We narrowed to 20,535 results for: Kin
-
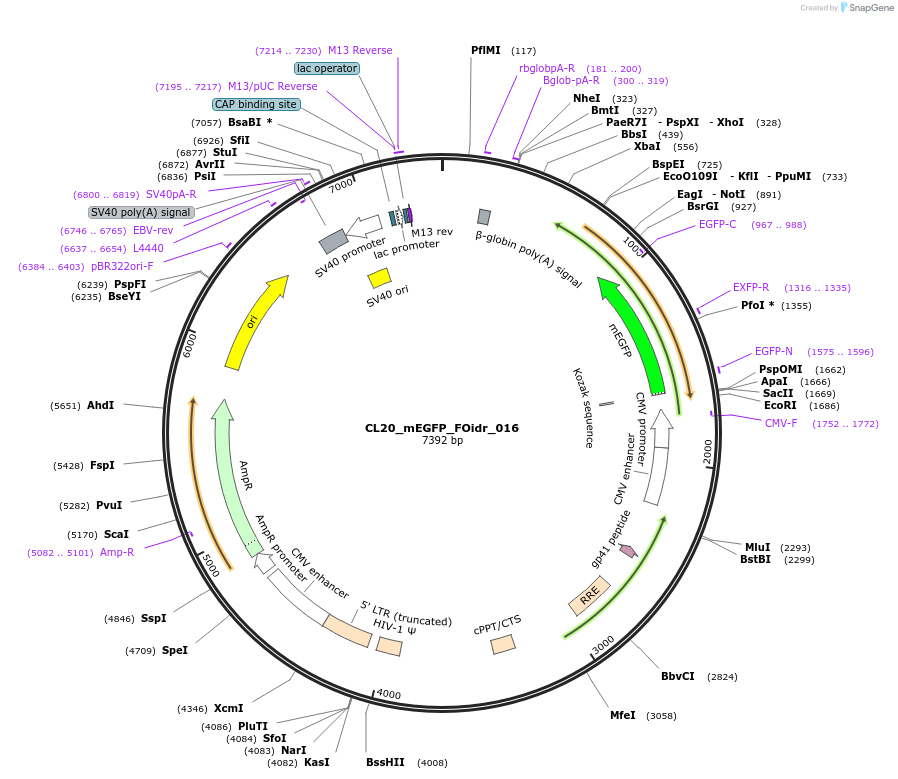
Plasmid#244568PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), FOidr_016, derived from human fusion protein ATP5B_VWA3BDepositorInsertFOidr_016
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
CL20_mEGFP_FOidr_013
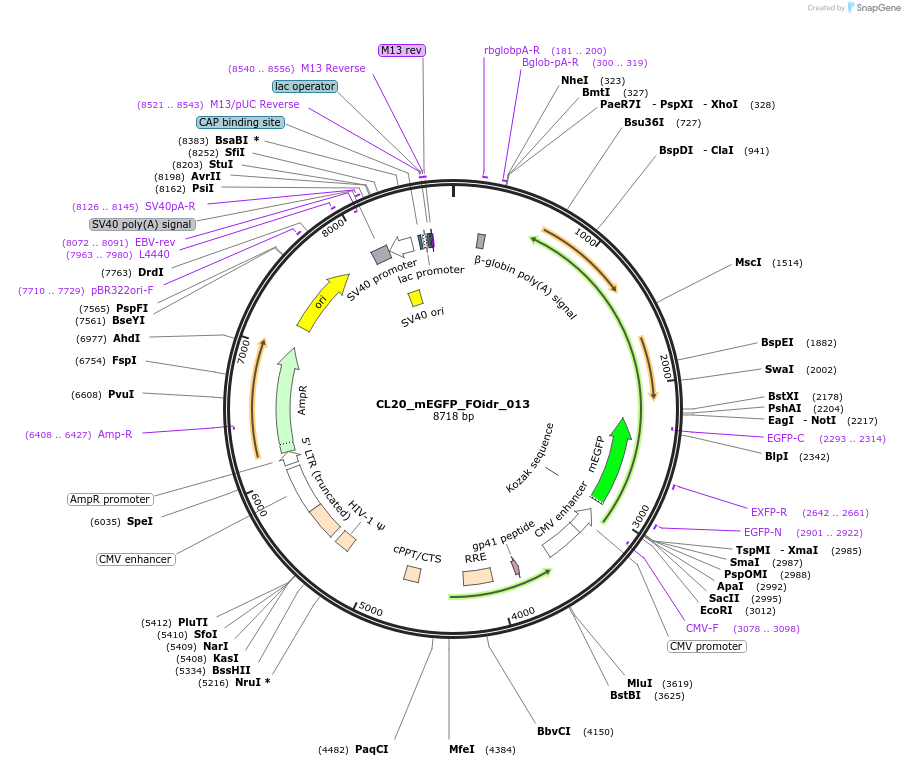
Plasmid#244565PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), FOidr_013, derived from human fusion protein ATF7IP_JAK2DepositorInsertFOidr_013
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
CL20_mEGFP_FOidr_001
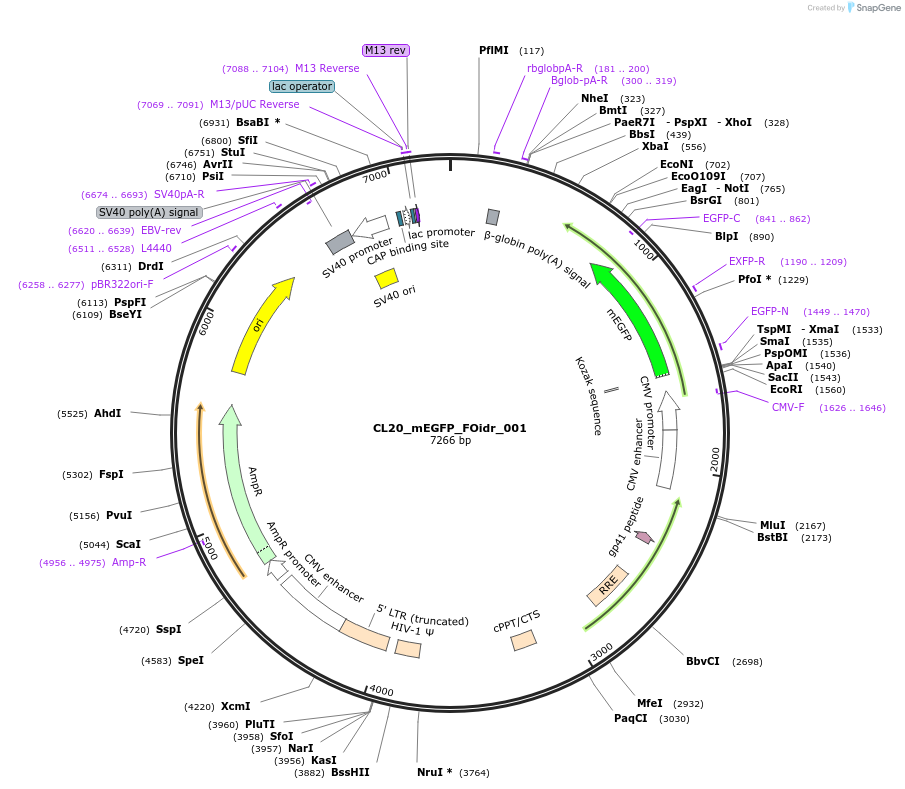
Plasmid#244553PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), FOidr_001, derived from human fusion protein ABR_DUXADepositorInsertFOidr_001
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
hPSD-95_PDZ3
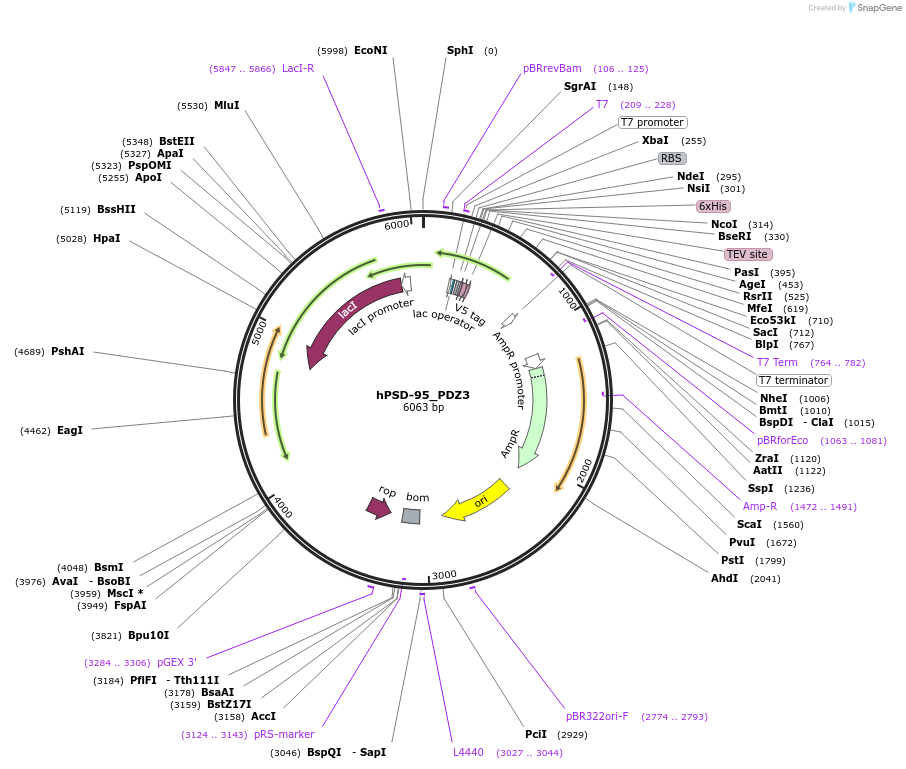
Plasmid#245905PurposeBacterial expression of PSD-95 PDZ3 domain (302-402)DepositorAvailable SinceOct. 23, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
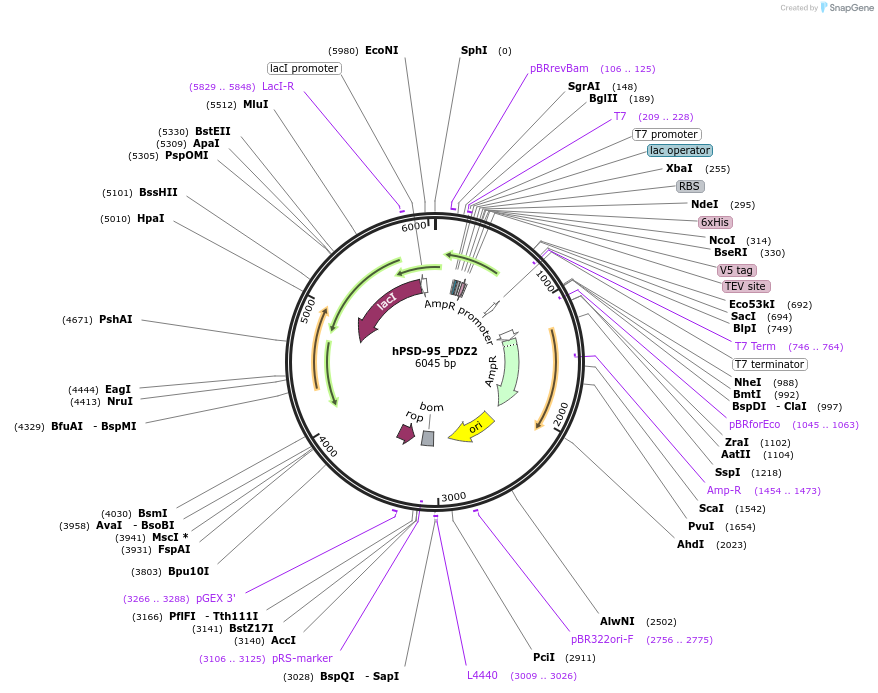
hPSD-95_PDZ2
Plasmid#245904PurposeBacterial expression of PSD-95 PDZ2 domain (155-249)DepositorAvailable SinceOct. 23, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
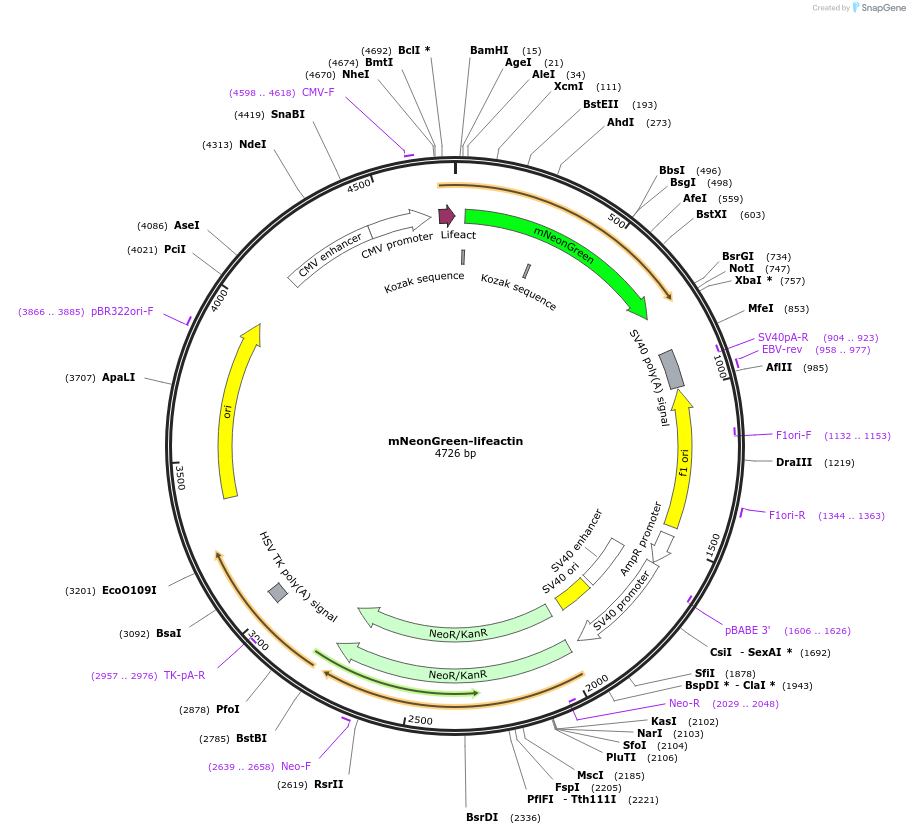
mNeonGreen-lifeactin
Plasmid#229607PurposeExpresses mNeonGreen-lifeactin fusion in mammalian cellsDepositorInsertmNeonGreen
ExpressionMammalianAvailable SinceOct. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
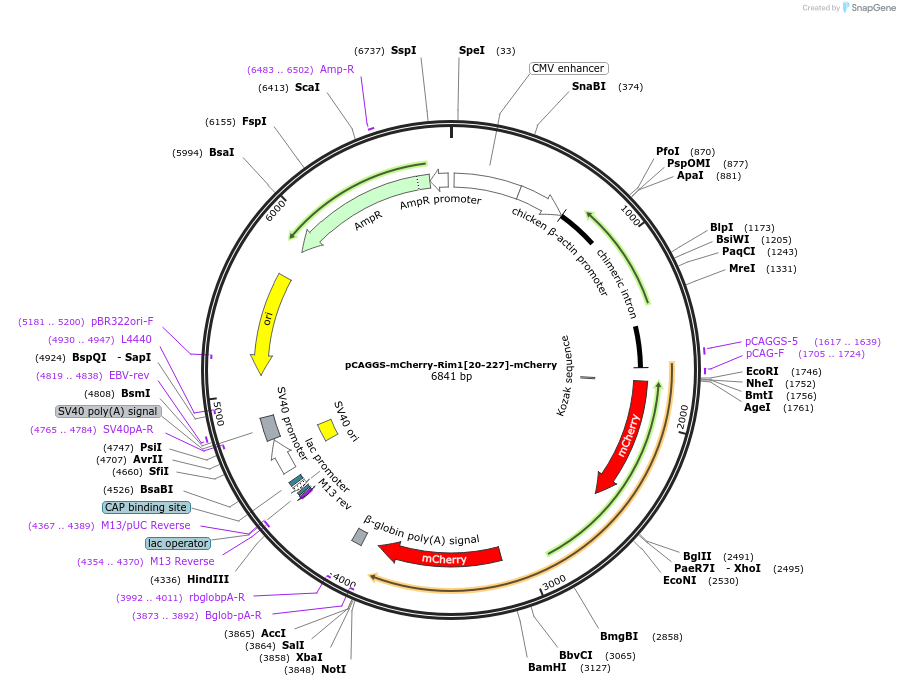
pCAGGS-mCherry-Rim1[20-227]-mCherry
Plasmid#246282PurposeRab10 sensor acceptorDepositorAvailable SinceOct. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
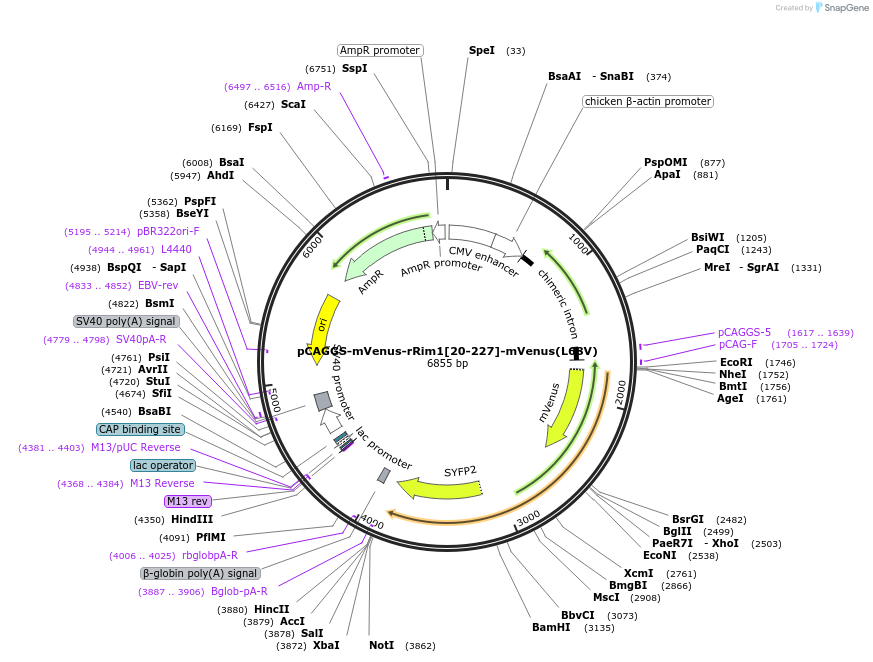
pCAGGS-mVenus-rRim1[20-227]-mVenus(L68V)
Plasmid#246280PurposeRab10 sensor acceptorDepositorAvailable SinceOct. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
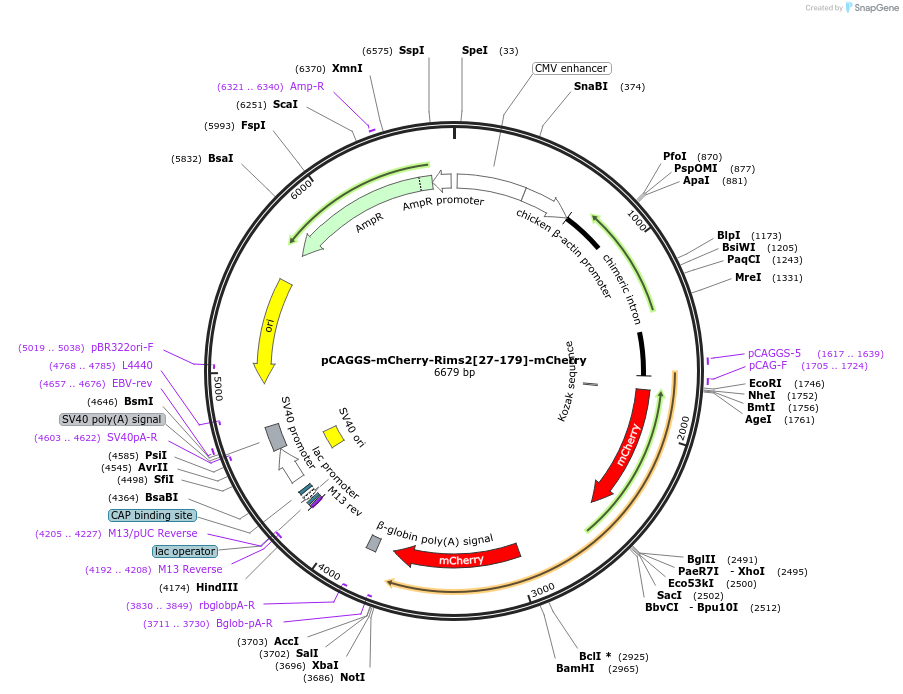
pCAGGS-mCherry-Rims2[27-179]-mCherry
Plasmid#246271PurposeRab8 sensor acceptorDepositorAvailable SinceOct. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
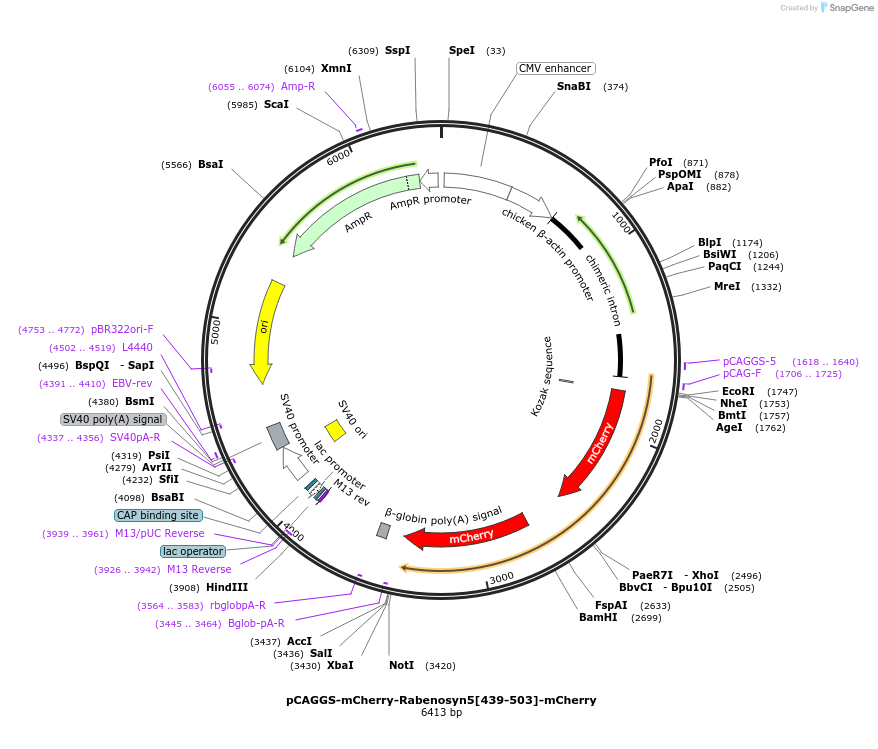
pCAGGS-mCherry-Rabenosyn5[439-503]-mCherry
Plasmid#246251PurposeRab4 sensor acceptorDepositorAvailable SinceOct. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
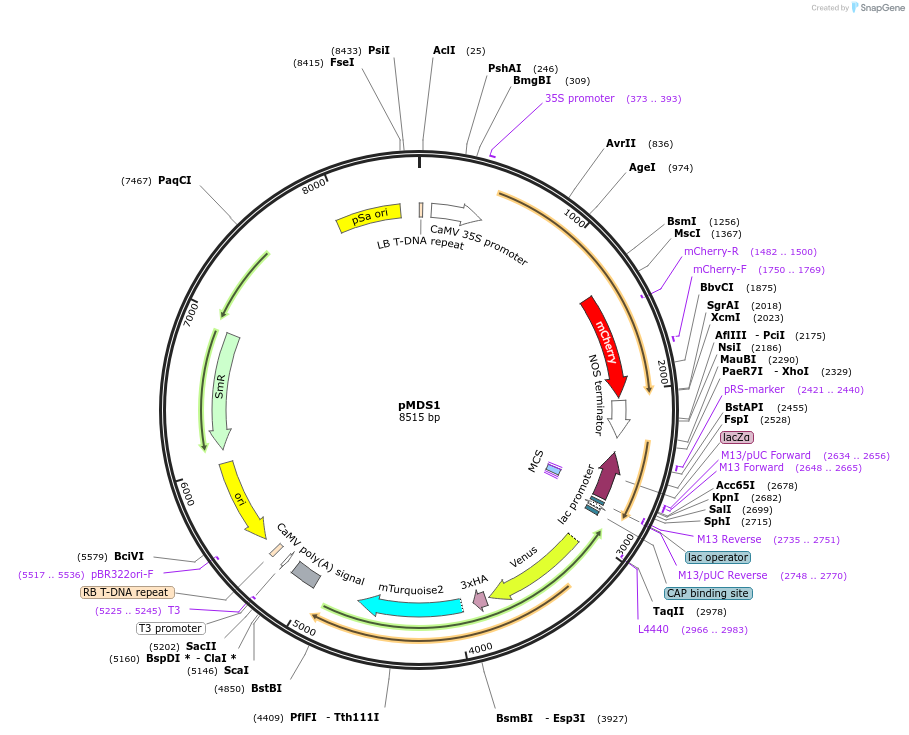
pMDS1
Plasmid#239460PurposeFor simultaneous monitoring of both transcription and translation of genes in plant tissues: C-terminus fusionDepositorTypeEmpty backboneTagsVENUS:3xHA:2A:mTurqN7ExpressionBacterial and PlantAvailable SinceOct. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
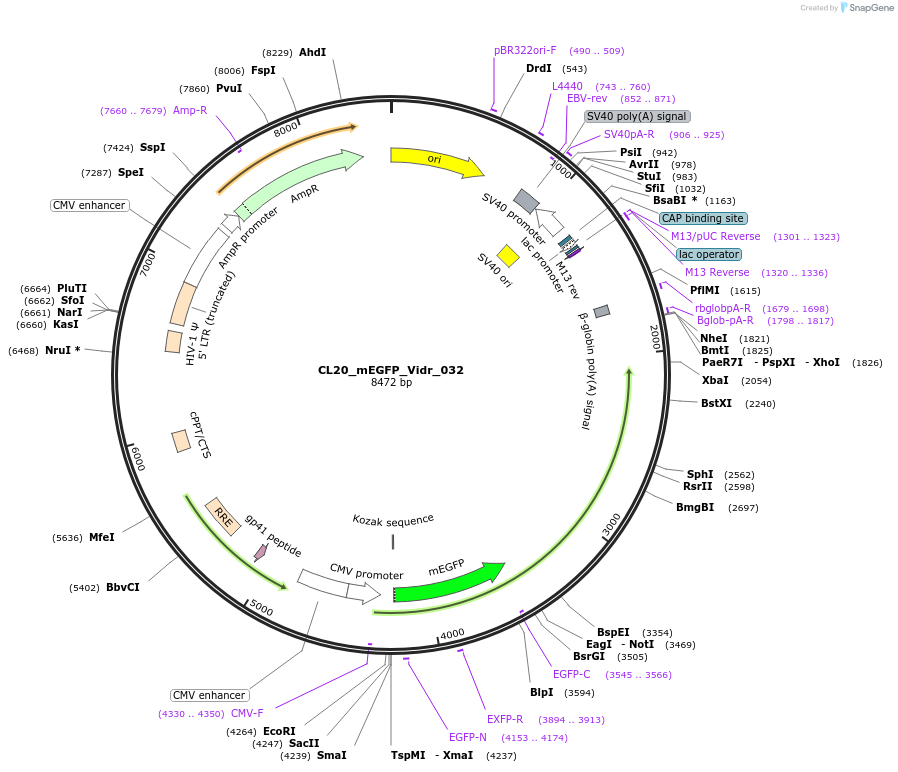
CL20_mEGFP_Vidr_032
Plasmid#244799PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_032, derived from human protein F90AEDepositorInsertVidr_032
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
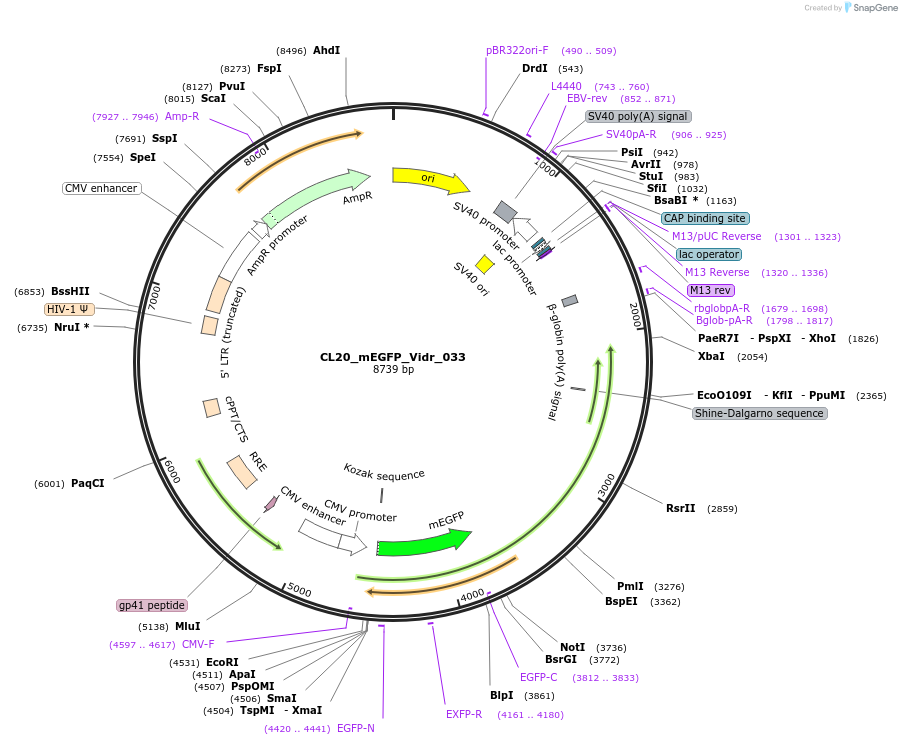
CL20_mEGFP_Vidr_033
Plasmid#244800PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_033, derived from human protein IRS4DepositorInsertVidr_033
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
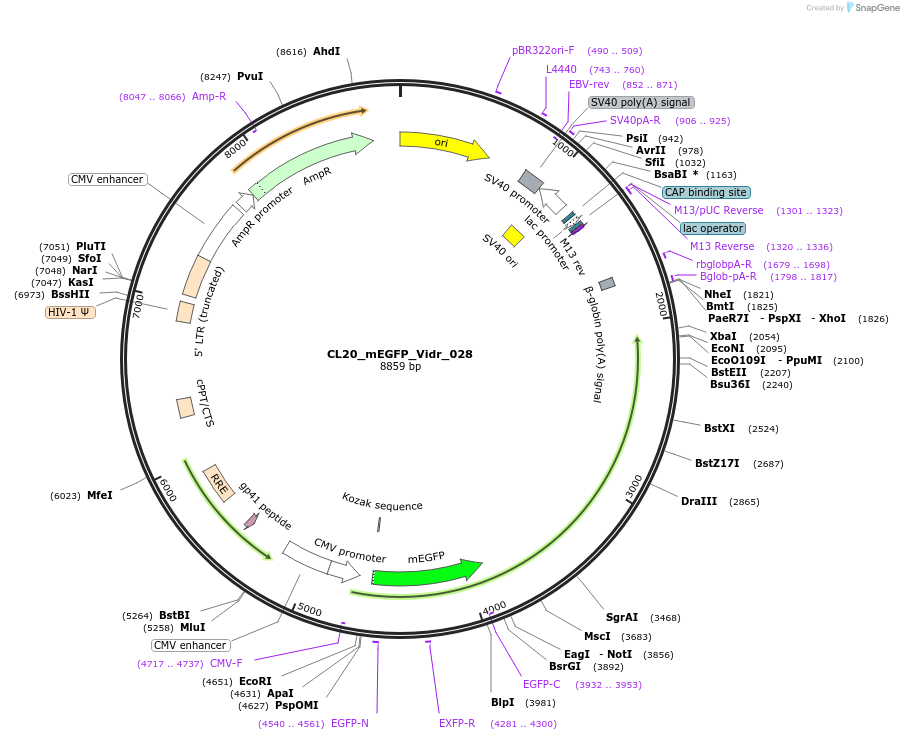
CL20_mEGFP_Vidr_028
Plasmid#244795PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_028, derived from human protein R3HD2DepositorInsertVidr_028
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
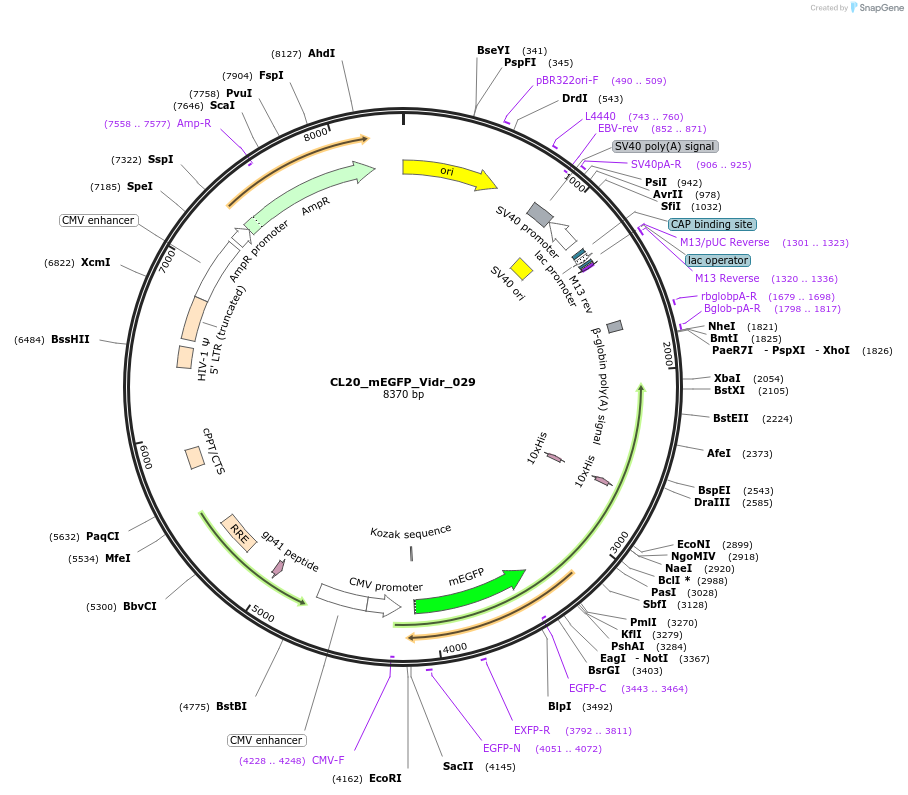
CL20_mEGFP_Vidr_029
Plasmid#244796PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_029, derived from human protein DLGP3DepositorInsertVidr_029
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
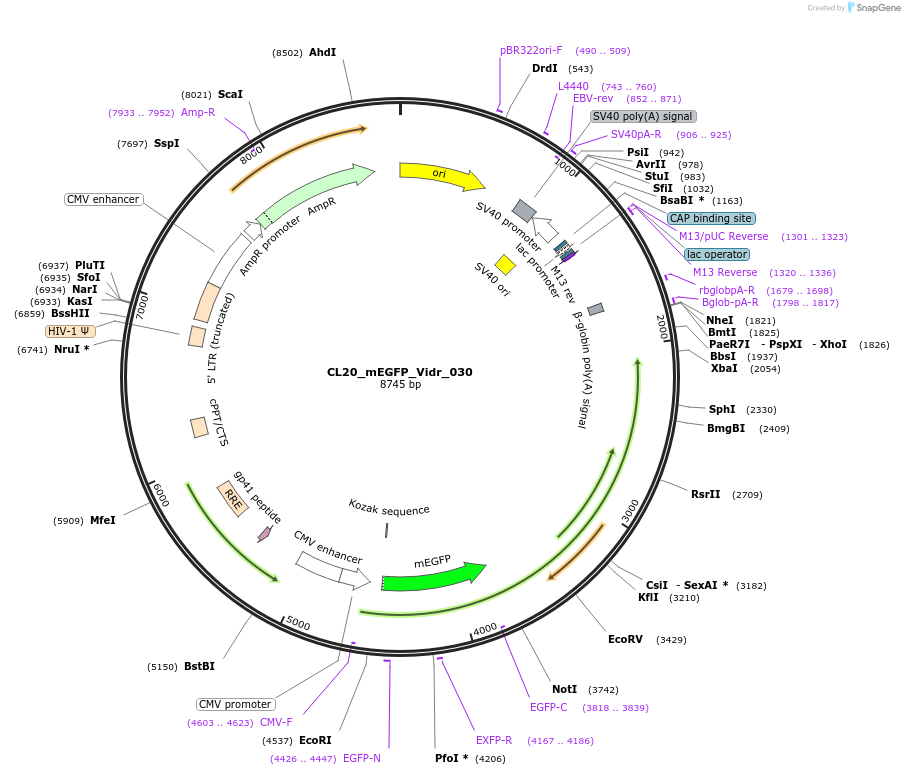
CL20_mEGFP_Vidr_030
Plasmid#244797PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_030, derived from human protein ARI1ADepositorInsertVidr_030
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
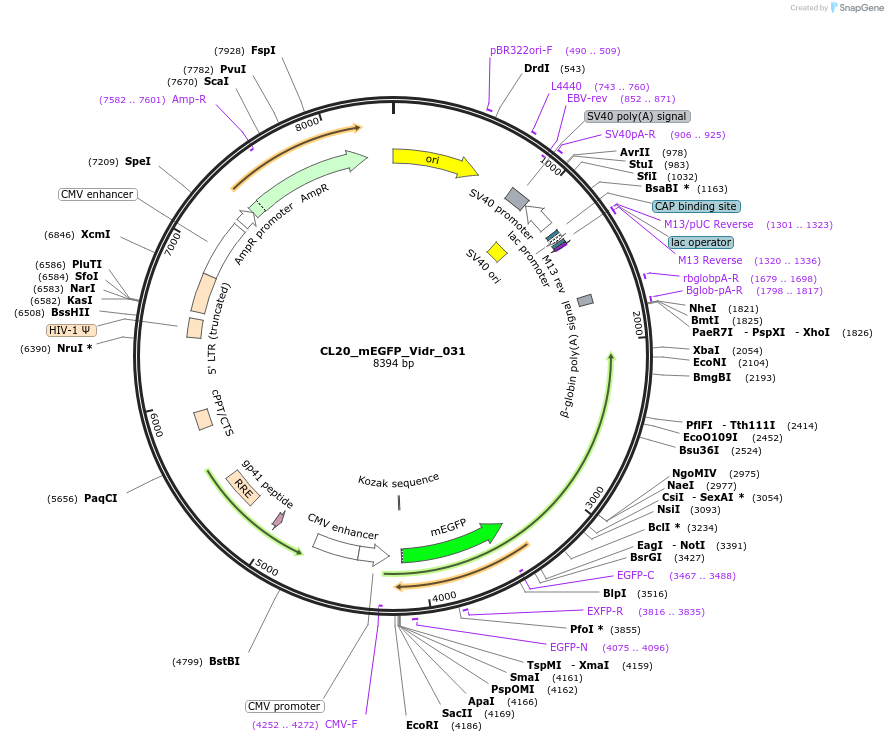
CL20_mEGFP_Vidr_031
Plasmid#244798PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_031, derived from human protein MD12LDepositorInsertVidr_031
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
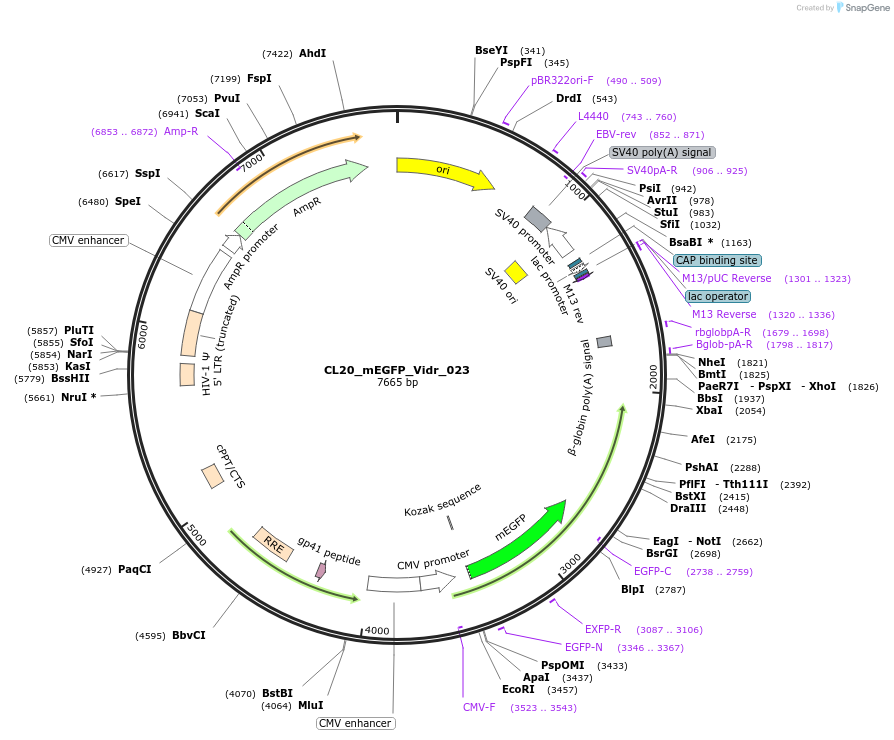
CL20_mEGFP_Vidr_023
Plasmid#244790PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_023, derived from human protein SPOC1DepositorInsertVidr_023
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
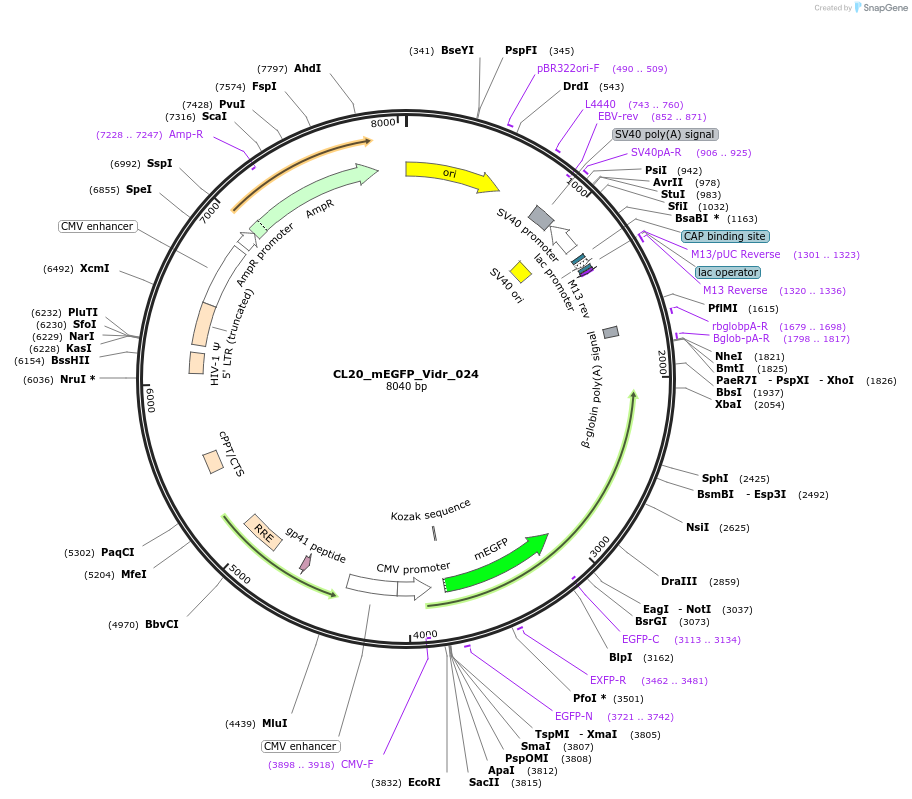
CL20_mEGFP_Vidr_024
Plasmid#244791PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_024, derived from human protein S31A1DepositorInsertVidr_024
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
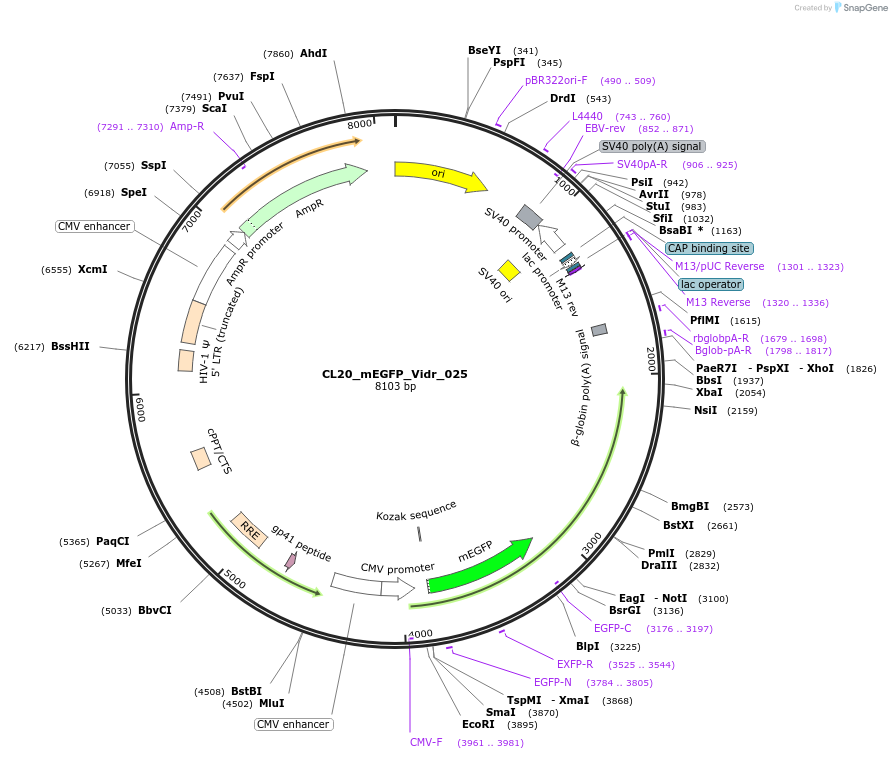
CL20_mEGFP_Vidr_025
Plasmid#244792PurposeExpress mEGFP-tagged intrinsically disordered protein (IDR), Vidr_025, derived from human protein NKAPDepositorInsertVidr_025
Tagsmonomeric EGFPExpressionMammalianMutationUnmutatedAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only