We narrowed to 14,493 results for: NTS;
-
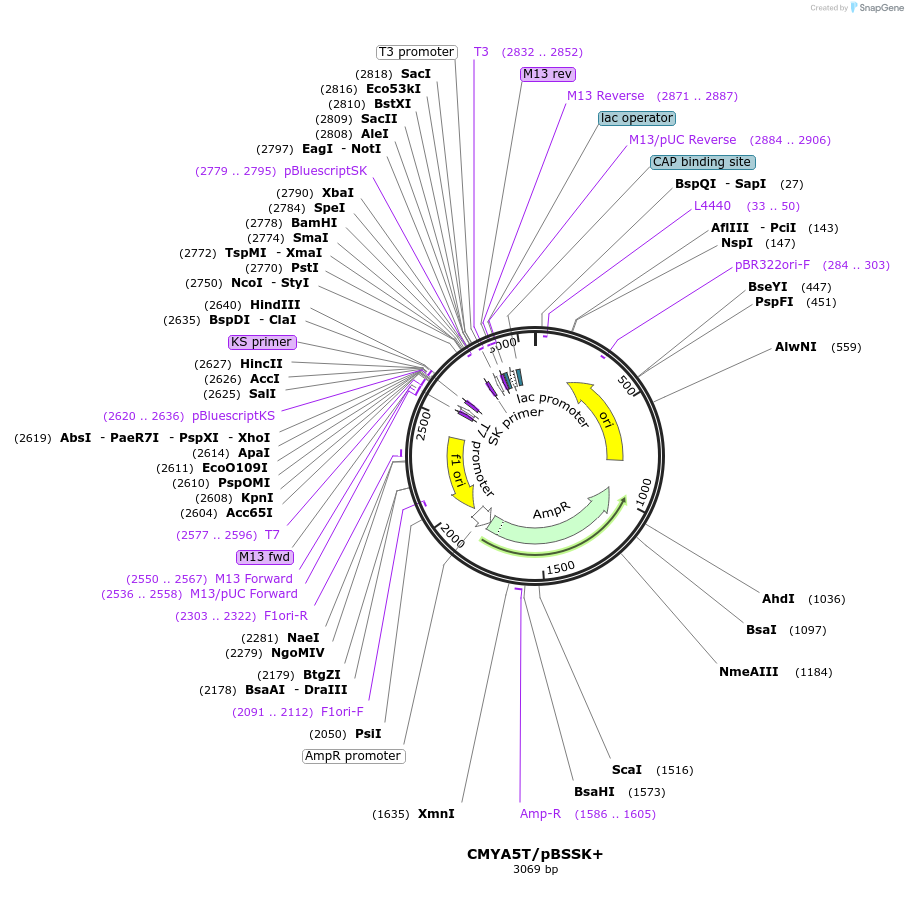
Plasmid#97206PurposeCMYA5 (NM_153610, nt 12205 to 12279) T allele (rs10043986) in pBSSK+DepositorAvailable SinceAug. 3, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits
-
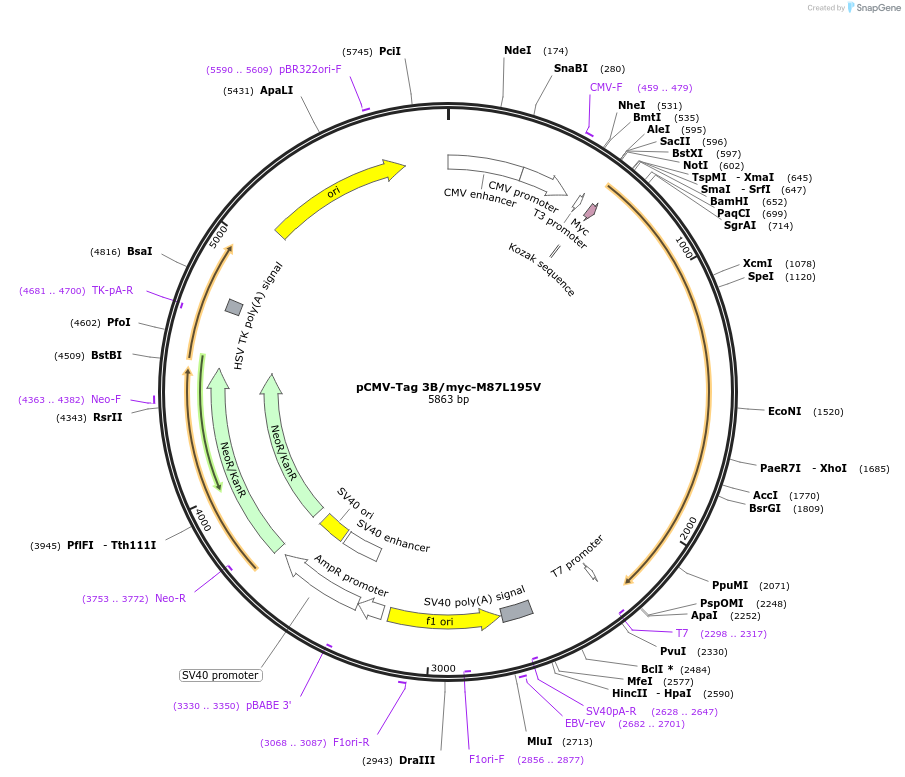
pCMV-Tag 3B/myc-M87L195V
Plasmid#87721PurposeExpresses myc-M87L195V spastin isoform in mammalian cellsDepositorInsertSPAST (SPAST Human)
TagsMycExpressionMammalianMutationM87 isoform (deletion of nt 1-490 - 5'UTR an…PromoterCMVAvailable SinceMarch 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
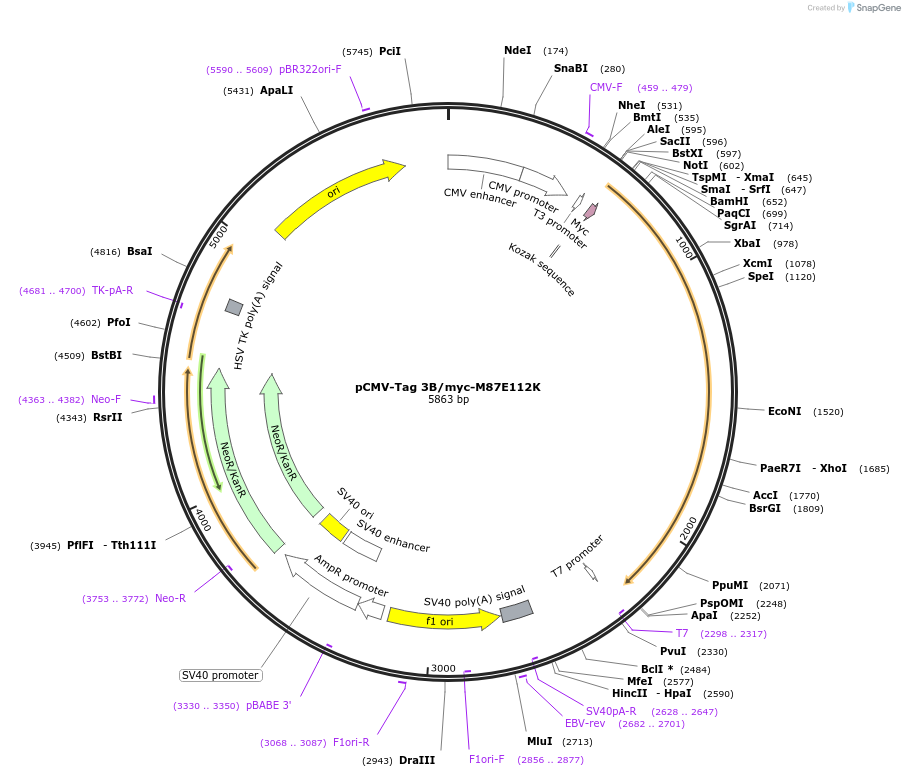
pCMV-Tag 3B/myc-M87E112K
Plasmid#87720PurposeExpresses myc-M87E112K spastin isoform in mammalian cellsDepositorInsertSPAST (SPAST Human)
TagsMycExpressionMammalianMutationM87 isoform (deletion of nt 1-490 - 5'UTR an…PromoterCMVAvailable SinceMarch 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
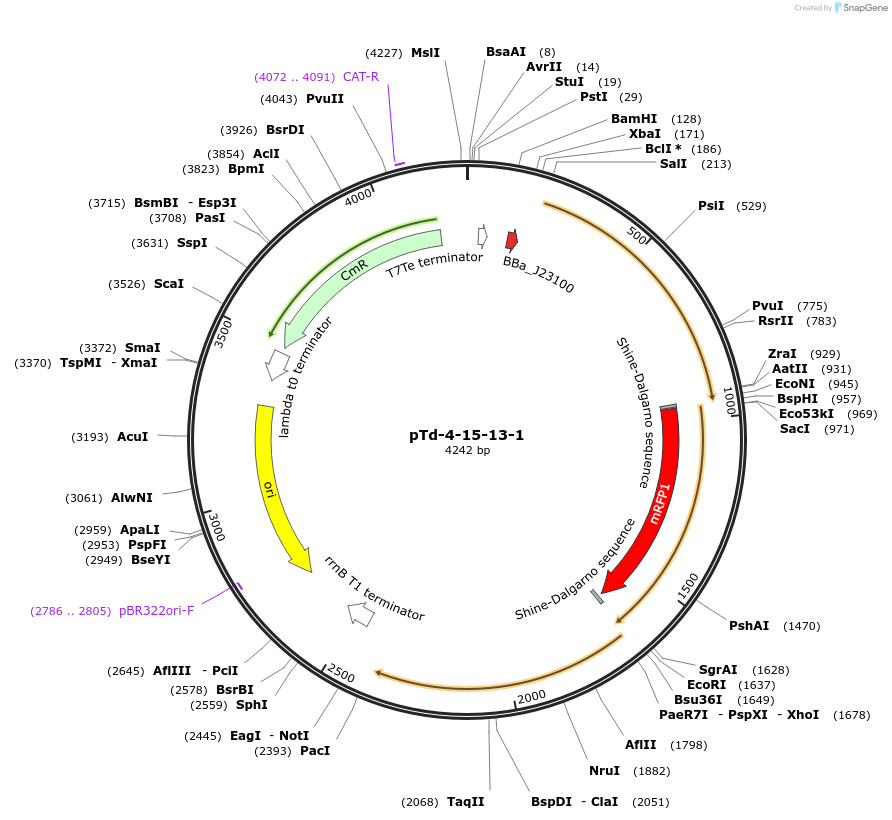
pTd-4-15-13-1
Plasmid#63826PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-2
Plasmid#63827PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-3
Plasmid#63828PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-4
Plasmid#63829PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-5
Plasmid#63830PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-6
Plasmid#63831PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-7
Plasmid#63832PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-8
Plasmid#63833PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-9
Plasmid#63834PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pTd-4-15-13-11
Plasmid#63836PurposeThis plasmid is designed to validate the biophysical model of translational couplingDepositorInsertCFP, mRFP1 and GFPmut3b
ExpressionBacterialMutationThe intergenic distance between each gene is -4 n…PromoterJ23100Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
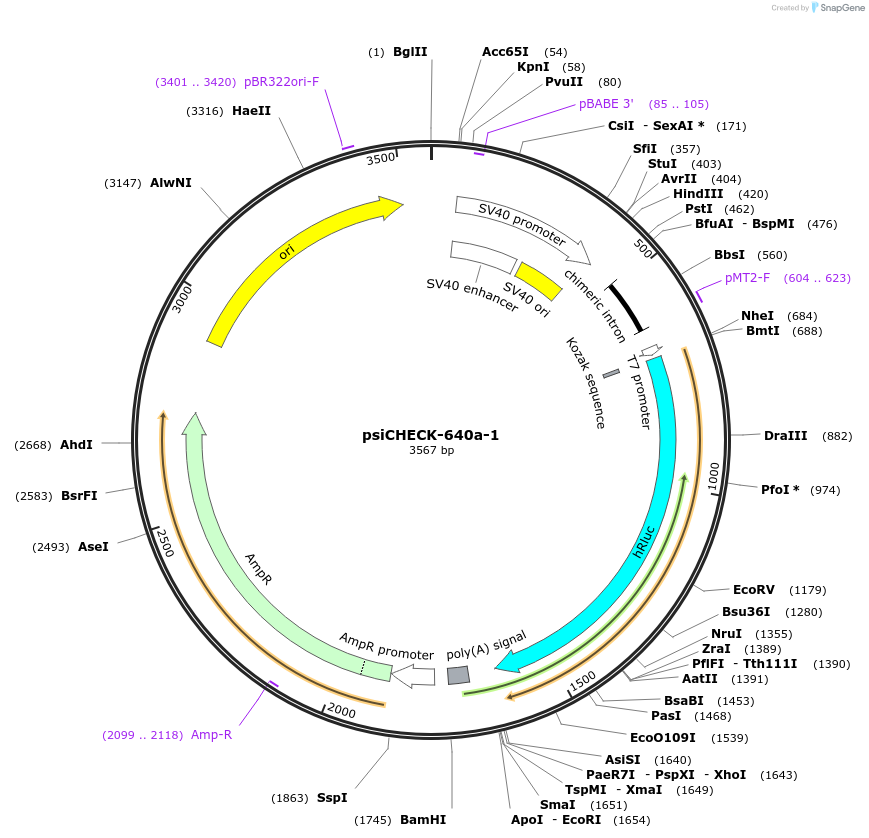
psiCHECK-640a-1
Plasmid#42092DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 820-842 nt segment of ZEB1 mRNA (NM_…Available SinceApril 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
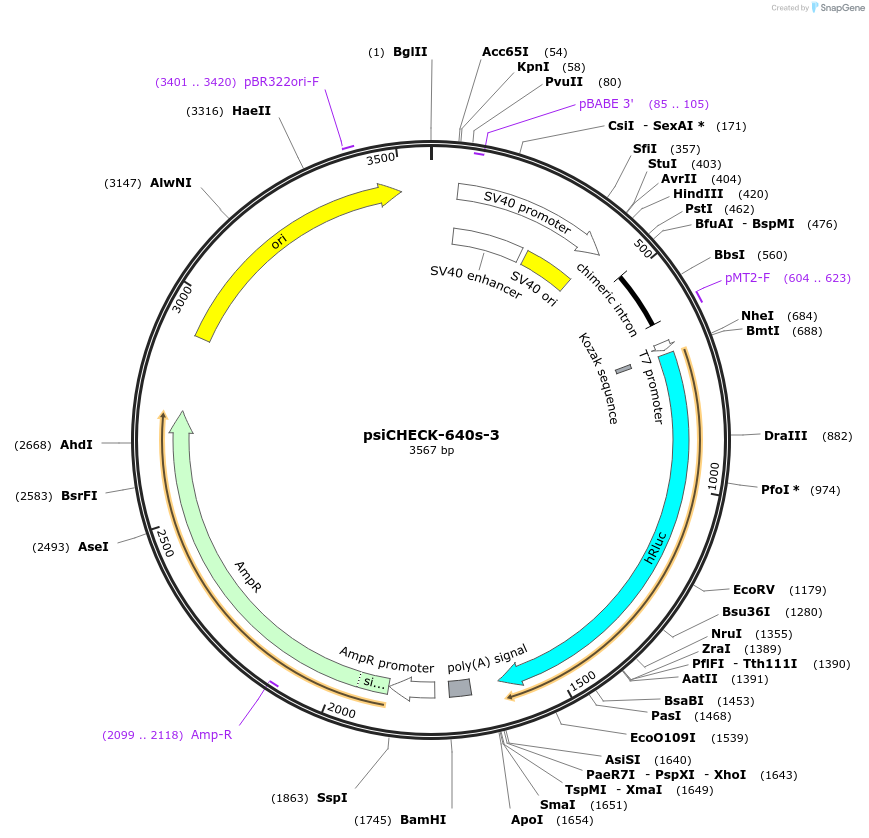
psiCHECK-640s-3
Plasmid#42091DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 4565-4587 nt segment of ZEB1 mRNA (N…Available SinceApril 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
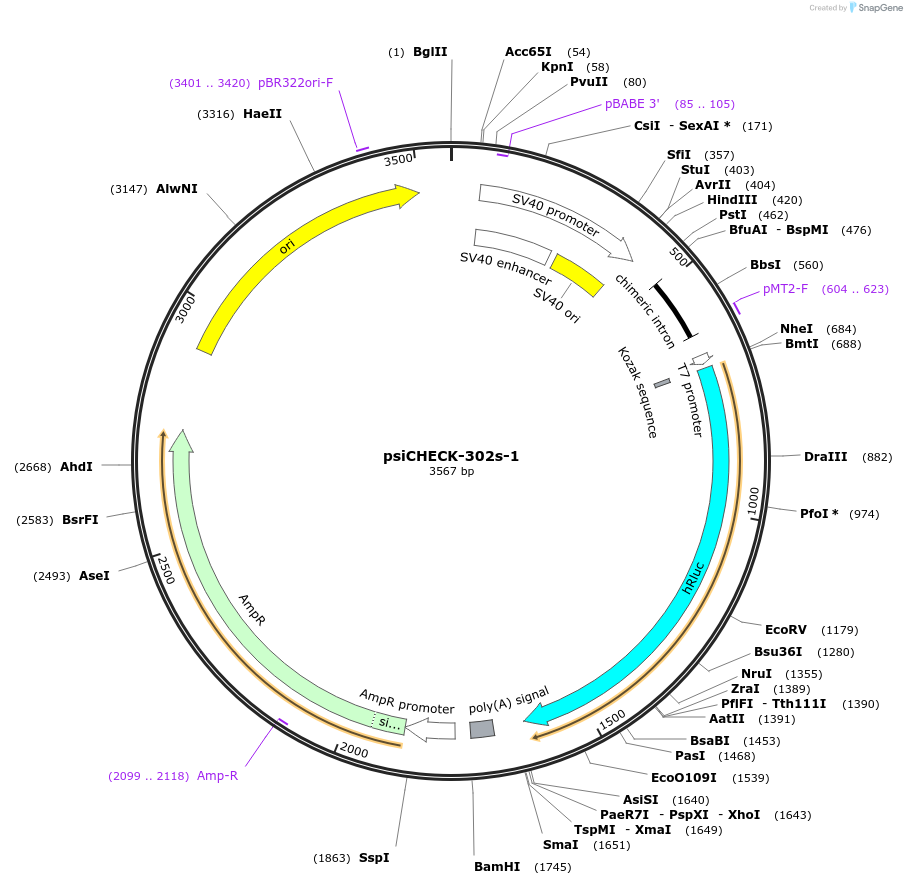
psiCHECK-302s-1
Plasmid#42087DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 4146-4168 nt segment of ZEB1 mRNA (N…Available SinceApril 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
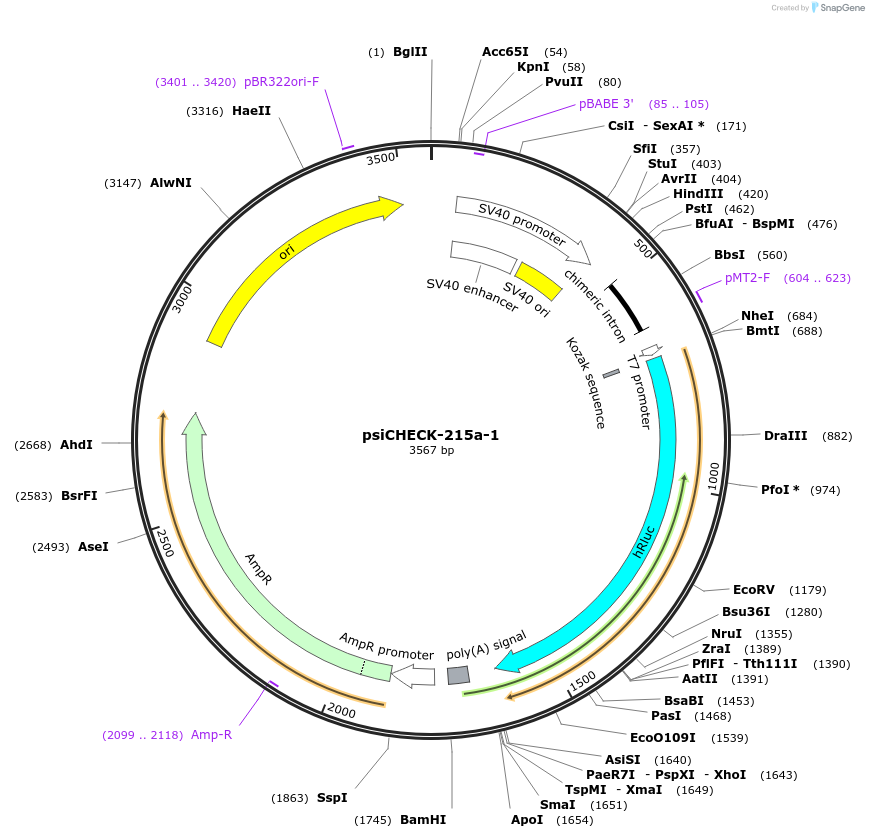
psiCHECK-215a-1
Plasmid#42085DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 1259-1281 nt segment of ZEB1 mRNA (N…Available SinceMarch 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
psiCHECK-640s-2
Plasmid#42090DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 1956-1981 nt segment of ZEB1 mRNA (N…Available SinceFeb. 27, 2013AvailabilityAcademic Institutions and Nonprofits only -
psiCHECK-640s-1
Plasmid#42089DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 1419-1441 nt segment of ZEB1 mRNA (N…Available SinceFeb. 27, 2013AvailabilityAcademic Institutions and Nonprofits only -
psiCHECK-302a-1
Plasmid#42088DepositorInsertZEB1 (ZEB1 Human)
UseLuciferaseExpressionMammalianMutationcontains the 4055-4078 nt segment of ZEB1 mRNA (N…Available SinceFeb. 27, 2013AvailabilityAcademic Institutions and Nonprofits only