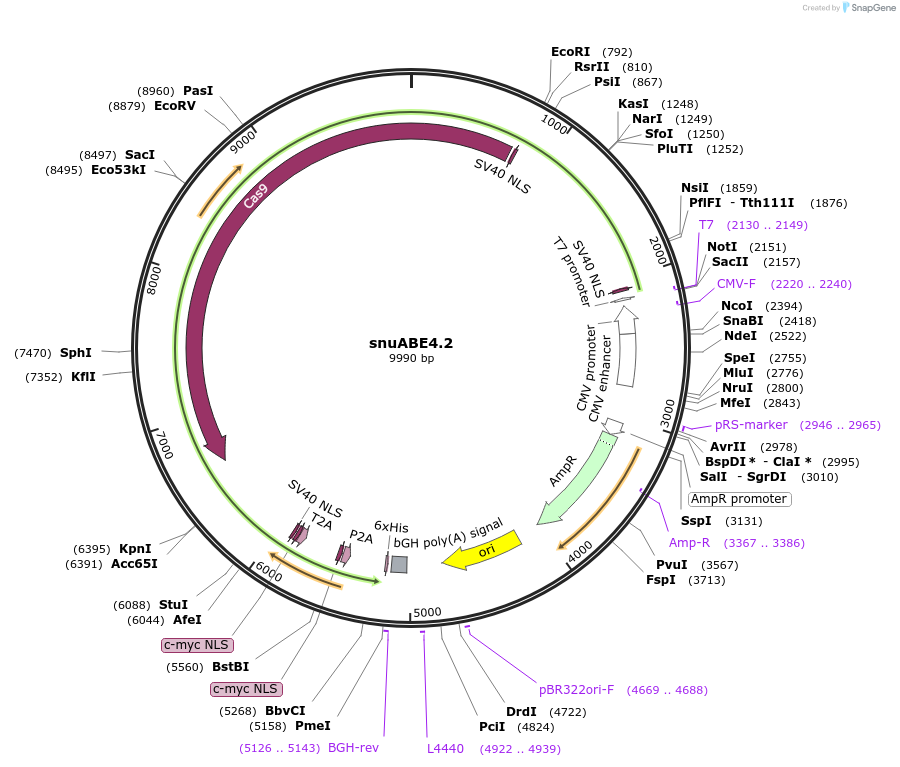
snuABE4.2
Plasmid#246014PurposesnuABE4.1 with MLH1-SB fused to the C-terminus via P2A.DepositorInsertPediculus humanus ADAR deaminase domain (E438Q, I536S, E637F mutant)
TagsMLH1-SBExpressionMammalianMutationE438Q, I536S, E637F, with 10 amino acids truncati…PromoterCMVAvailable SinceNov. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
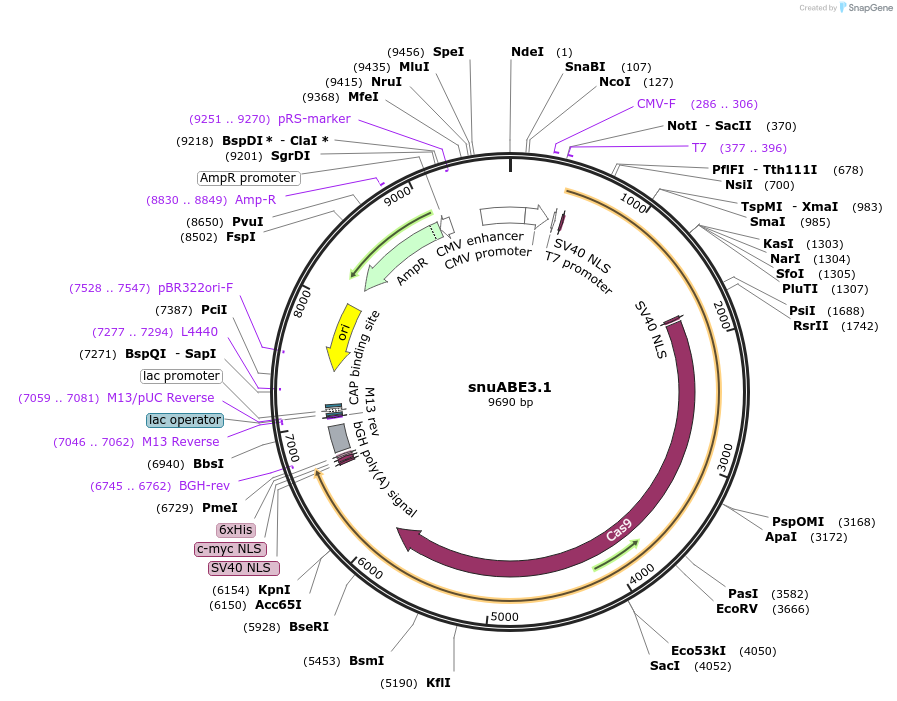
snuABE3.1
Plasmid#246011PurposesnuABE3.1 composed of PhADAR(E438Q) fused to SpCas9 nickase [R221K, N394K, H840A] via an SV40 linker and further fused to La protein.DepositorInsertPediculus humanus ADAR deaminase domain (E438Q mutant)
ExpressionMammalianMutationE438QPromoterCMVAvailable SinceNov. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
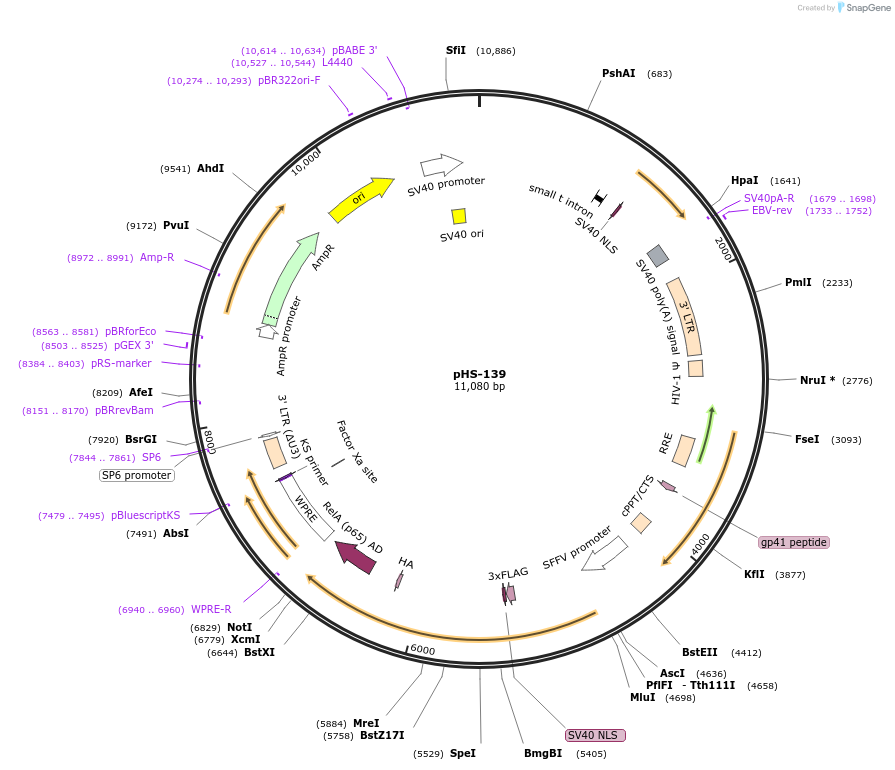
pHS-139
Plasmid#195461PurposeLentiviral Integratable, GZV-inducible expression of synZiFTR: ZF10-p65DepositorInsertpSFFV - ZF10 - NS3 - p65
UseLentiviralExpressionMammalianAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
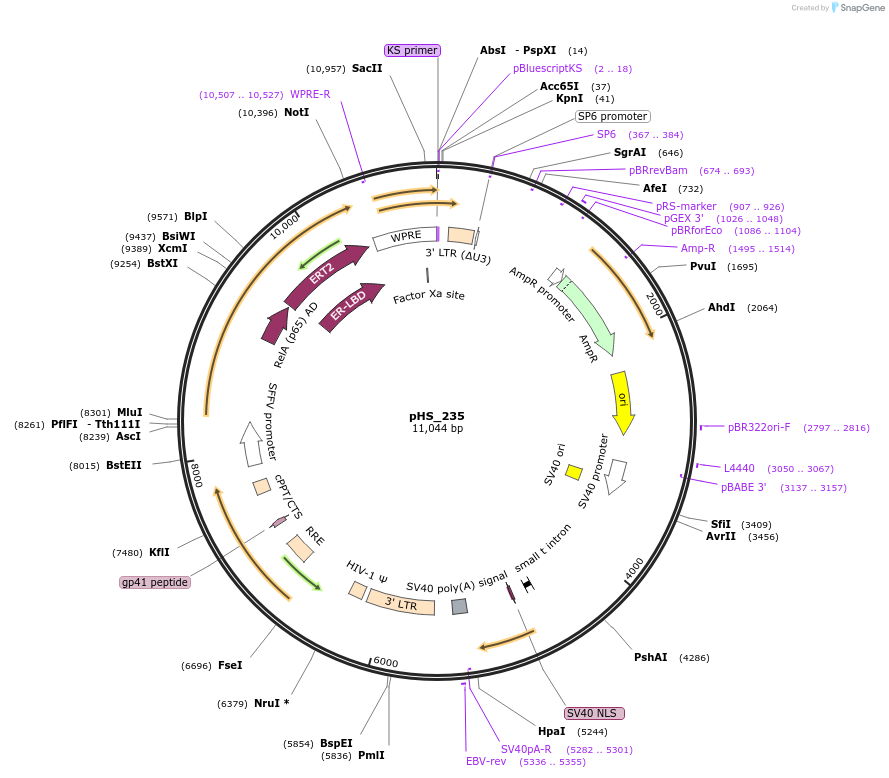
pHS_235
Plasmid#195460PurposeLentiviral Integratable, 4OHT/TMX-inducible expression of synZiFTR: ZF3-p65DepositorInsertpSFFV - ZF3 - p65 - ERT2
UseLentiviralExpressionMammalianAvailable SinceMarch 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
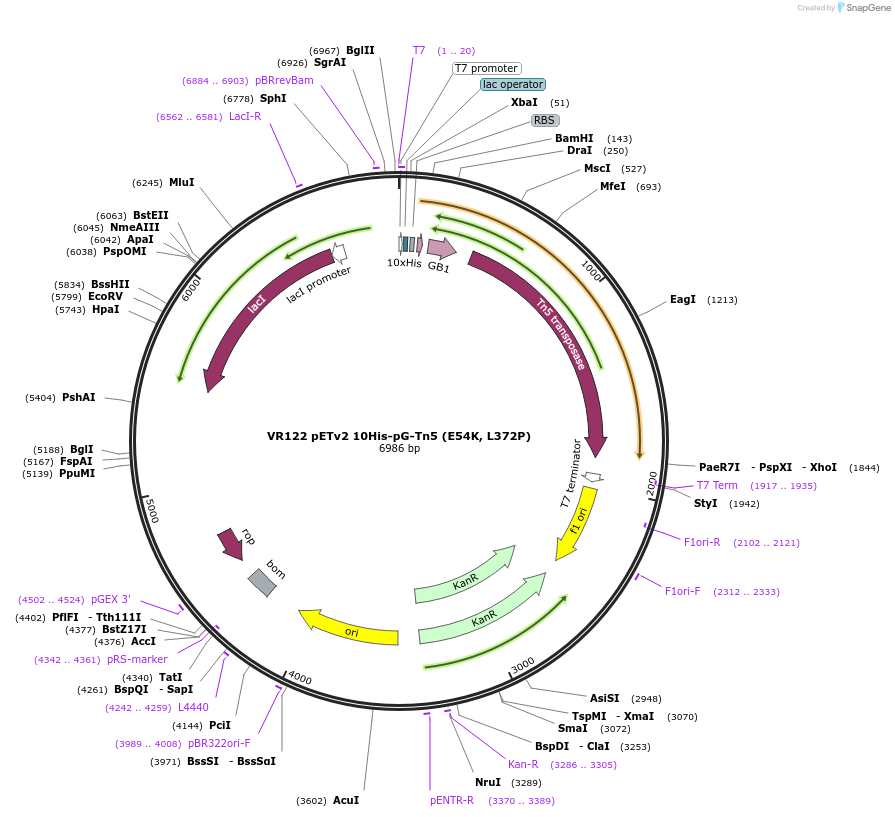
VR122 pETv2 10His-pG-Tn5 (E54K, L372P)
Plasmid#198467PurposeE. coli protein expression plasmid encoding hyperactive Tn5 transposase fused to protein G, in DH5α cloning strain.DepositorInsert10His-pG-Tn5 (E54K, L372P)
Tags10His-pGExpressionBacterialMutationE54K, L372PPromoterConsensus φ10 T7 PromoterAvailable SinceApril 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
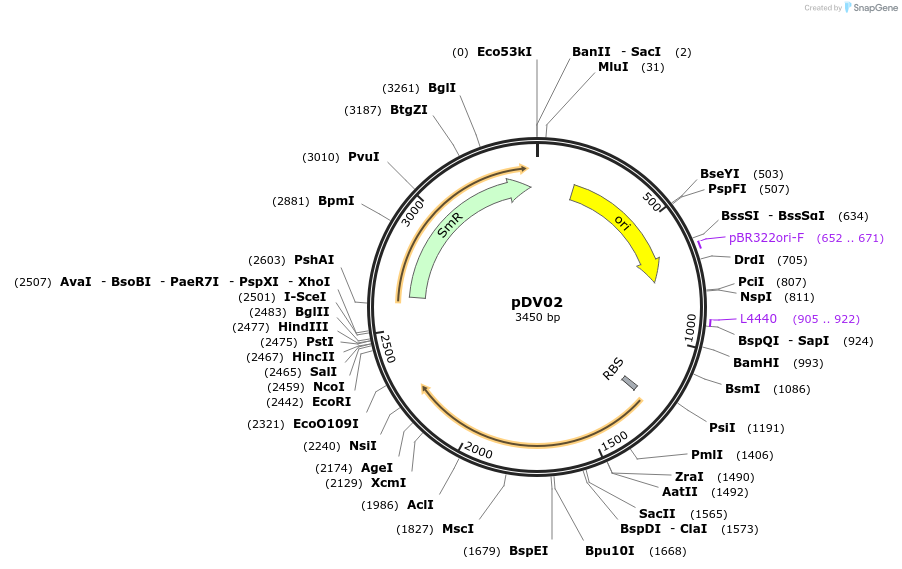
pDV02
Plasmid#212156PurposePlasmid expressing secondary alcohol dehydrogenase under constitutive promoter PgyrA control.DepositorInsertsecondary alcohol dehydrogenase
ExpressionBacterialPromoterPgyrAAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
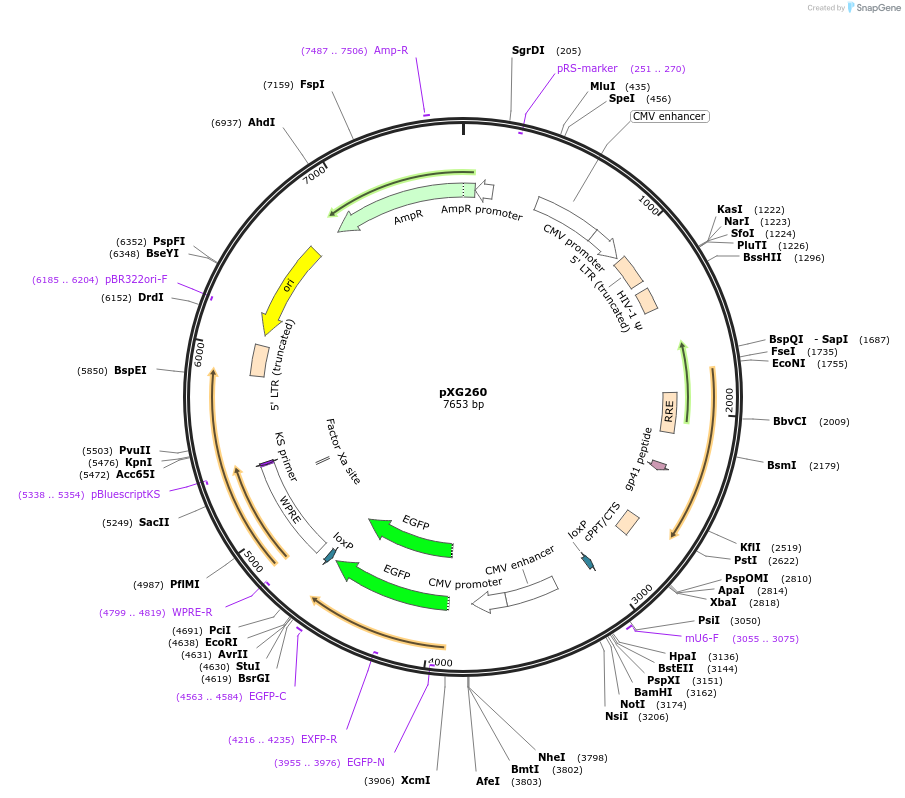
pXG260
Plasmid#141282PurposeCMV:EGFP (as control for pXG237)DepositorInsertEGFP
UseLentiviralPromoterCMVAvailable SinceSept. 19, 2020AvailabilityAcademic Institutions and Nonprofits only