We narrowed to 11,082 results for: CHL
-
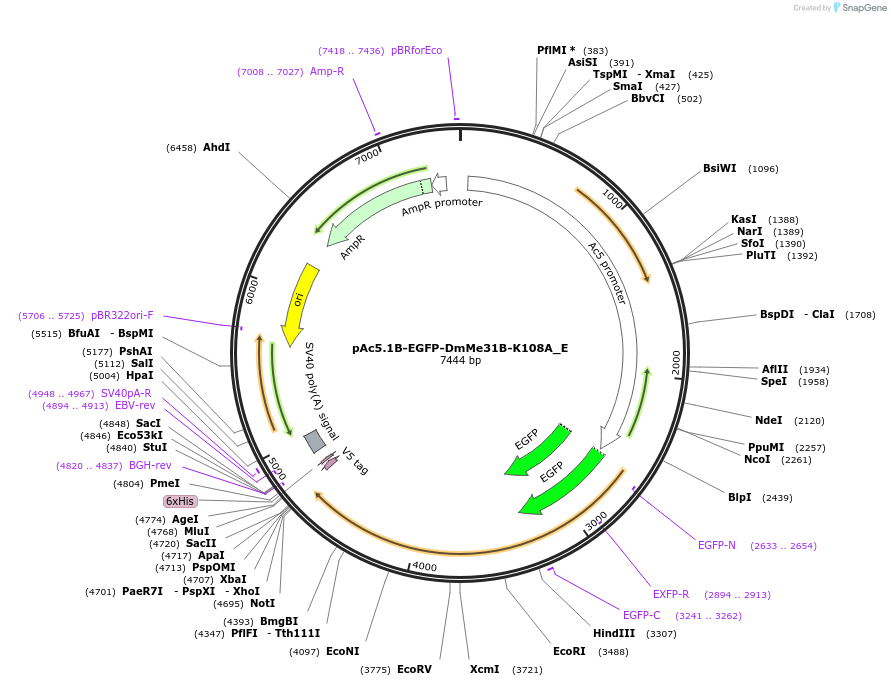
Plasmid#146197PurposeInsect Expression of DmMe31B-K108ADepositorInsertDmMe31B-K108A (me31B Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-EGFP-DmMe31B-mut1_E
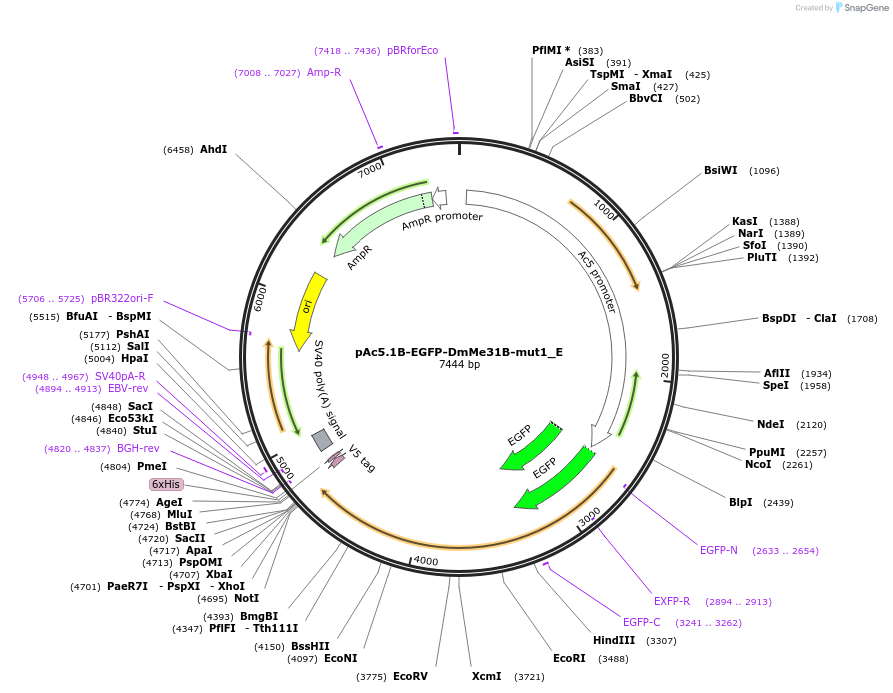
Plasmid#146198PurposeInsect Expression of DmMe31B-mut1DepositorInsertDmMe31B-mut1 (me31B Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmMe31B-E208Q_E
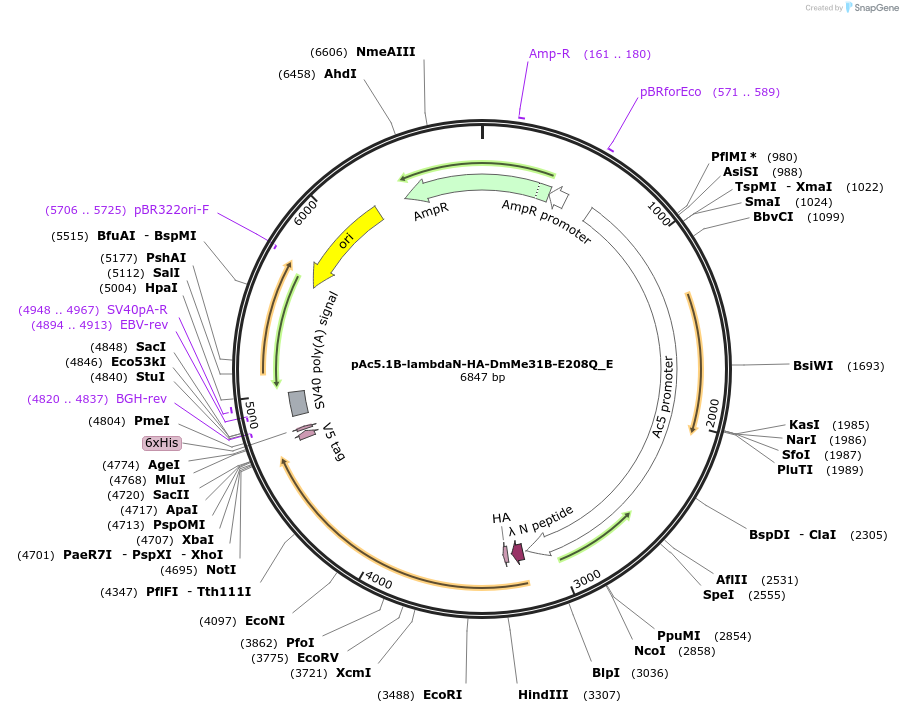
Plasmid#146201PurposeInsect Expression of DmMe31B-E208QDepositorInsertDmMe31B-E208Q (me31B Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAc5.1B-lambdaN-HA-DmMe31B-K108A_E
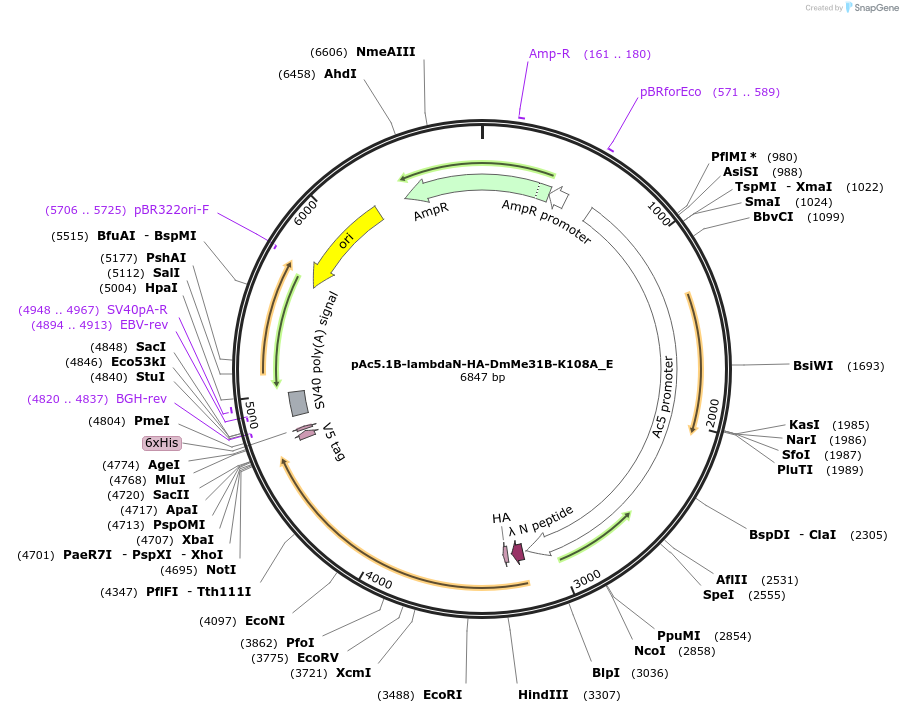
Plasmid#146202PurposeInsect Expression of DmMe31B-K108ADepositorInsertDmMe31B-K108A (me31B Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
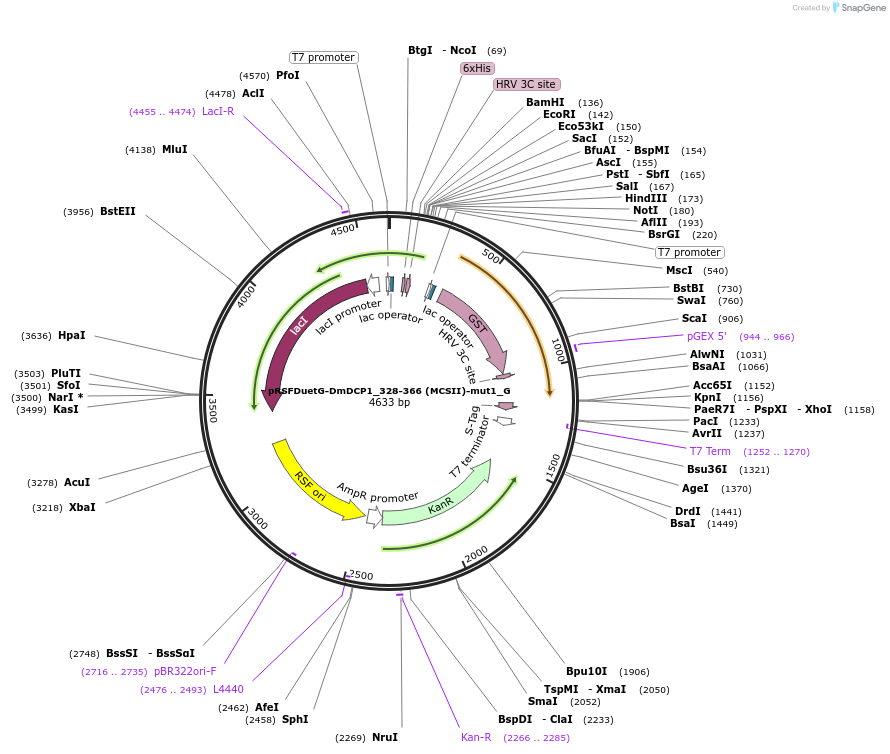
pRSFDuetG-DmDCP1_328-366 (MCSII)-mut1_G
Plasmid#146403PurposeBacterial Expression of DmDCP1_328-366-mut1DepositorInsertDmDCP1_328-366-mut1 (Dcp1 Fly)
ExpressionBacterialAvailable SinceAug. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
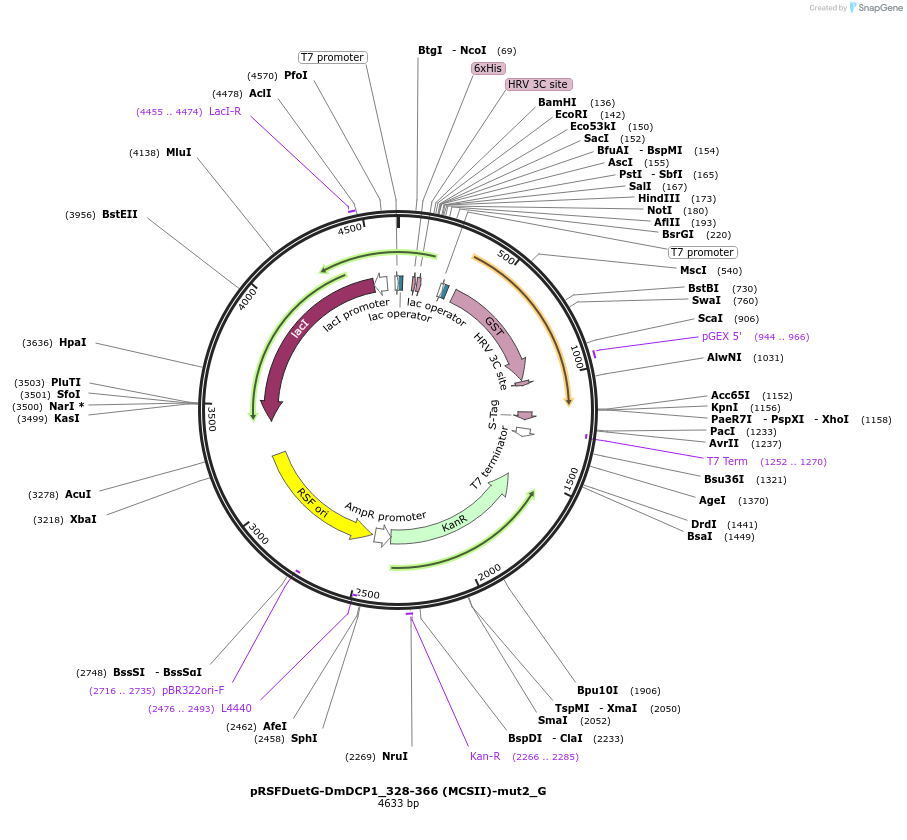
pRSFDuetG-DmDCP1_328-366 (MCSII)-mut2_G
Plasmid#146404PurposeBacterial Expression of DmDCP1_328-366-mut2DepositorInsertDmDCP1_328-366-mut2 (Dcp1 Fly)
ExpressionBacterialAvailable SinceAug. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
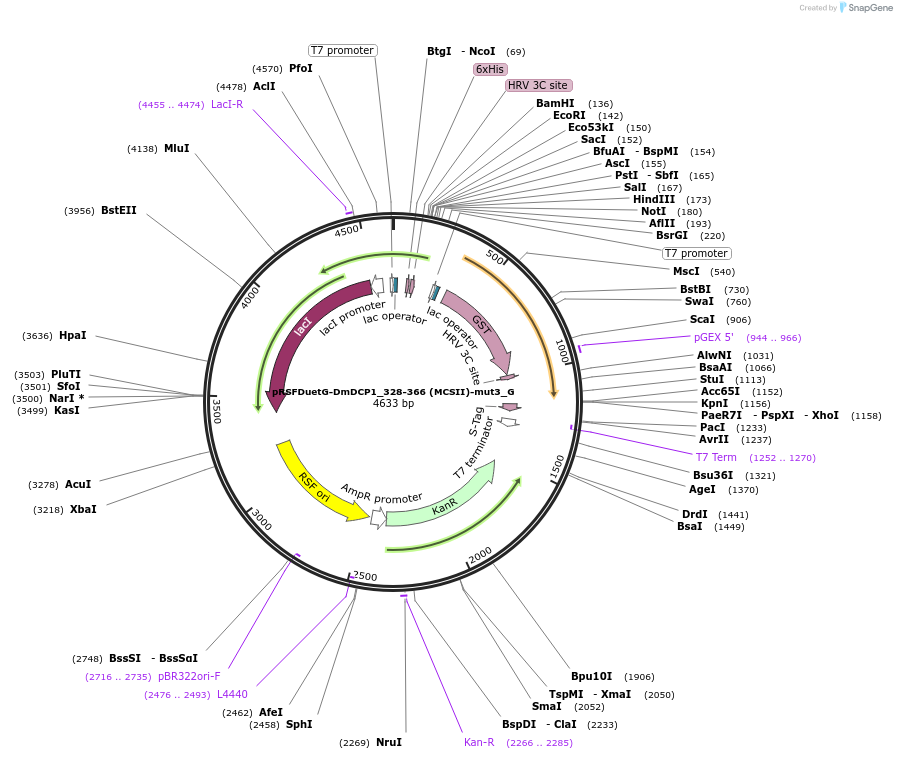
pRSFDuetG-DmDCP1_328-366 (MCSII)-mut3_G
Plasmid#146405PurposeBacterial Expression of DmDCP1_328-366-mut3DepositorInsertDmDCP1_328-366-mut3 (Dcp1 Fly)
ExpressionBacterialAvailable SinceAug. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
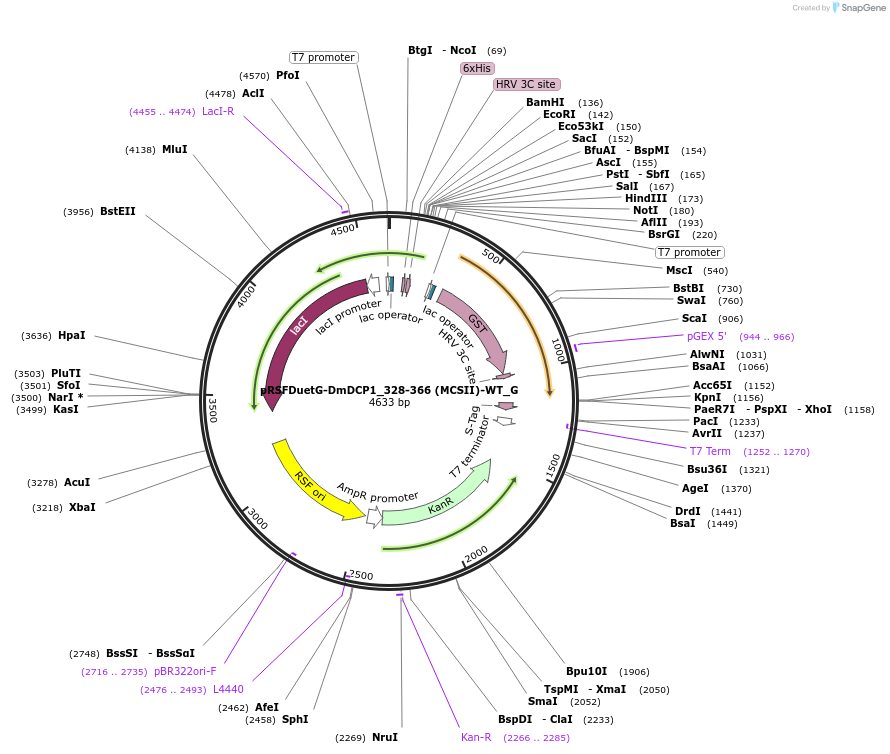
pRSFDuetG-DmDCP1_328-366 (MCSII)-WT_G
Plasmid#146406PurposeBacterial Expression of DmDCP1_328-366DepositorInsertDmDCP1_328-366 (Dcp1 Fly)
ExpressionBacterialAvailable SinceAug. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
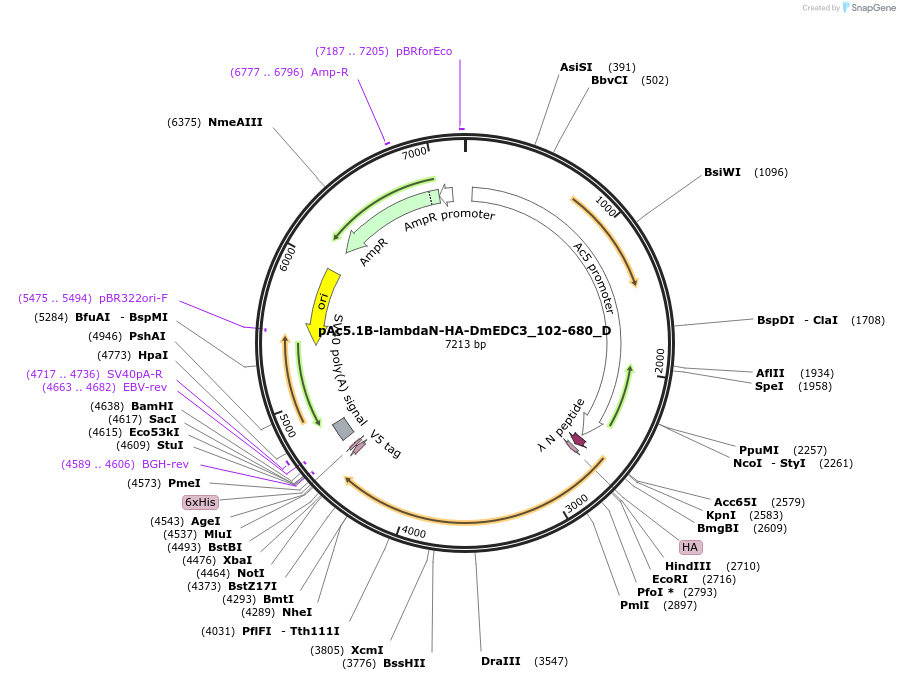
pAc5.1B-lambdaN-HA-DmEDC3_102-680_D
Plasmid#146124PurposeInsect Expression of DmEDC3_102-680DepositorInsertDmEDC3_102-680 (Edc3 Fly)
ExpressionInsectMutationone silent mutation A456G and three mutations N16…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
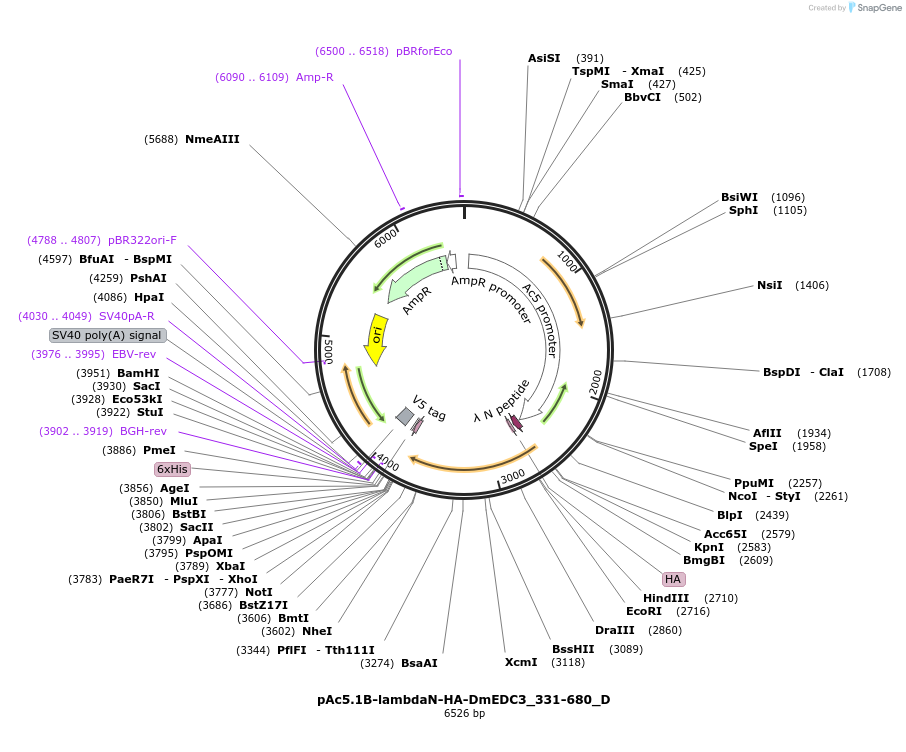
pAc5.1B-lambdaN-HA-DmEDC3_331-680_D
Plasmid#146125PurposeInsect Expression of DmEDC3_331-680DepositorInsertDmEDC3_331-680 (Edc3 Fly)
ExpressionInsectMutationone silent mutation A456G and three mutations N16…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
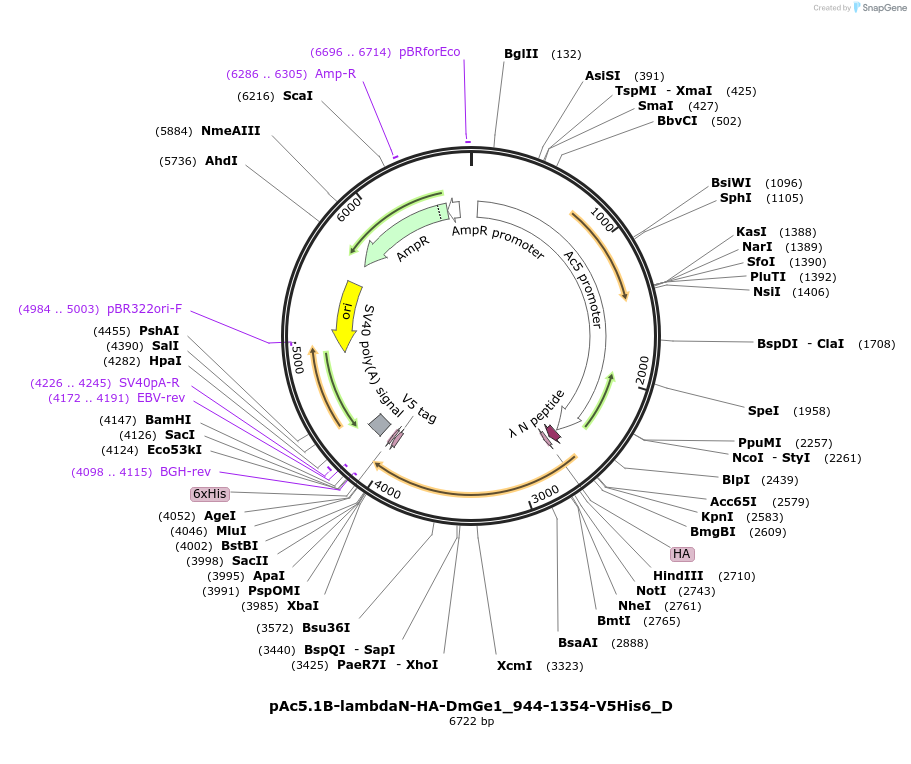
pAc5.1B-lambdaN-HA-DmGe1_944-1354-V5His6_D
Plasmid#146136PurposeInsect Expression of DmGe1_944-1354DepositorInsertDmGe1_944-1354 (Ge-1 Fly)
ExpressionInsectAvailable SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
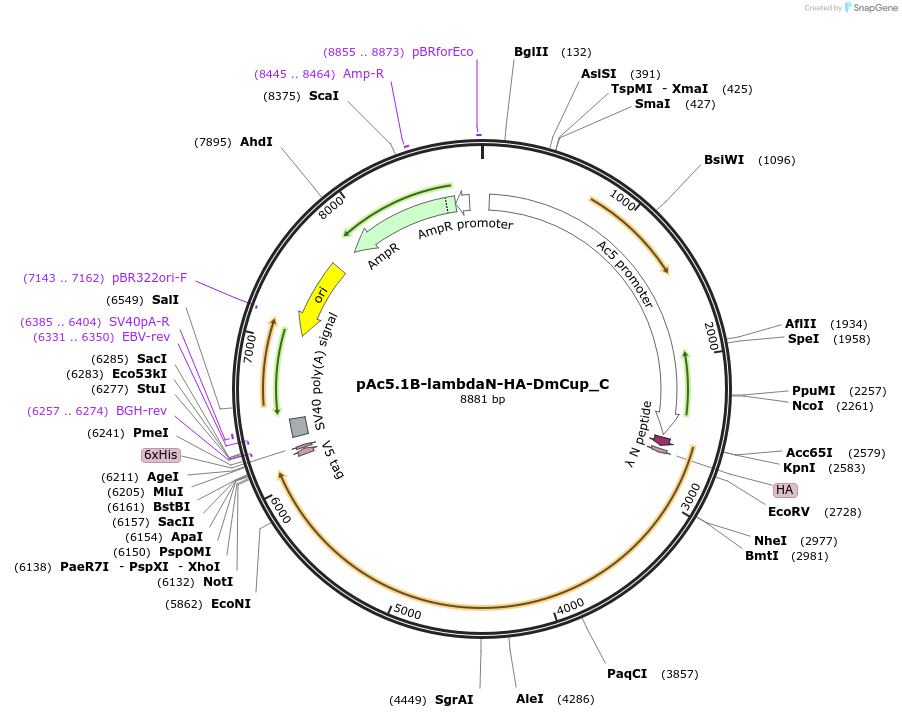
pAc5.1B-lambdaN-HA-DmCup_C
Plasmid#146064PurposeInsect Expression of DmCupDepositorInsertDmCup (cup Fly)
ExpressionInsectMutationTwo silent mutations compared to the sequence giv…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
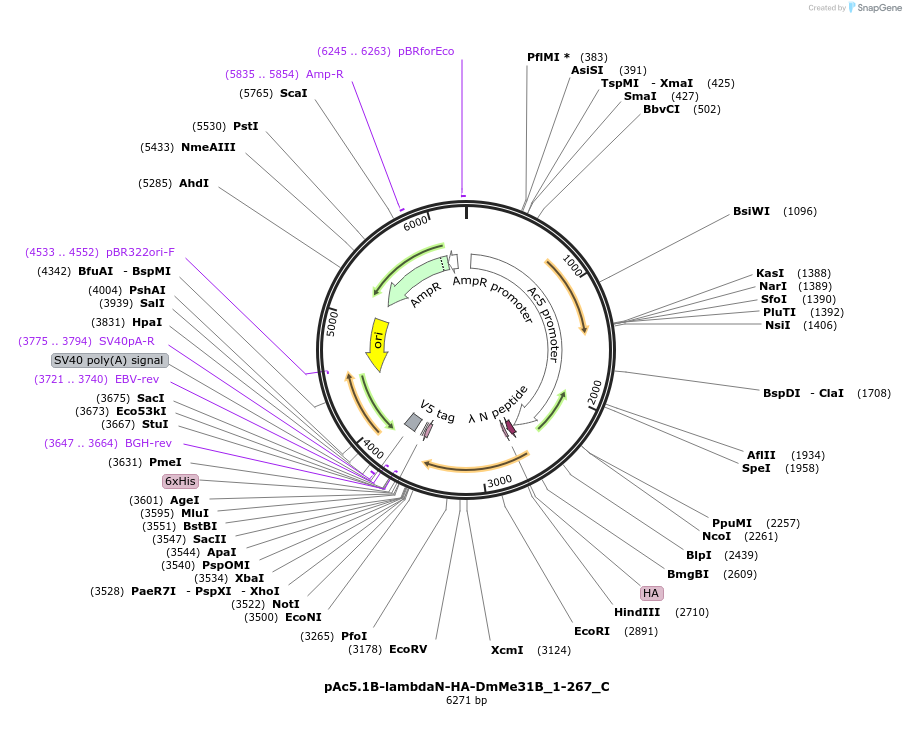
pAc5.1B-lambdaN-HA-DmMe31B_1-267_C
Plasmid#146065PurposeInsect Expression of DmMe31B_1-267DepositorInsertDmMe31B_1-267 (me31B Fly)
ExpressionInsectAvailable SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
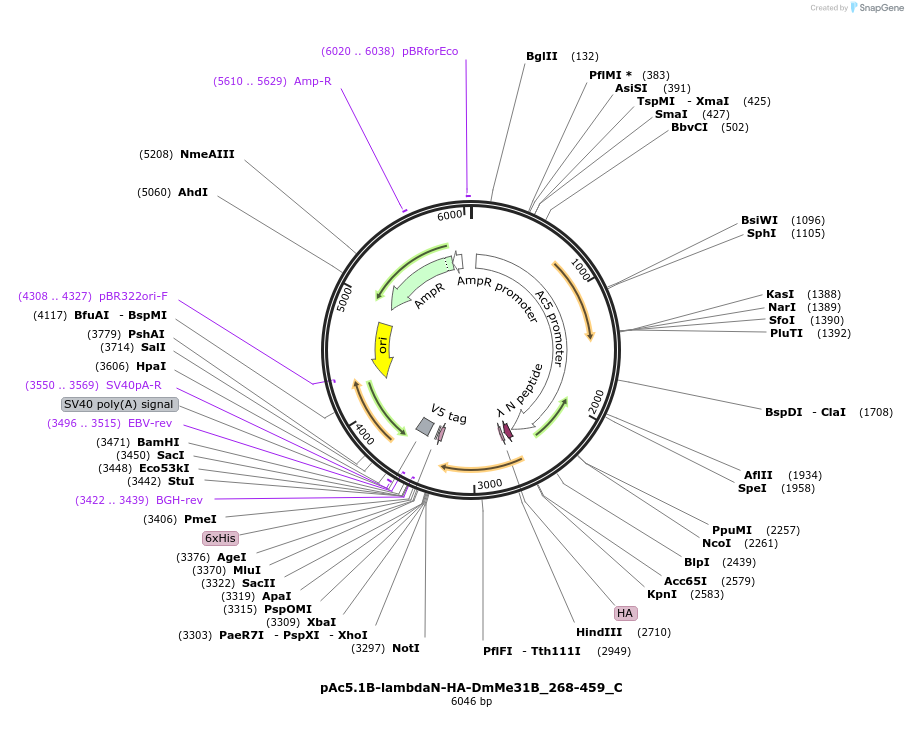
pAc5.1B-lambdaN-HA-DmMe31B_268-459_C
Plasmid#146066PurposeInsect Expression of DmMe31B_268-459DepositorInsertDmMe31B_268-459 (me31B Fly)
ExpressionInsectAvailable SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
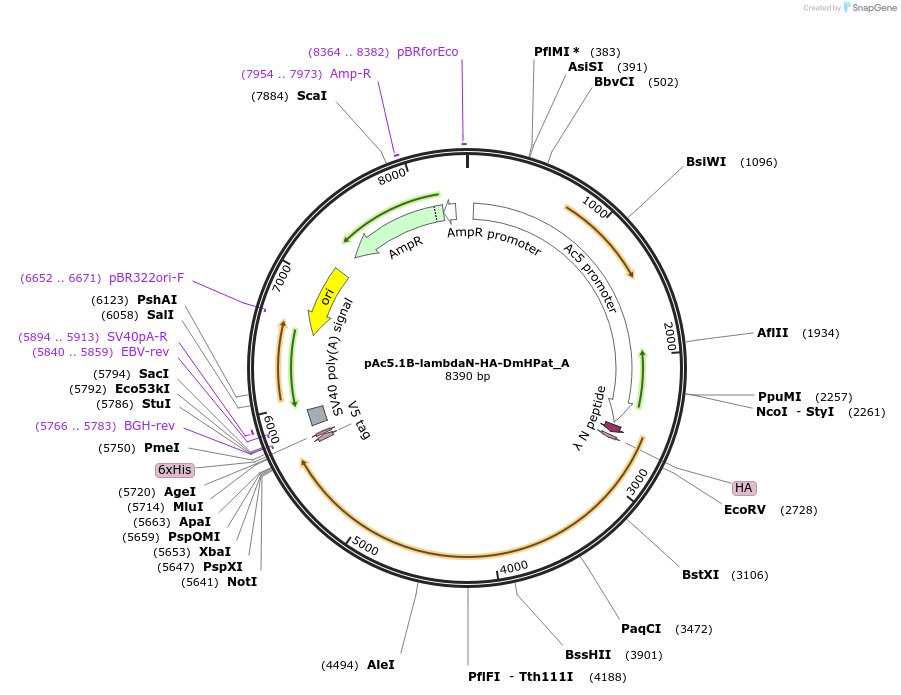
pAc5.1B-lambdaN-HA-DmHPat_A
Plasmid#145893PurposeInsect Expression of DmHPatDepositorInsertDmHPat (Patr-1 Fly)
ExpressionInsectMutationthree silent mutations compared to the sequence g…Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
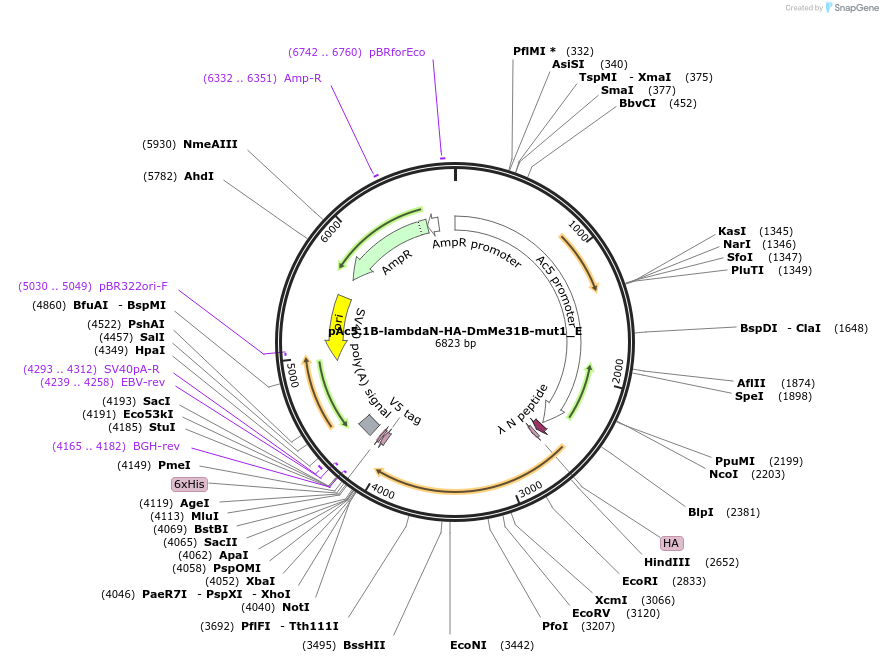
pAc5.1B-lambdaN-HA-DmMe31B-mut1_E
Plasmid#146203PurposeInsect Expression of DmMe31B-mut1DepositorInsertDmMe31B-mut1 (me31B Fly)
ExpressionInsectAvailable SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
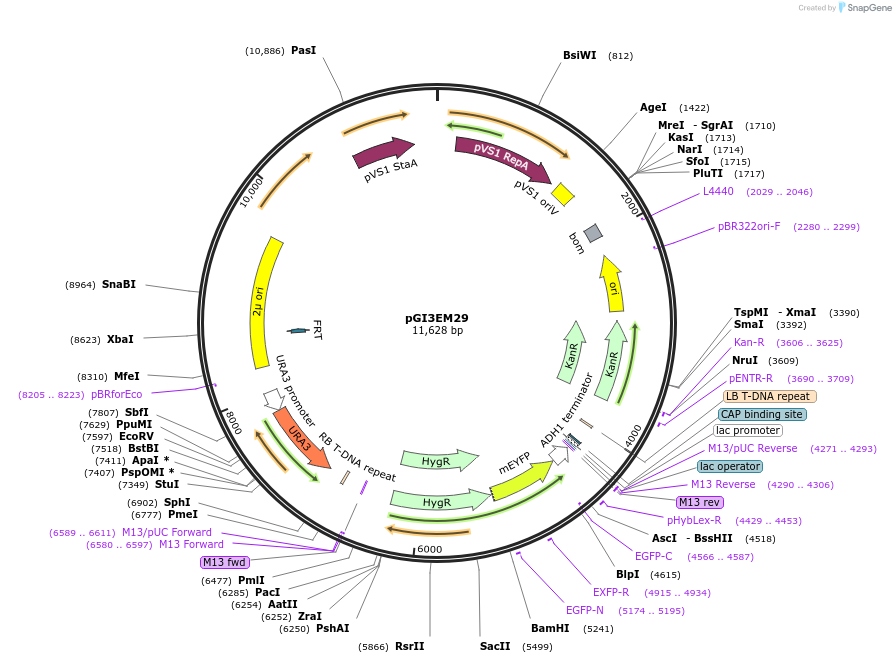
pGI3EM29
Plasmid#135489PurposeAgrobacterium T-DNA with Spun H2Bpr driving hph-mCitrineDepositorInserthph SpunH2A/Bpr hph-mCitrine
TagsmCitrineExpressionBacterial and YeastPromoterSpunH2A/Bpr (divergent)Available SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
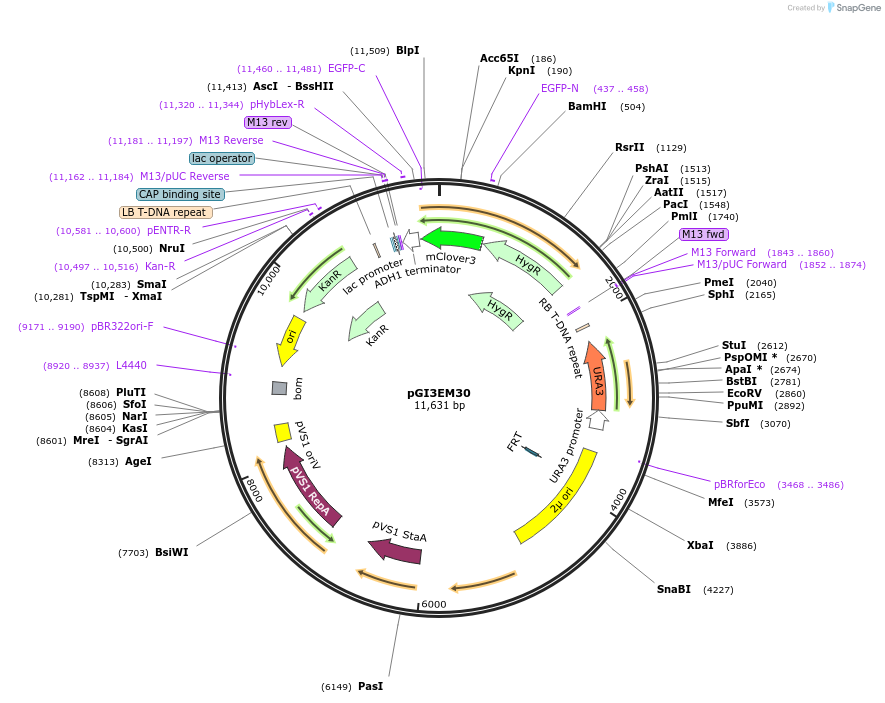
pGI3EM30
Plasmid#135490PurposeAgrobacterium T-DNA with Spun H2Bpr driving hph-mClover3DepositorInserthph SpunH2A/Bpr hph-mClover3
TagsmClover3ExpressionBacterial and YeastPromoterSpunH2A/Bpr (divergent)Available SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
FUP95Q15AGW (D5)
Plasmid#74028Purposelentiviral expression of Psd95-Q15A-EGFP fusionDepositorAvailable SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
FUP95E17RGW (D6)
Plasmid#74029Purposelentiviral expression of Psd95-E17R-EGFP fusionDepositorAvailable SinceMarch 31, 2016AvailabilityAcademic Institutions and Nonprofits only