We narrowed to 1,811 results for: FAST
-
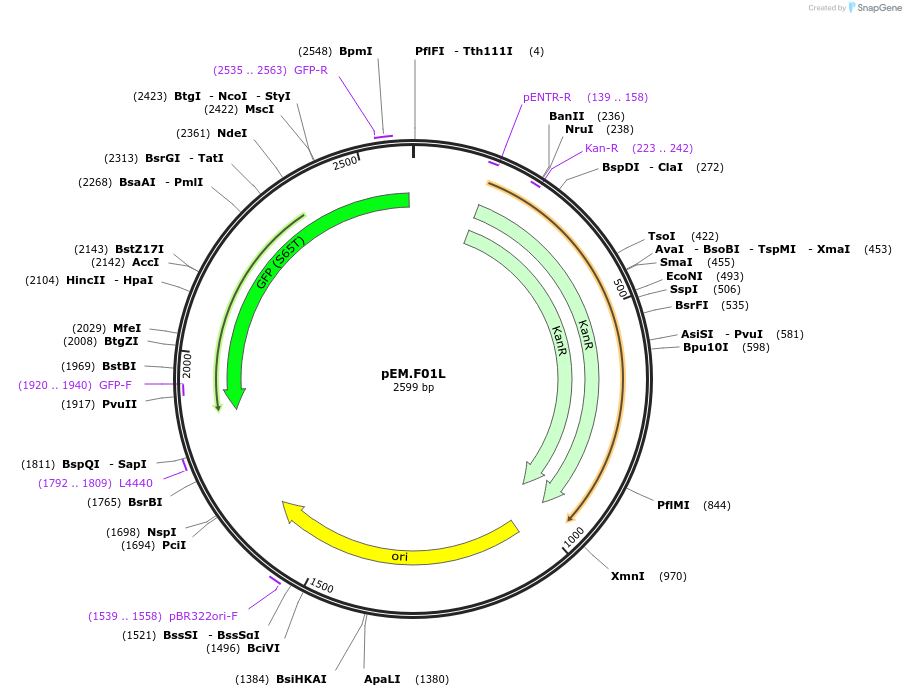
Plasmid#198659PurposeEasy-MISE toolkit pEM-plasmid containing GFP coding sequence with CD protruding endsDepositorInsertGFP
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
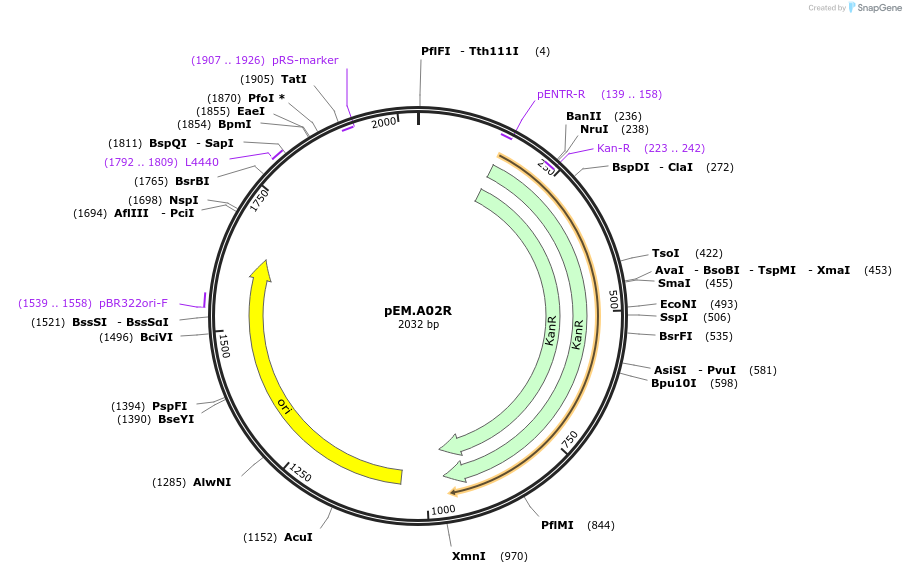
pEM.A02R
Plasmid#198658PurposeEasy-MISE toolkit pEM-plasmid containing a spacer sequence with FL protruding endsDepositorInsertAdaptor
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
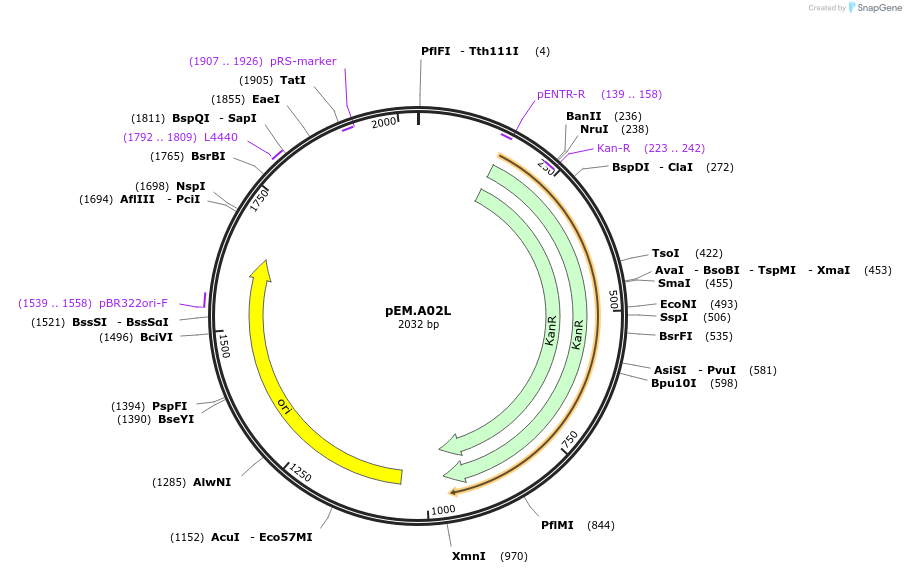
pEM.A02L
Plasmid#198657PurposeEasy-MISE toolkit pEM-plasmid containing a spacer sequence with BF protruding endsDepositorInsertAdaptor
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
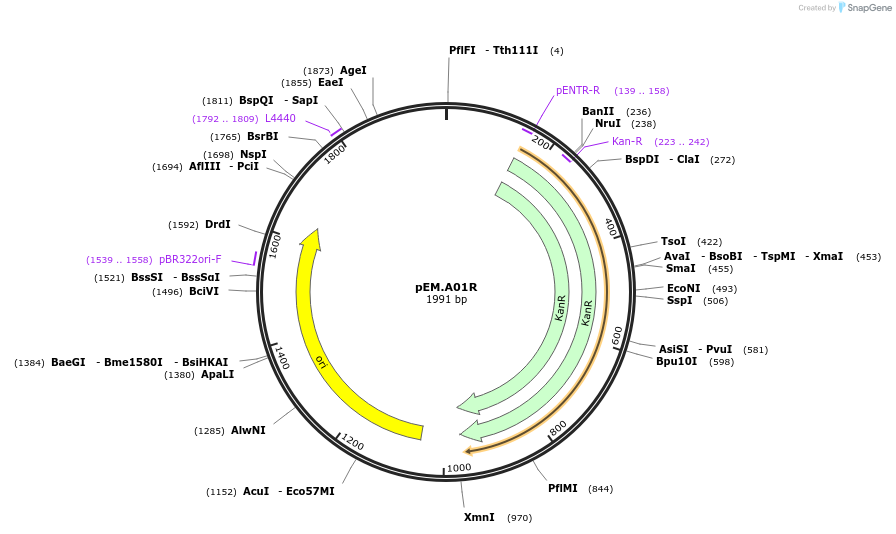
pEM.A01R
Plasmid#198656PurposeEasy-MISE toolkit pEM-plasmid containing a spacer sequence with HI protruding endsDepositorInsertAdaptor
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
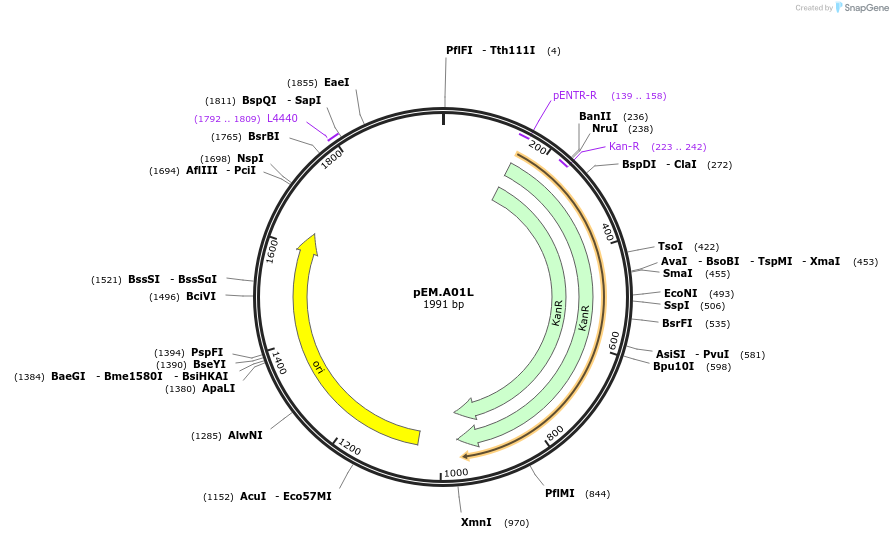
pEM.A01L
Plasmid#198655PurposeEasy-MISE toolkit pEM-plasmid containing a spacer sequence with CD protruding endsDepositorInsertAdaptor
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
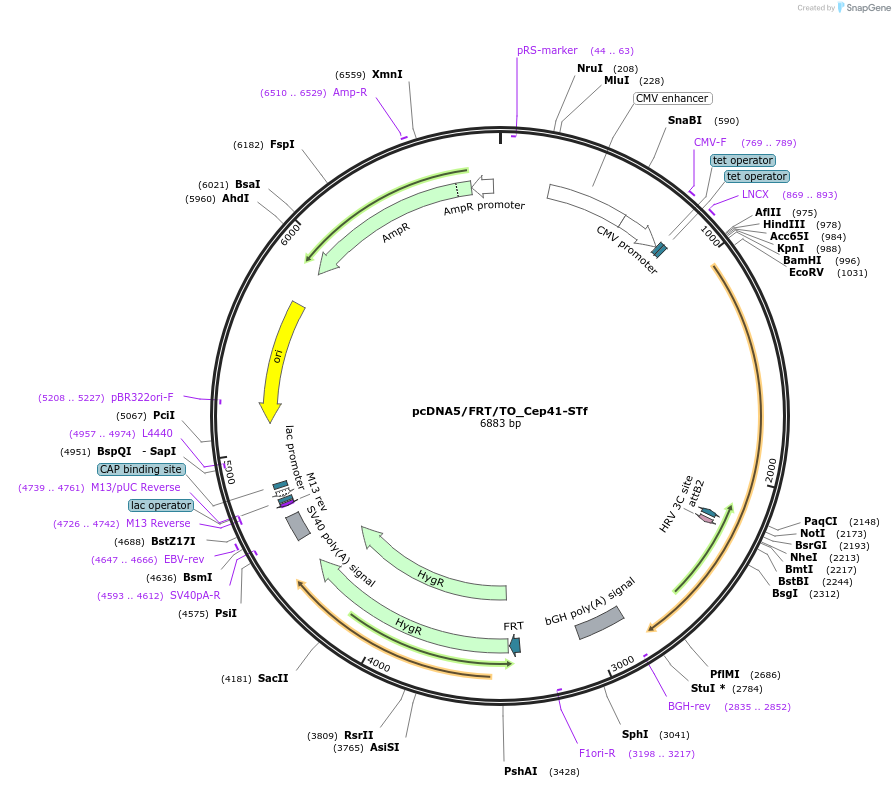
pcDNA5/FRT/TO_Cep41-STf
Plasmid#182010PurposeCMV overexpression of SNAP-tag fast in mammalian cell lines associated to microtubulesDepositorInsertCep41-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
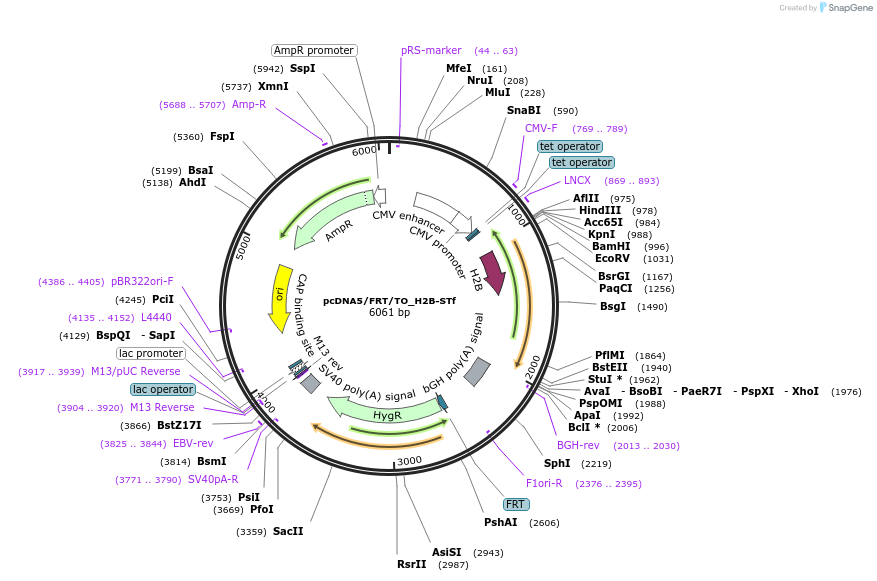
pcDNA5/FRT/TO_H2B-STf
Plasmid#181998PurposeCMV overexpression of SNAP-tag fast fusion to H2B for nuclear localization in mammalian cell linesDepositorInsertH2B-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
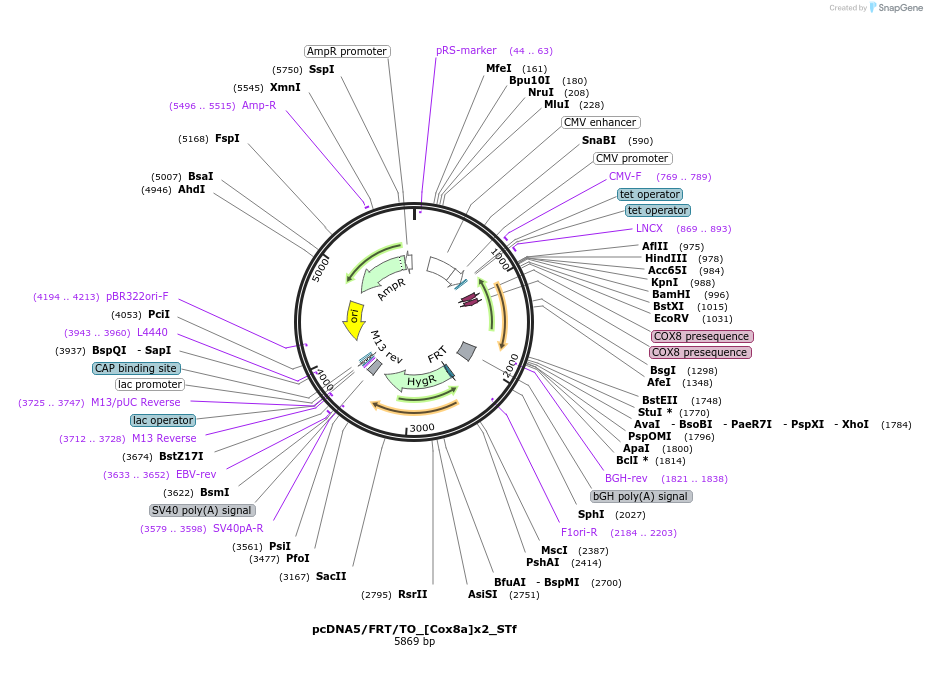
pcDNA5/FRT/TO_[Cox8a]x2_STf
Plasmid#182003PurposeCMV overexpression of SNAP-tag fast in the mitochondrial matrix of mammalian cell linesDepositorInsert[Cox8a]x2_STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
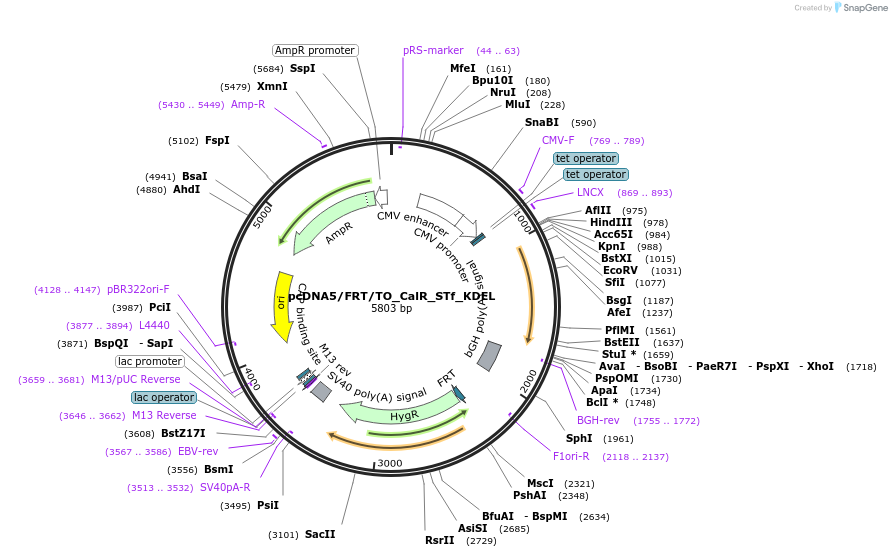
pcDNA5/FRT/TO_CalR_STf_KDEL
Plasmid#182004PurposeCMV overexpression of SNAP-tag fast in the endoplasmic reticulum of mammalian cell linesDepositorInsertCalR_STf_KDEL
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
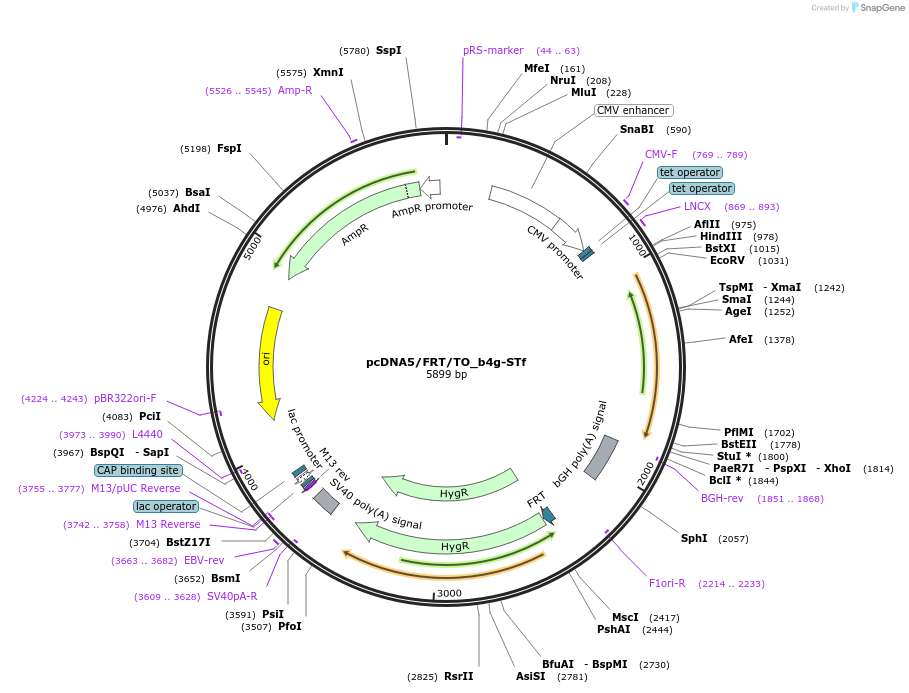
pcDNA5/FRT/TO_b4g-STf
Plasmid#182005PurposeCMV overexpression of SNAP-tag fast in the golgi apparatus of mammalian cell linesDepositorInsertb4g-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
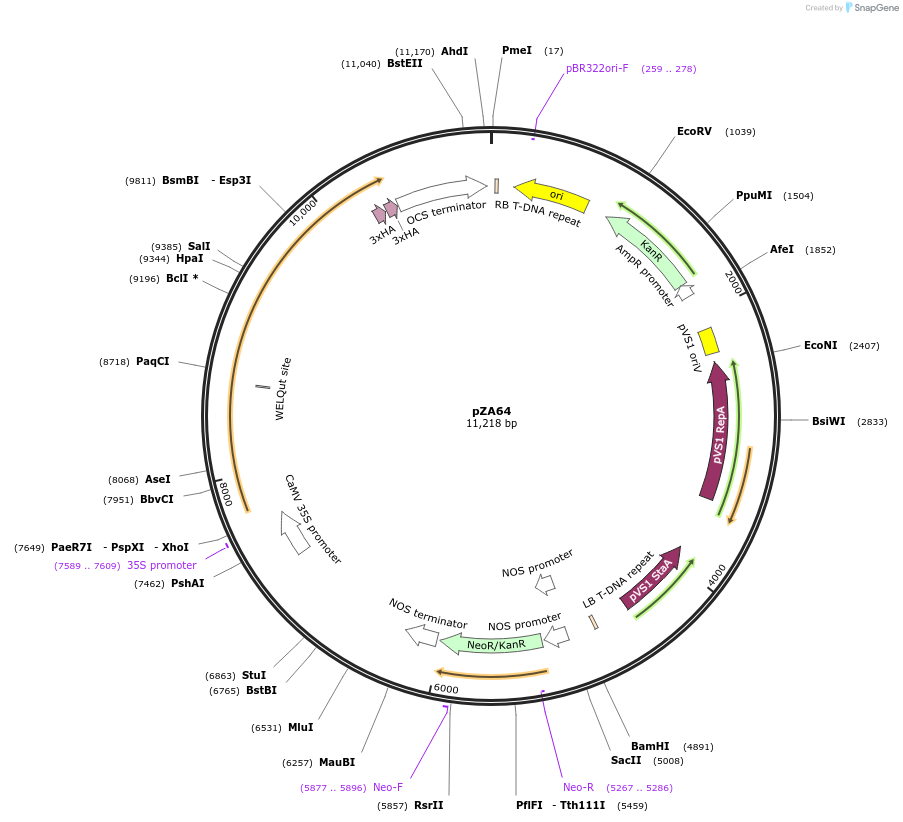
pZA64
Plasmid#174777PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E/D481V-6xHA, Kmr plant selection marker
Tags6xHAExpressionPlantMutationL17E/D481VAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
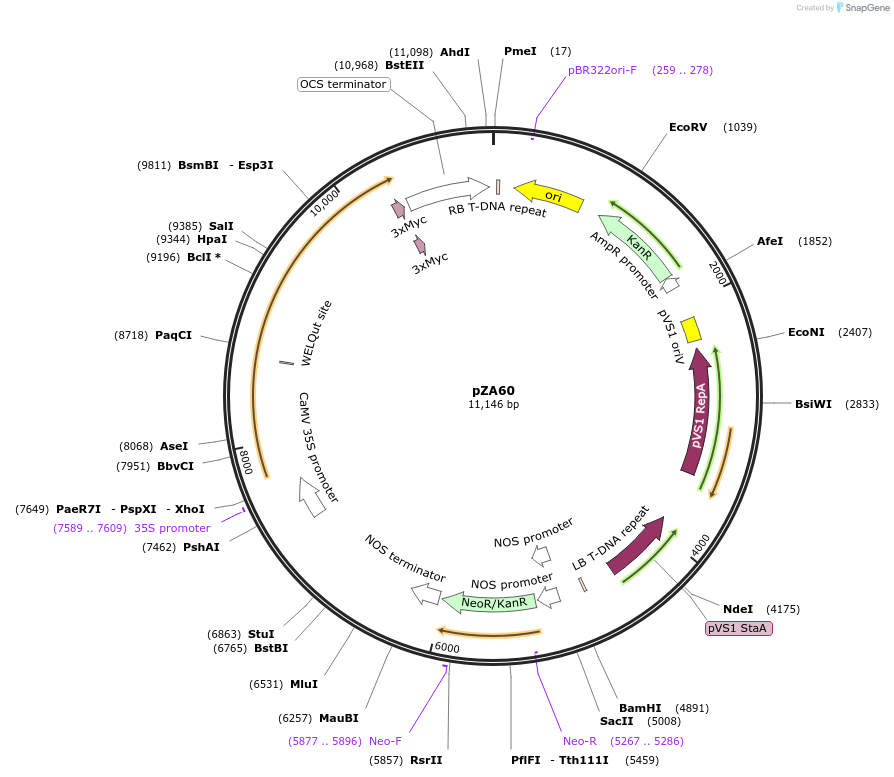
pZA60
Plasmid#174773PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E/D481V-4xMyc, Kmr plant selection marker
Tags4xMycExpressionPlantMutationL17E/D481VAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
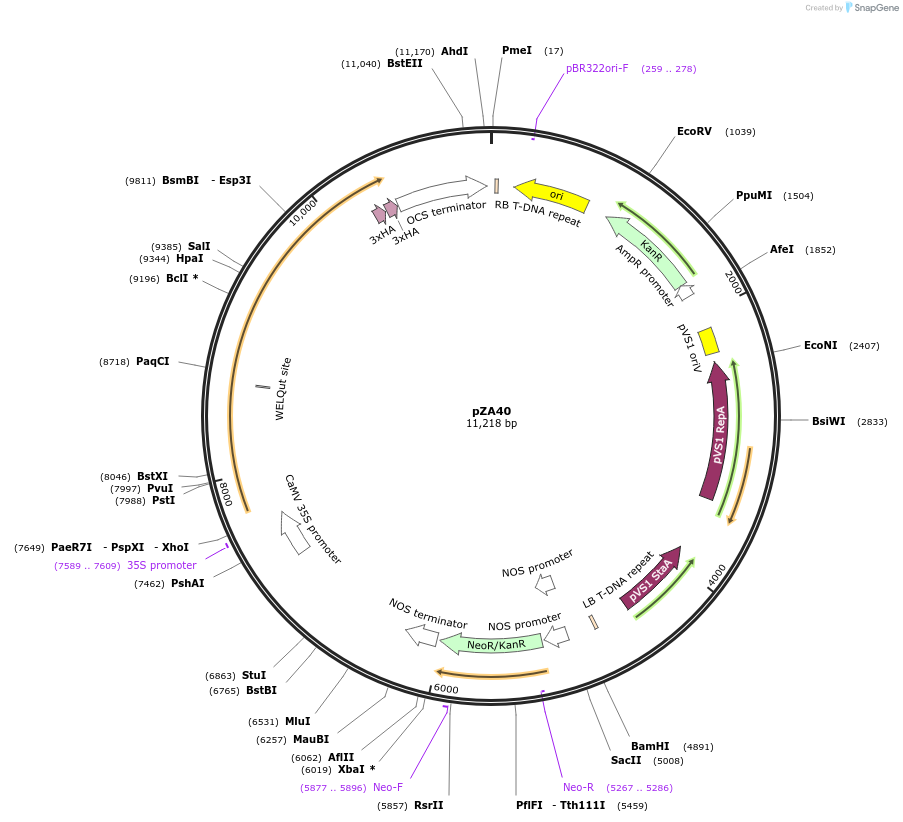
pZA40
Plasmid#158518PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17E, D481VAvailable SinceNov. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
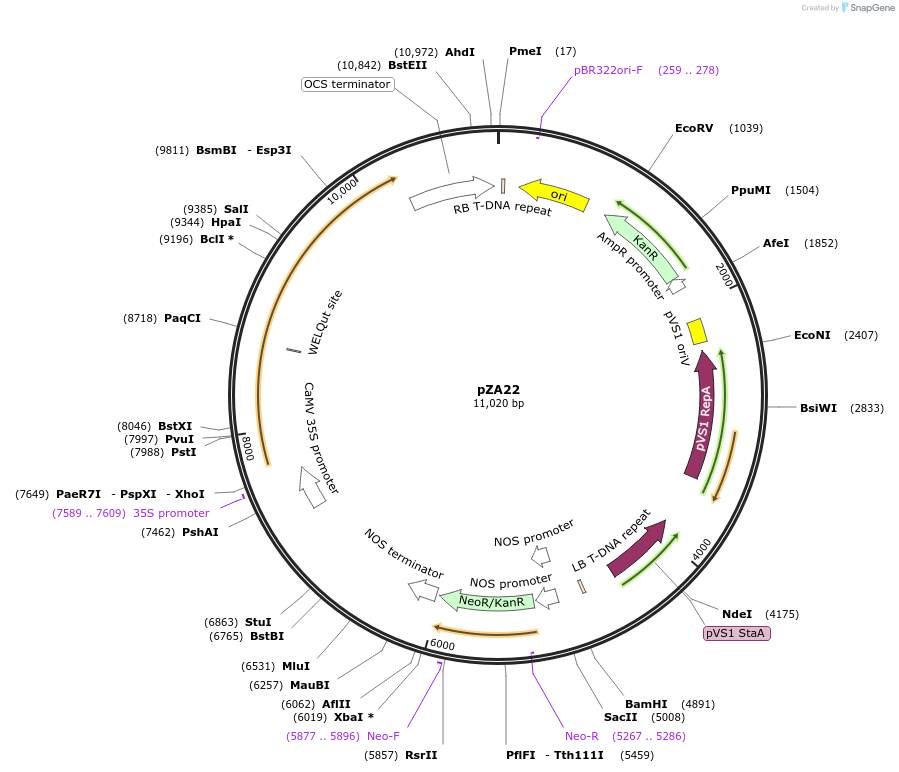
pZA22
Plasmid#158500PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V. Km plant selection marker.DepositorInsertZAR1
ExpressionPlantMutationD481VAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
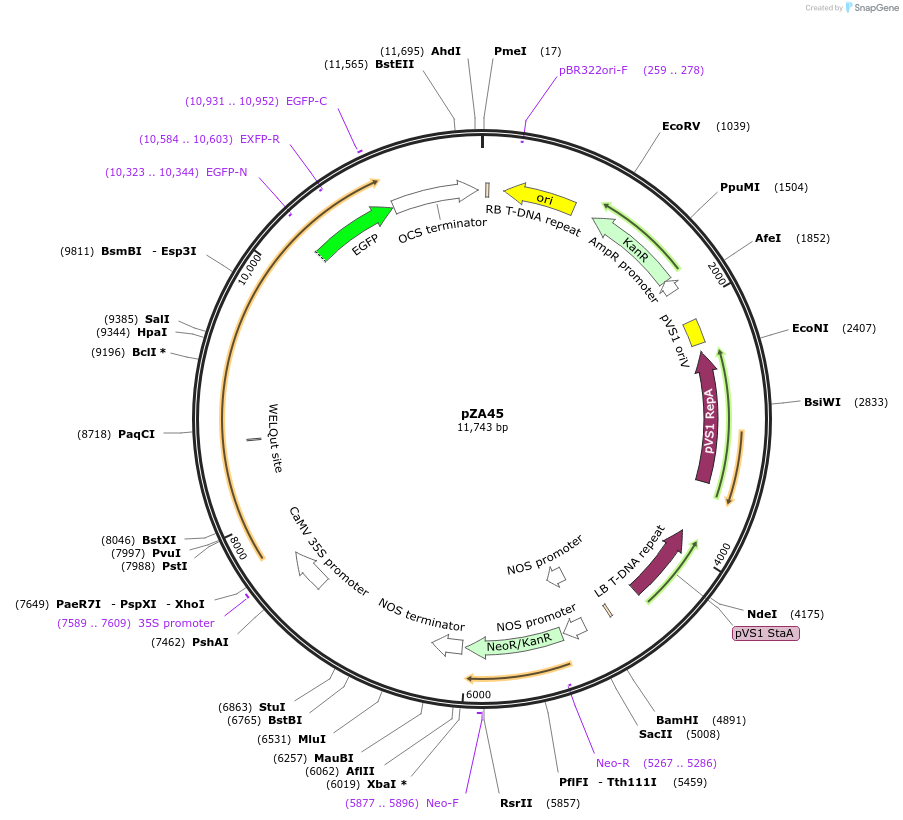
pZA45
Plasmid#158523PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantAvailable SinceSept. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
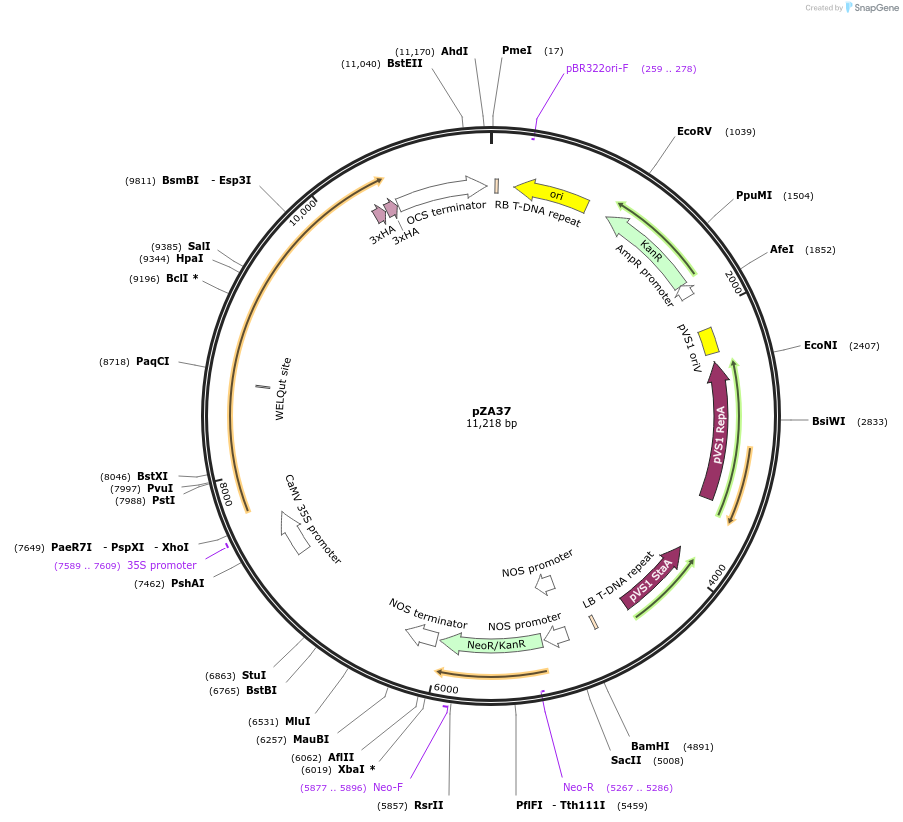
pZA37
Plasmid#158515PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
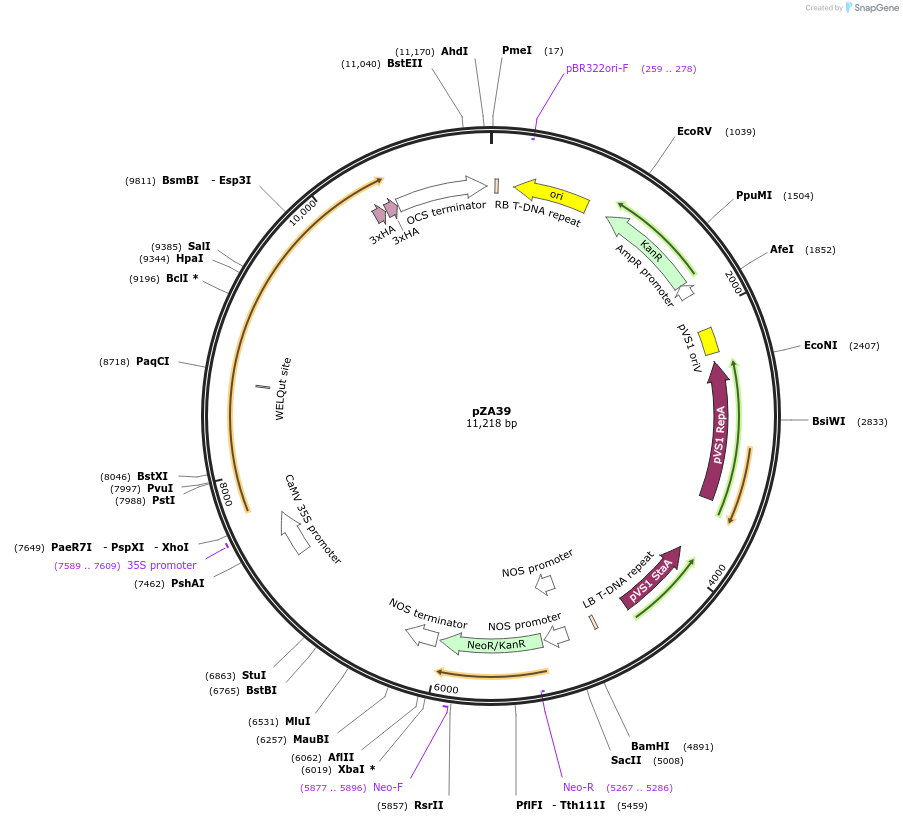
pZA39
Plasmid#158517PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17EAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
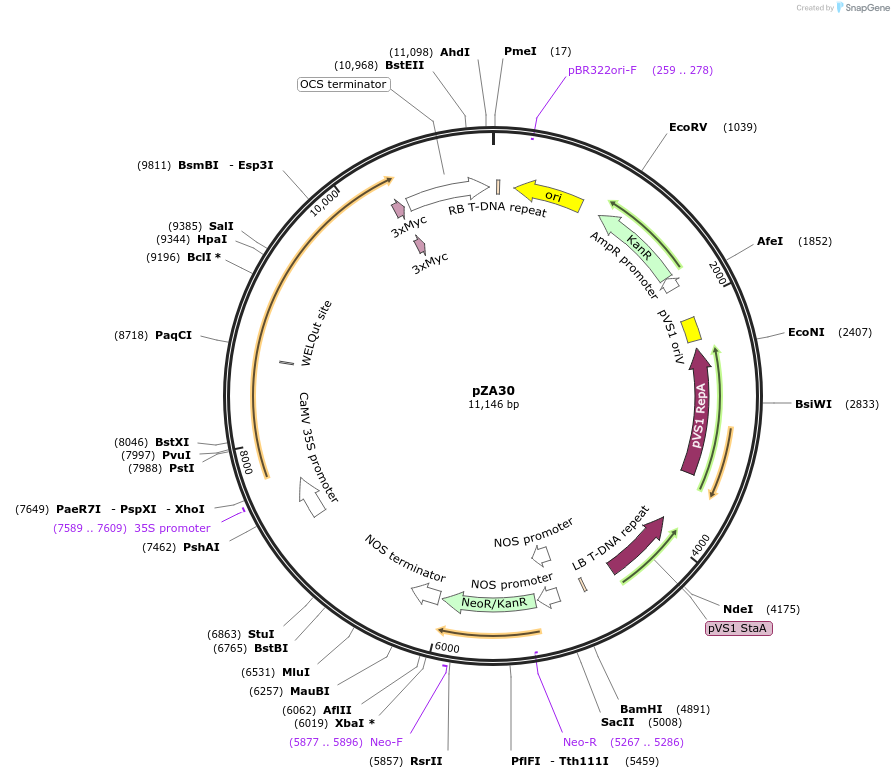
pZA30
Plasmid#158508PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
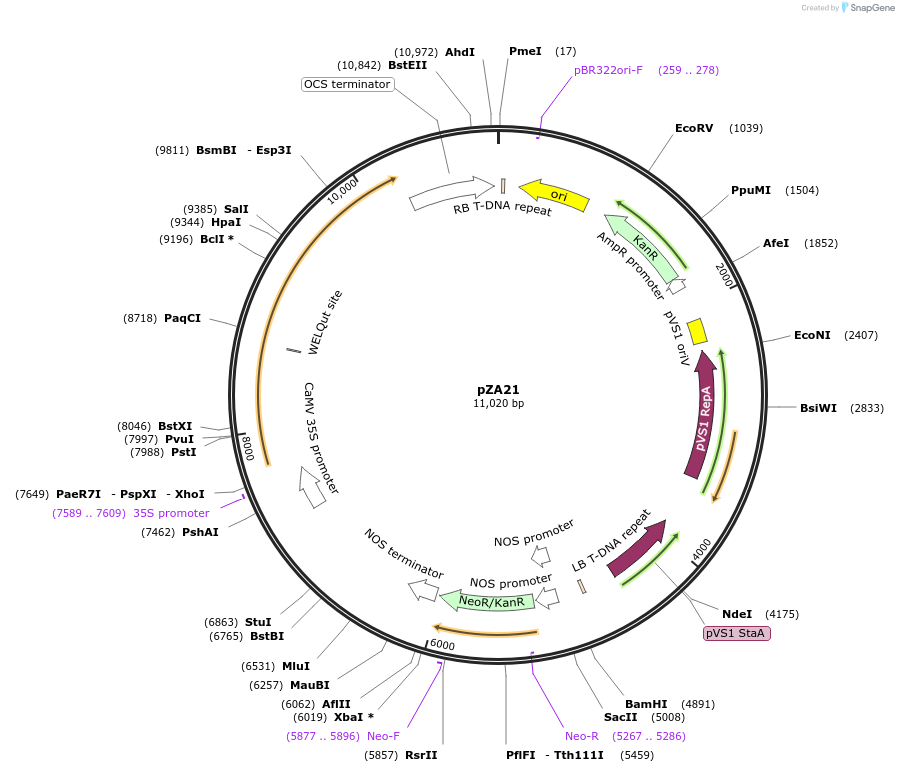
pZA21
Plasmid#158499PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1. Km plant selection marker.DepositorInsertZAR1
ExpressionPlantAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
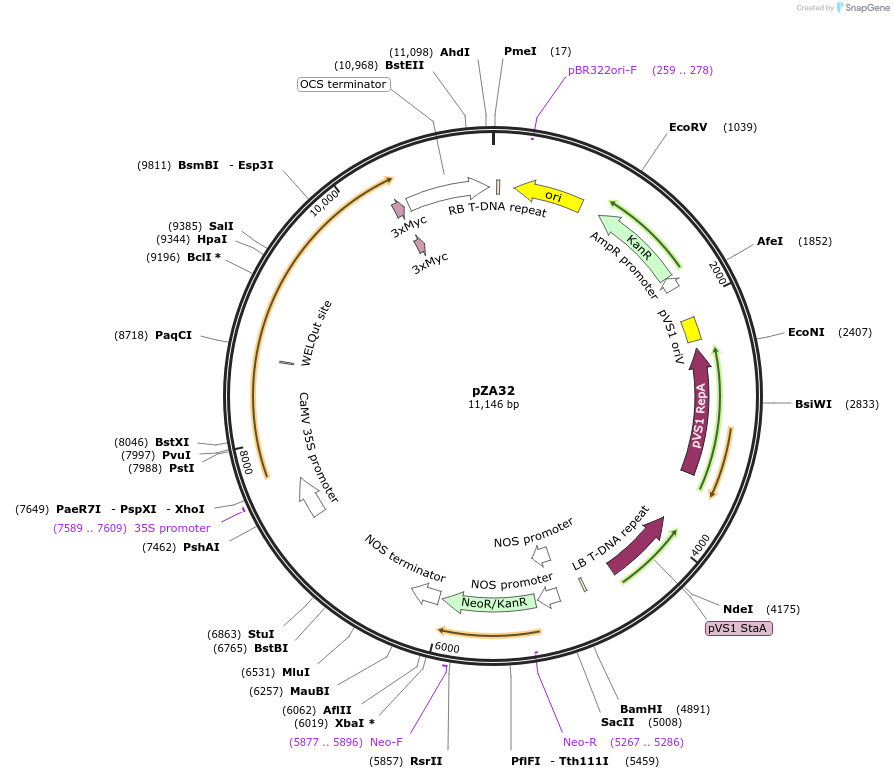
pZA32
Plasmid#158510PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only