We narrowed to 18,548 results for: MUT
-
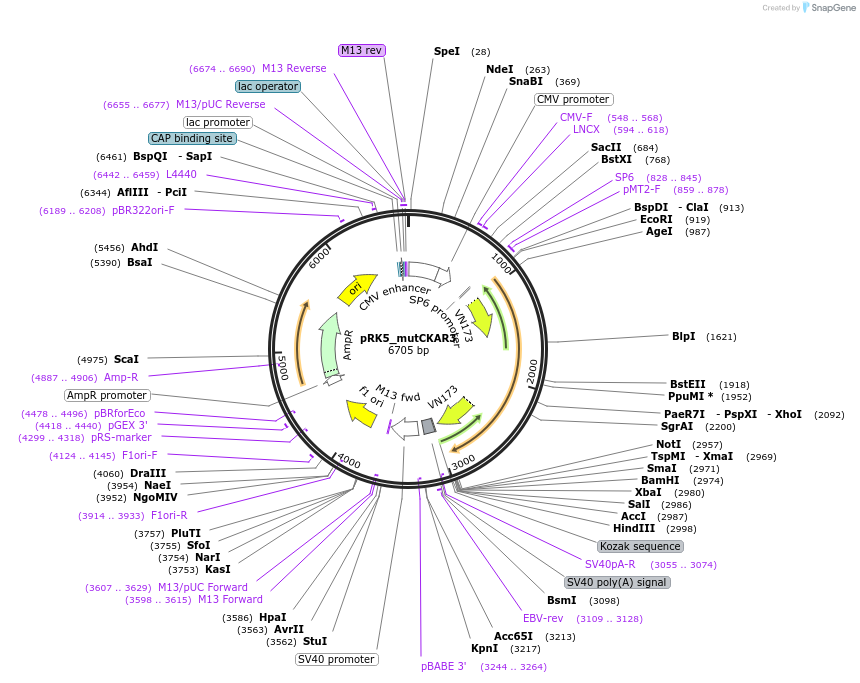
Plasmid#238412PurposeMammalian expression of a phosphorylation deficient (T to A) mutant of CKAR3 under a CMV promotor.DepositorInsertphosphorylation deficient mutant of CKAR3 (T to A)
ExpressionMammalianAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
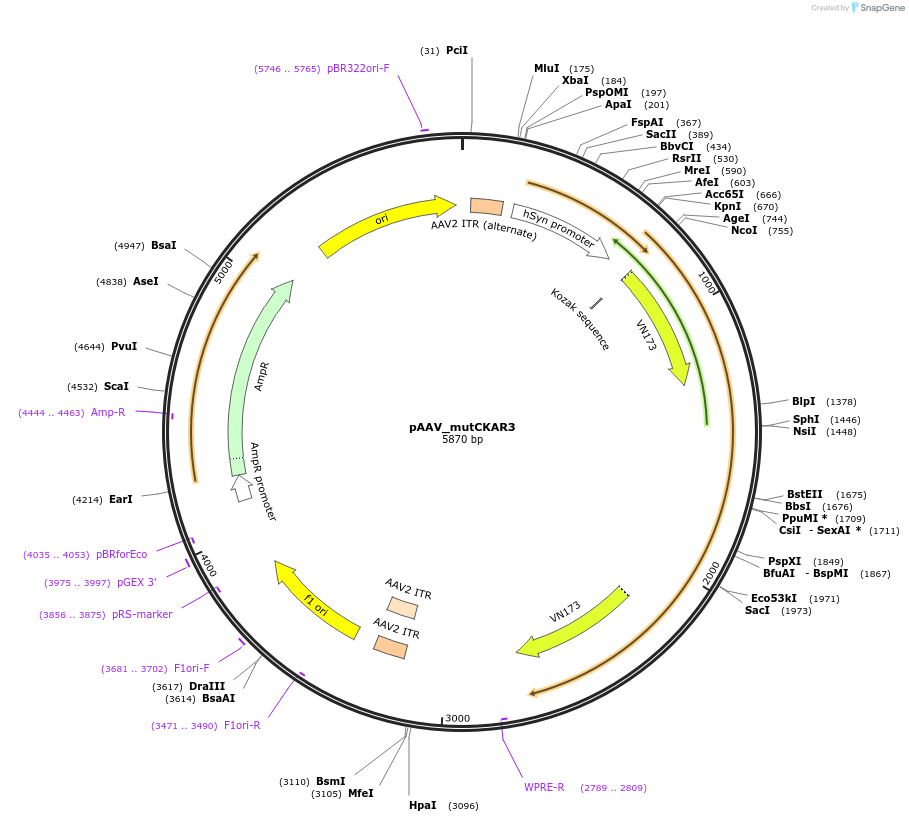
pAAV_mutCKAR3
Plasmid#238414PurposeAAV construct with CKAR3 mutant expression under a human synapsin promotor and WPRE3 translation enhancer.DepositorInsertphosphorylation deficient mutant of CKAR3 (T to A)
UseAAVExpressionMammalianAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
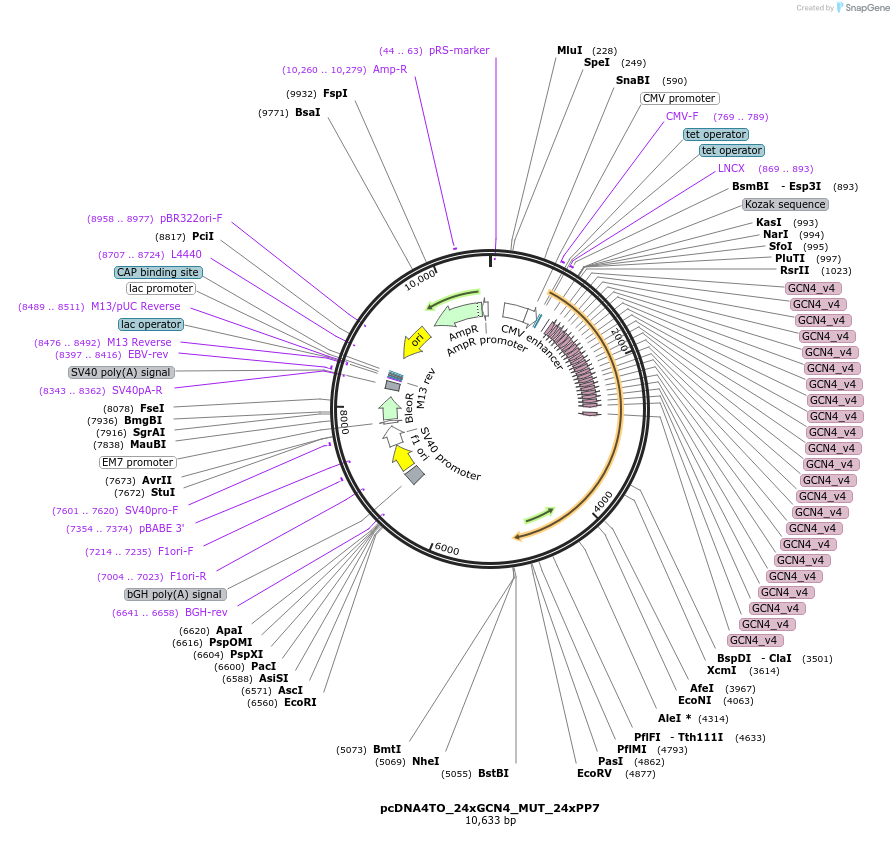
pcDNA4TO_24xGCN4_MUT_24xPP7
Plasmid#236093PurposeConstitutively encodes 24xGCN4-MUT-24xPP7 stem loops, for visualization of the Mut mRNA.DepositorInsert24xGCN4-MUT-24xPP7
ExpressionMammalianAvailable SinceApril 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
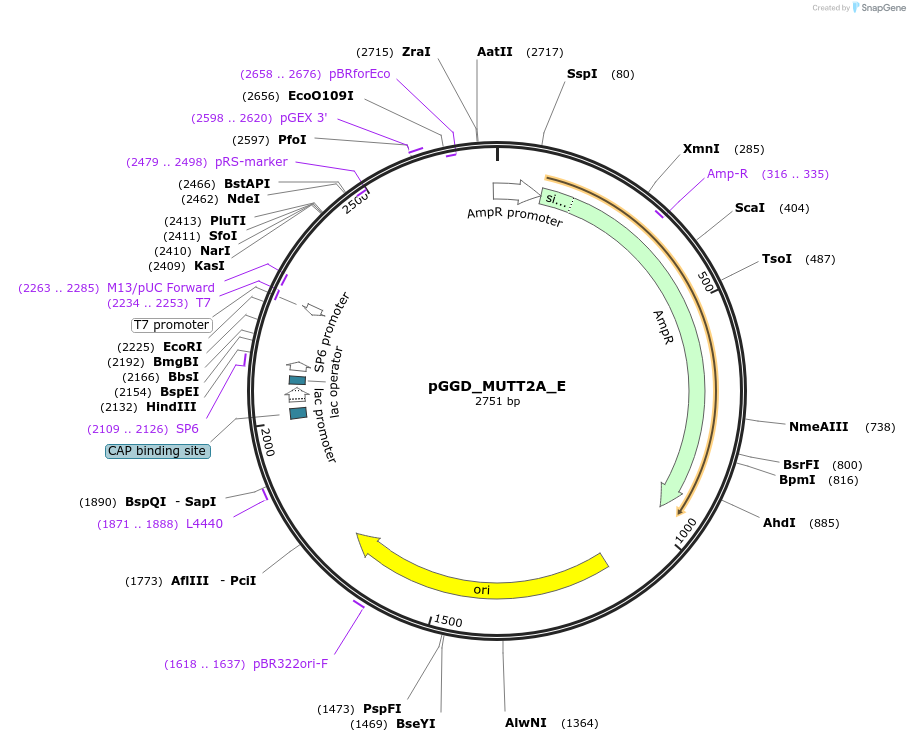
pGGD_MUTT2A_E
Plasmid#218899Purposegolden gate cloningDepositorInsertMUTT2A
UseSynthetic BiologyAvailable SinceMay 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
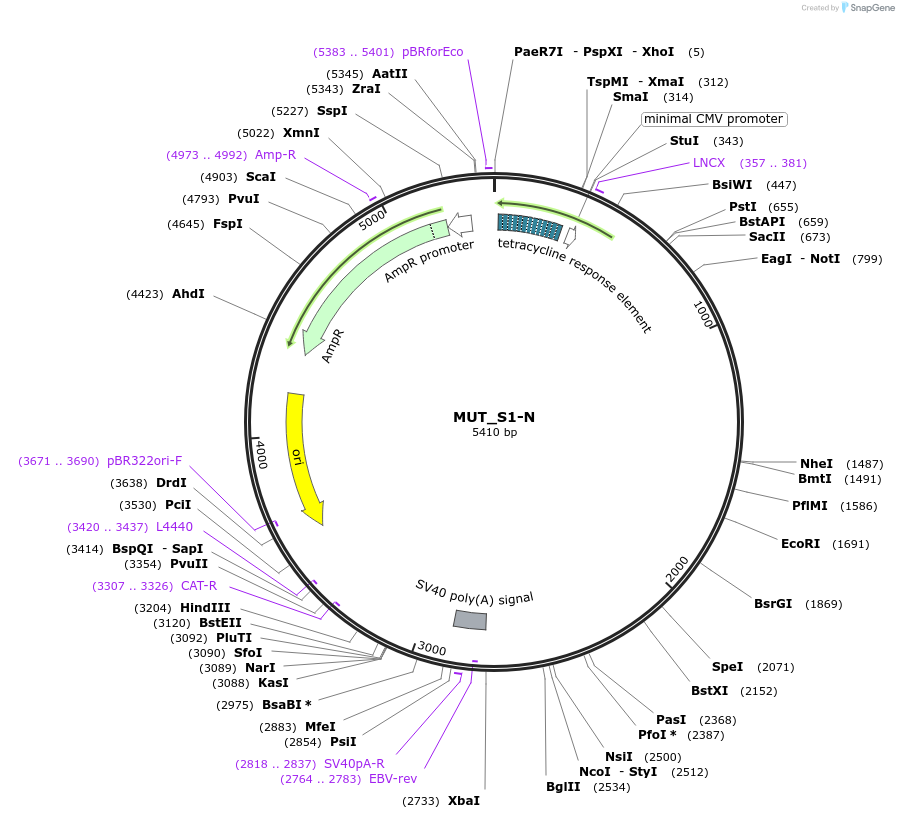
MUT_S1-N
Plasmid#218398Purpose3-exon minigene, with SRSF1 motif-rich middle exon, neutral region in second intron, and AG>GG mutation in final 3' splice siteDepositorInsertModified CHO DHFR minigene
ExpressionMammalianPromotertetracycline-responsive promoterAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
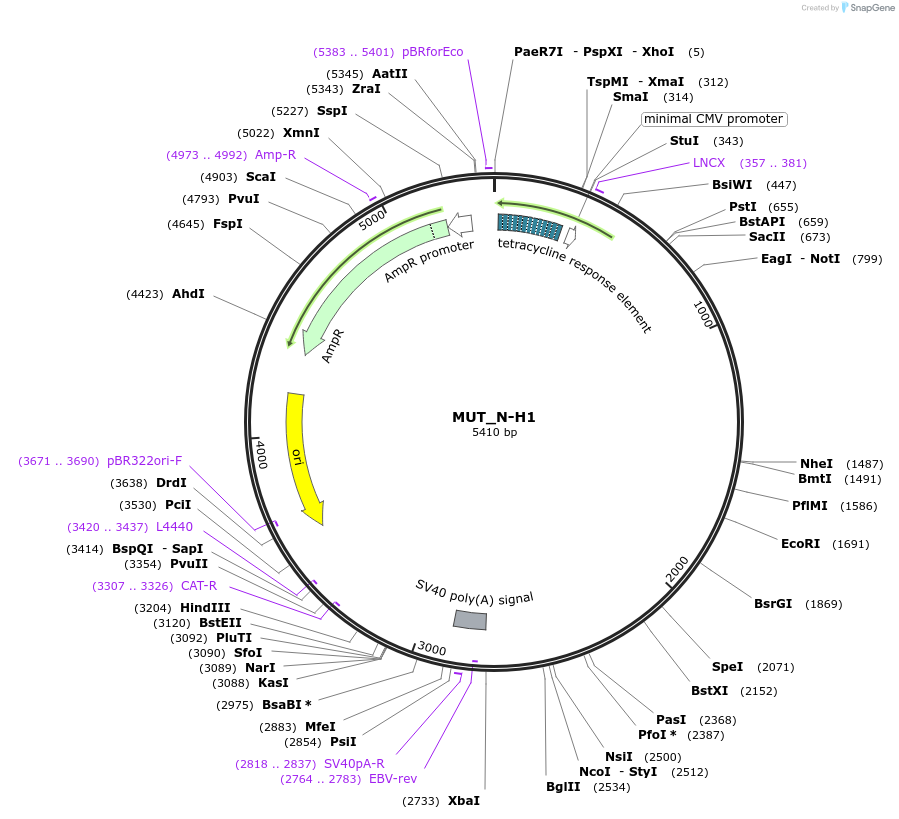
MUT_N-H1
Plasmid#218397Purpose3-exon minigene, with neutral middle exon, hnRNPA1 motif-rich region in second intron, and AG>GG mutation in final 3' splice siteDepositorInsertModified CHO DHFR minigene
ExpressionMammalianPromotertetracycline-responsive promoterAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
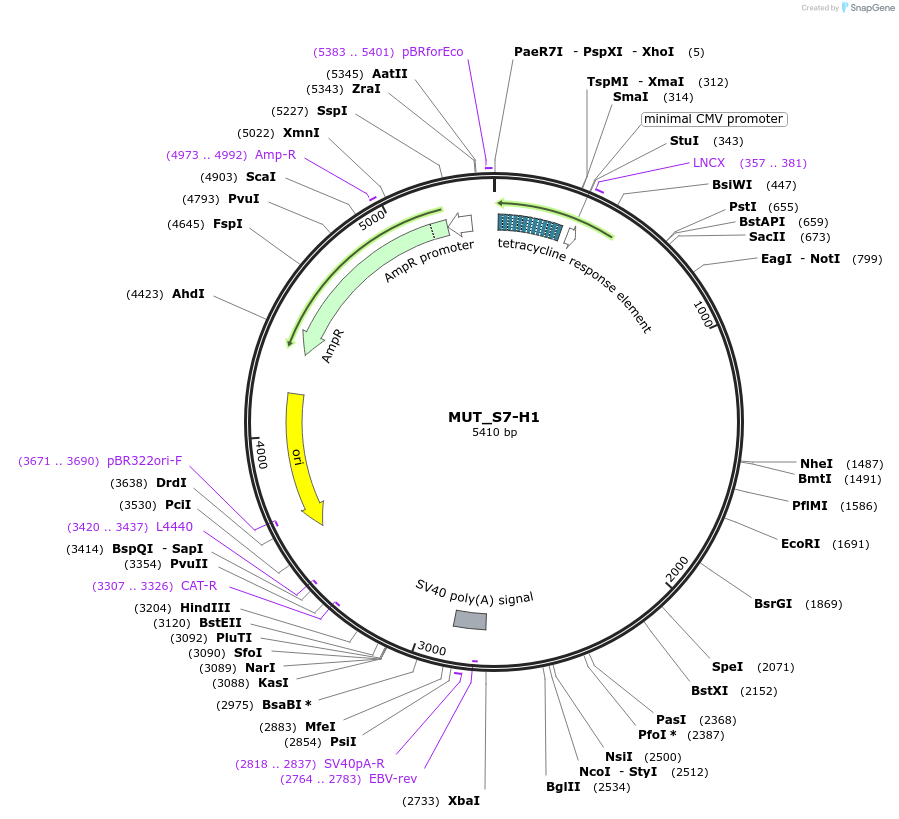
MUT_S7-H1
Plasmid#218396Purpose3-exon minigene, with SRSF7 motif-rich middle exon, hnRNPA1 motif-rich region in second intron, and AG>GG mutation in final 3' splice siteDepositorInsertModified CHO DHFR minigene
ExpressionMammalianPromotertetracycline-responsive promoterAvailable SinceMay 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
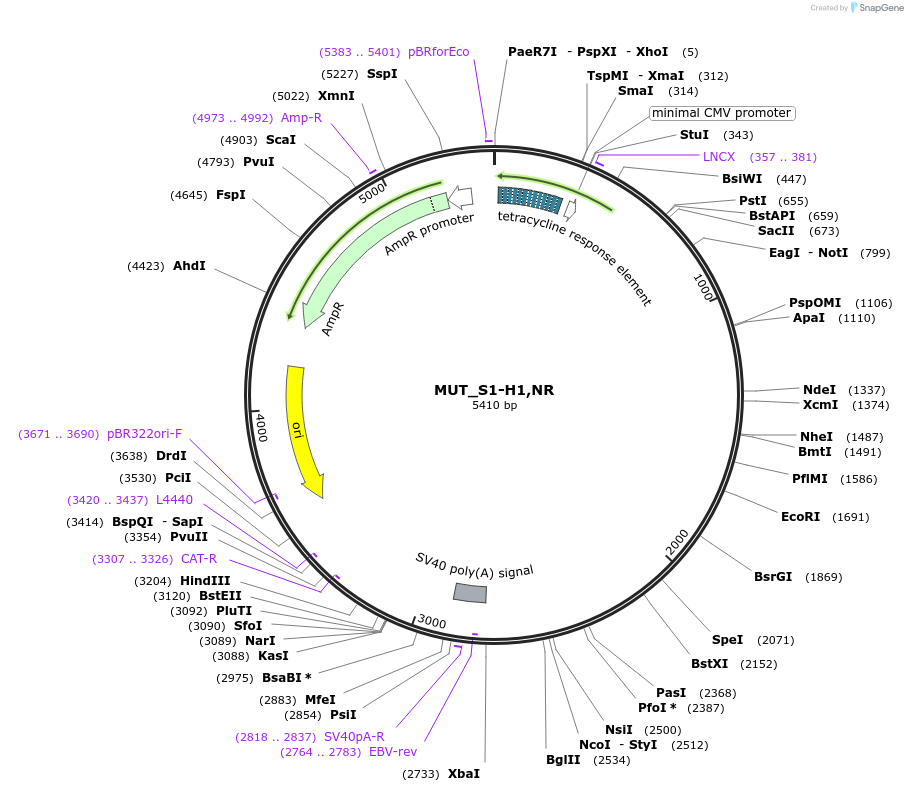
MUT_S1-H1,NR
Plasmid#218395Purpose3-exon minigene, with SRSF1 motif-rich middle exon, hnRNPA1 motif-rich region in second intron, both randomly mutated to avoid sequence repeats, and AG>GG mutation in final 3' splice siteDepositorInsertModified CHO DHFR minigene
ExpressionMammalianPromotertetracycline-responsive promoterAvailable SinceMay 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
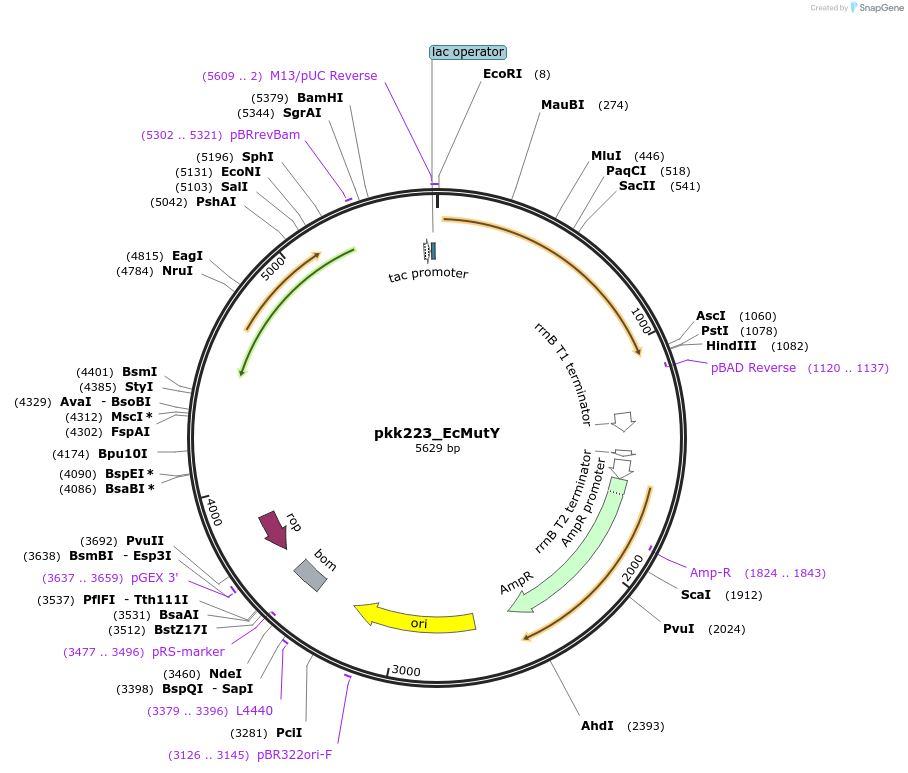
pkk223_EcMutY
Plasmid#213110Purposepkk223 with E coli MutY insertDepositorInsertMutY (adenine glycosylase) (mutY Escherichia coli)
ExpressionBacterialAvailable SinceApril 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
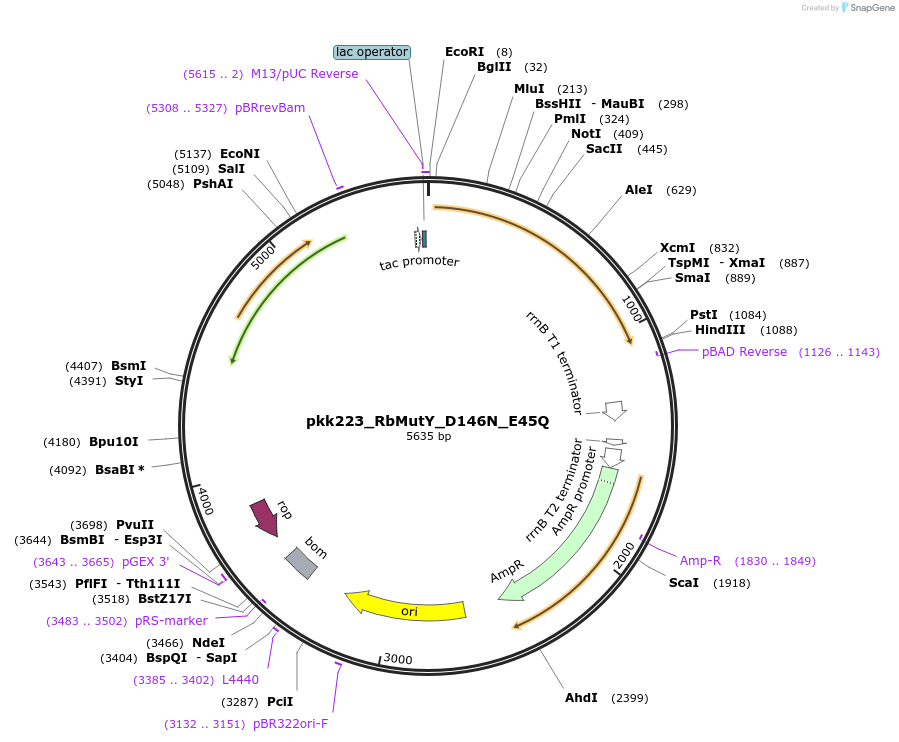
pkk223_RbMutY_D146N_E45Q
Plasmid#210798Purposepkk223 with Rhodobacteraceae MutY containing D146N and E45Q mutations to reduce catalytic activityDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationD146N_E45QAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
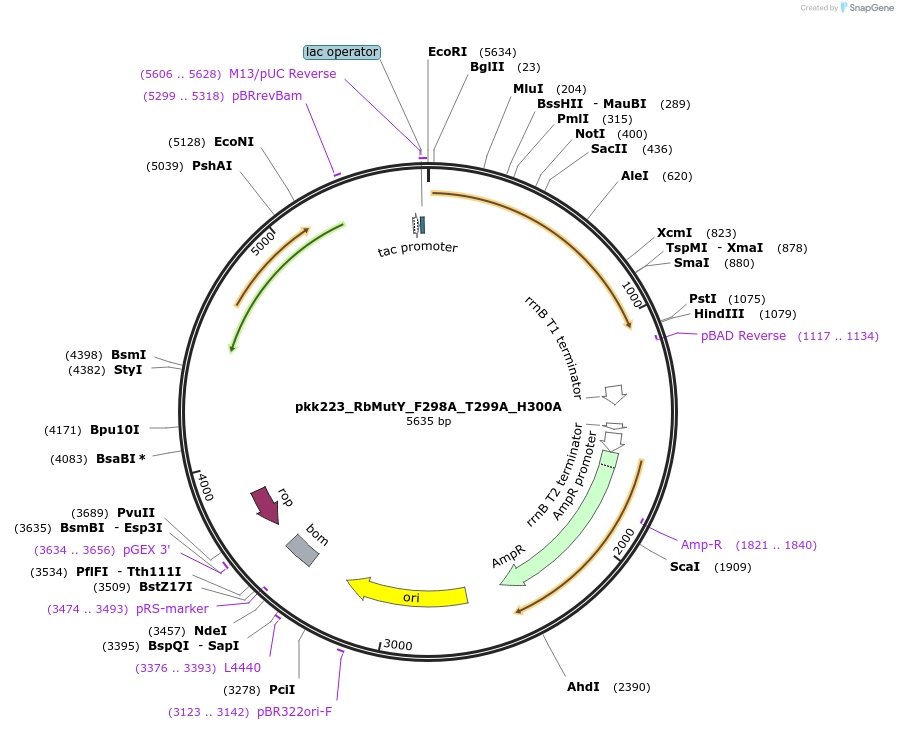
pkk223_RbMutY_F298A_T299A_H300A
Plasmid#210799Purposepkk223 with Rhodobacteraceae MutY containing F298A, S299A, and H300A mutations to reduce recognition of OG:ADepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationF298A_T299A_H300AAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
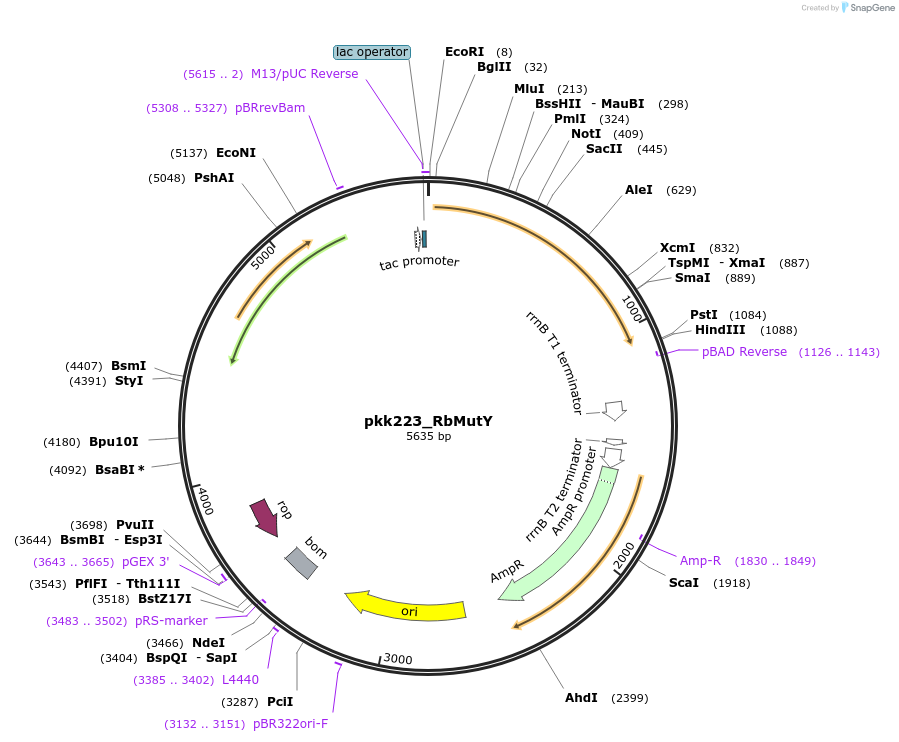
pkk223_RbMutY
Plasmid#210797Purposepkk223 with Rhodobacteraceae MutY insertDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
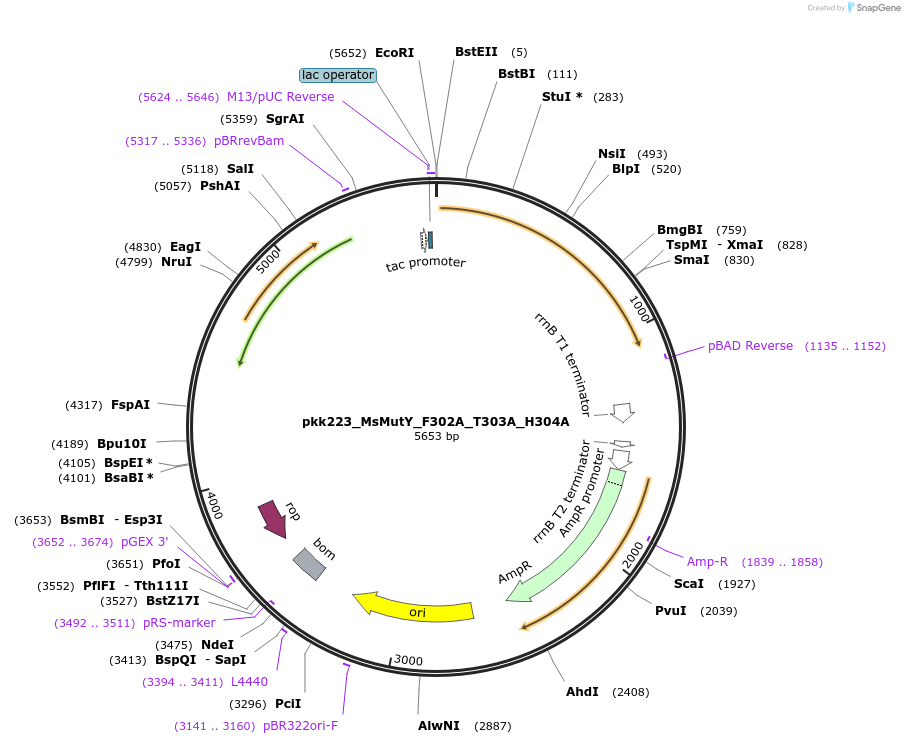
pkk223_MsMutY_F302A_T303A_H304A
Plasmid#210795Purposepkk223 with Marinosulfonomonas MutY containing F302A, T303A, and H304A mutations to reduce recognition of OG:ADepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationF302A_T303A_H304AAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
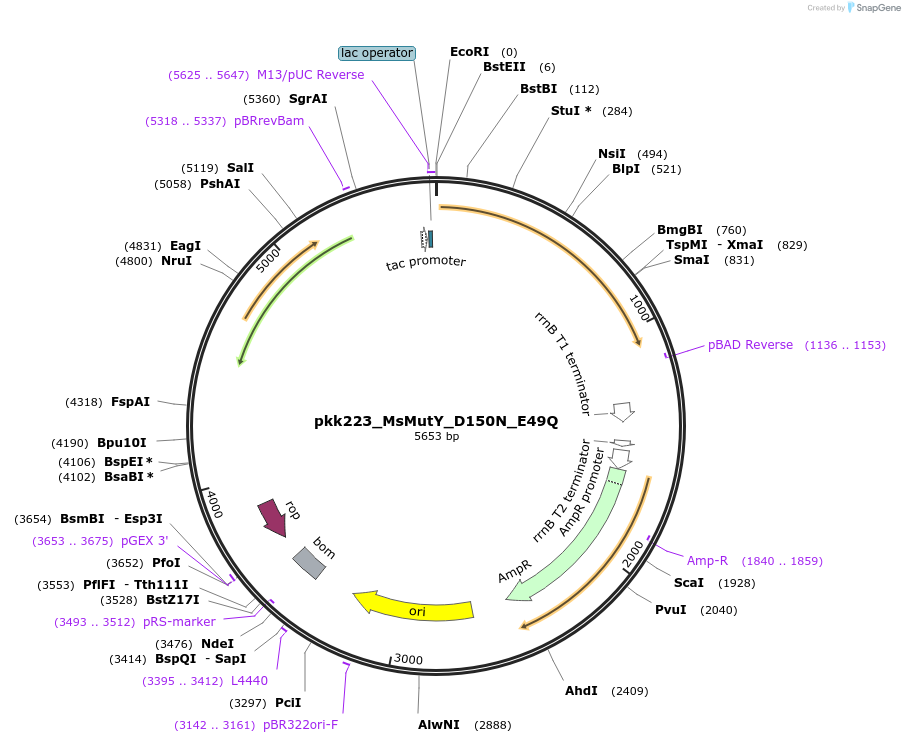
pkk223_MsMutY_D150N_E49Q
Plasmid#210794Purposepkk223 with Marinosulfonomonas MutY containing D150N and E49Q mutations to reduce catalytic activityDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationD150N_E49QAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
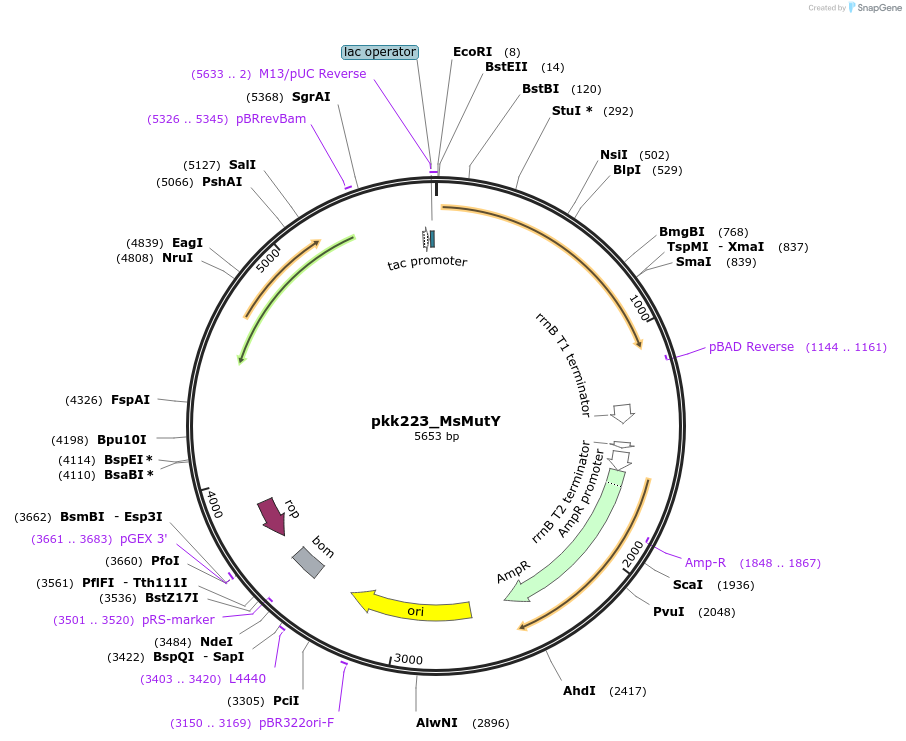
pkk223_MsMutY
Plasmid#210796Purposepkk223 with Marinosulfonomonas MutY insertDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
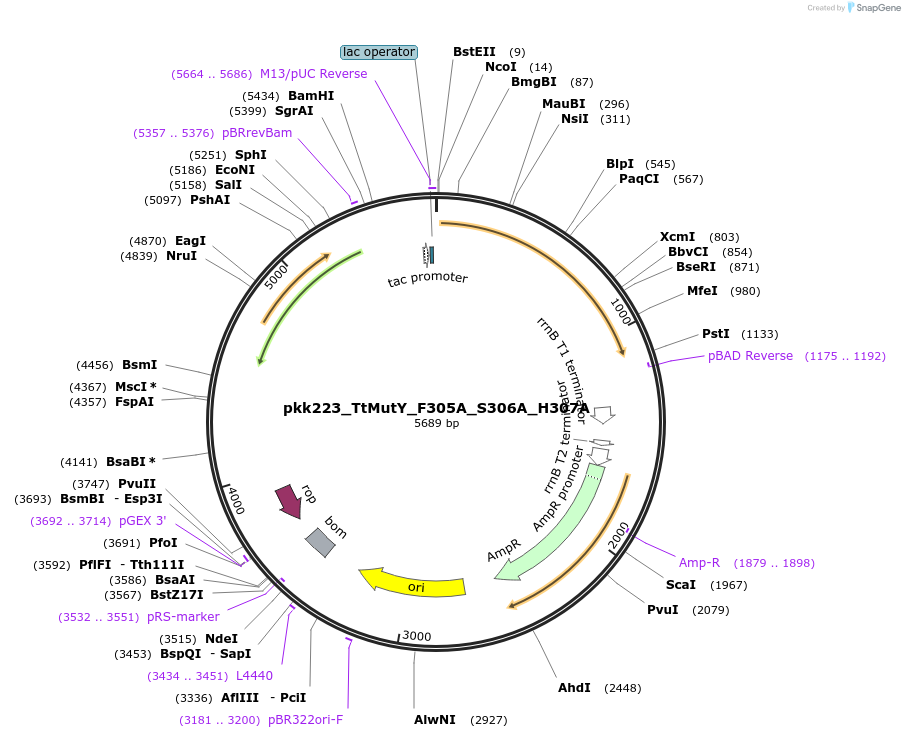
pkk223_TtMutY_F305A_S306A_H307A
Plasmid#210793Purposepkk223 with Thiotrichaceae MutY containing F305A, S306A, and H307A mutations to reduce recognition of OG:ADepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationF305A_S306A_H307AAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
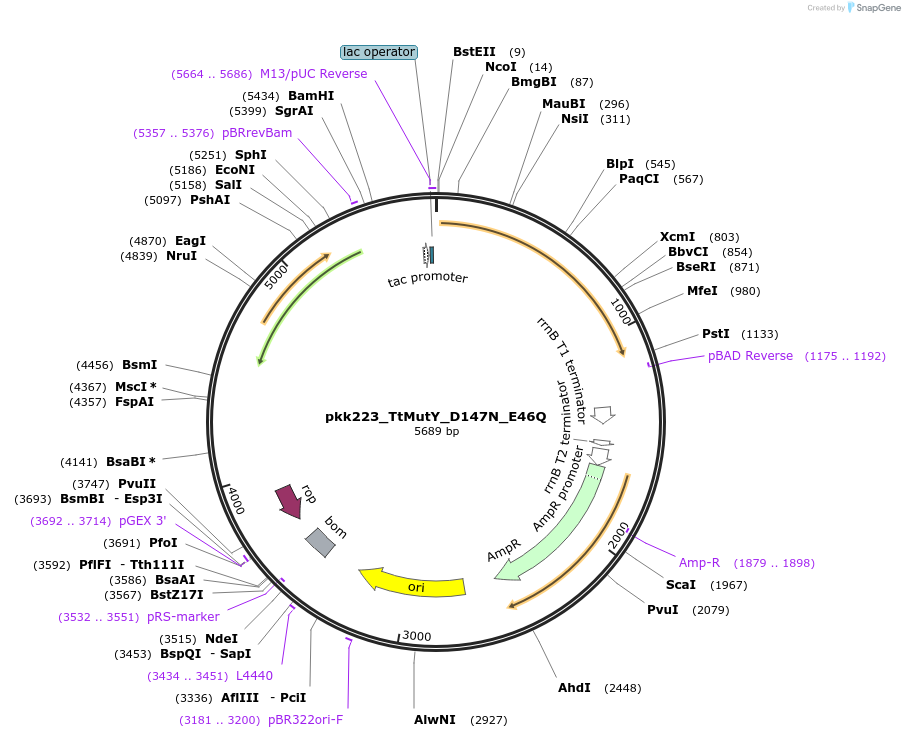
pkk223_TtMutY_D147N_E46Q
Plasmid#210792Purposepkk223 with Thiotrichaceae MutY containing D147N and E46Q mutations to reduce catalytic activityDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialMutationD147N_E46QAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
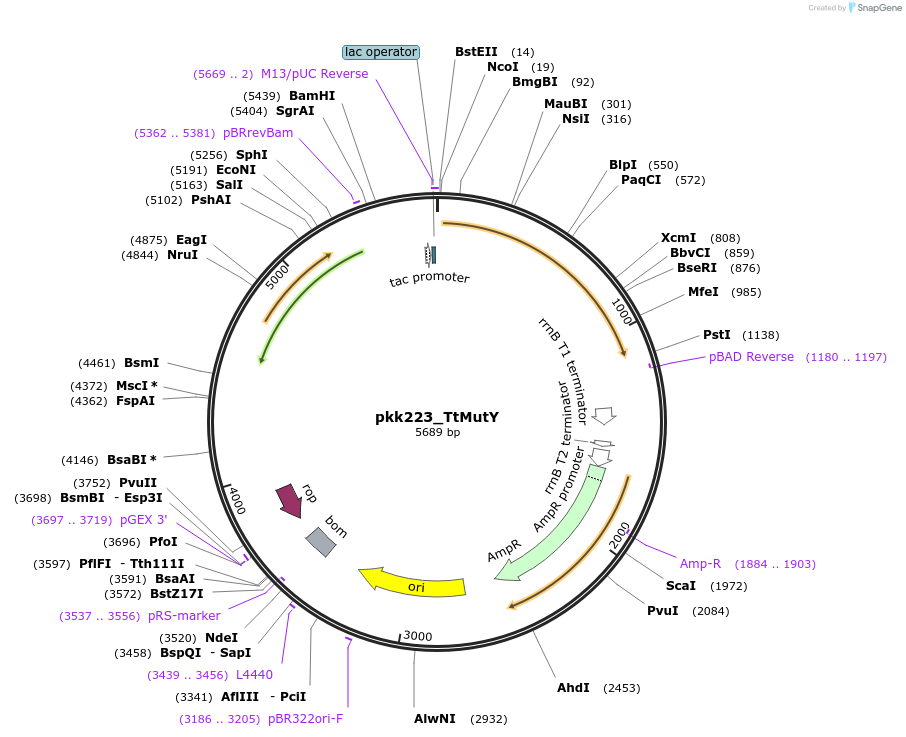
pkk223_TtMutY
Plasmid#210791Purposepkk223 with Thiotrichaceae MutY insertDepositorInsertMutY (adenine glycosylase)
ExpressionBacterialAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
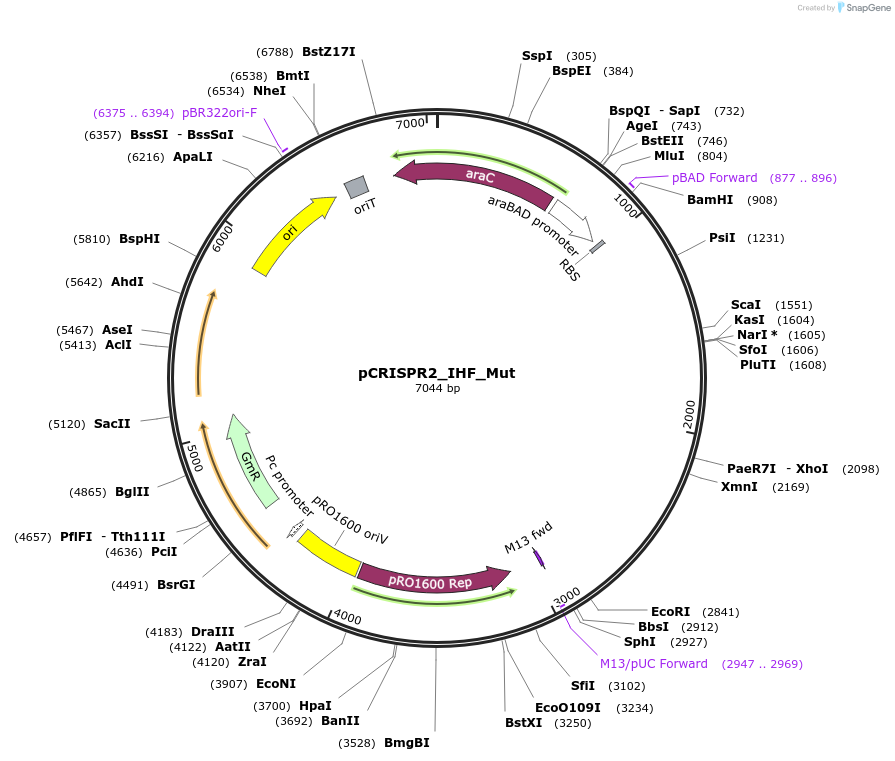
pCRISPR2_IHF_Mut
Plasmid#149396PurposeP. aeruginosa PA14 CRISPR2 locus, with a mutated IHFprox site, that should not be recognized by IHFDepositorInsertPseudomonas aeruginosa PA14 CRISPR2 locus
ExpressionBacterialMutationIHFprox site is mutated at key positions needed f…PromoterpBADAvailable SinceAug. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
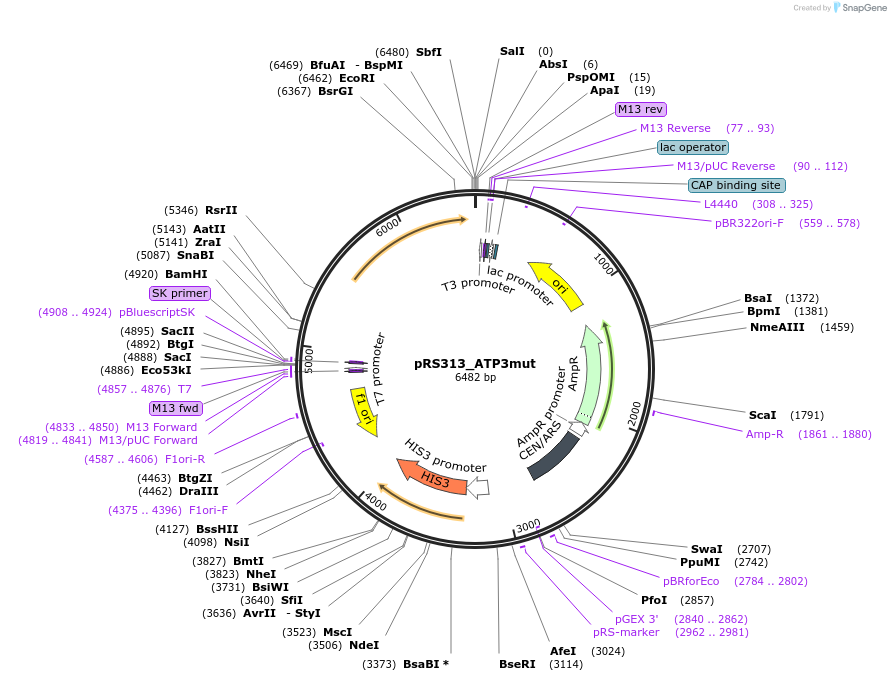
pRS313_ATP3mut
Plasmid#120256PurposeExpression of ATP3-67 in yeastDepositorInsertATP3 (ATP3 Budding Yeast)
ExpressionYeastMutationIsoleucine 304 changed to Asparagine; Alanine 307…PromoterATP3Available SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only