We narrowed to 12,767 results for: ache
-
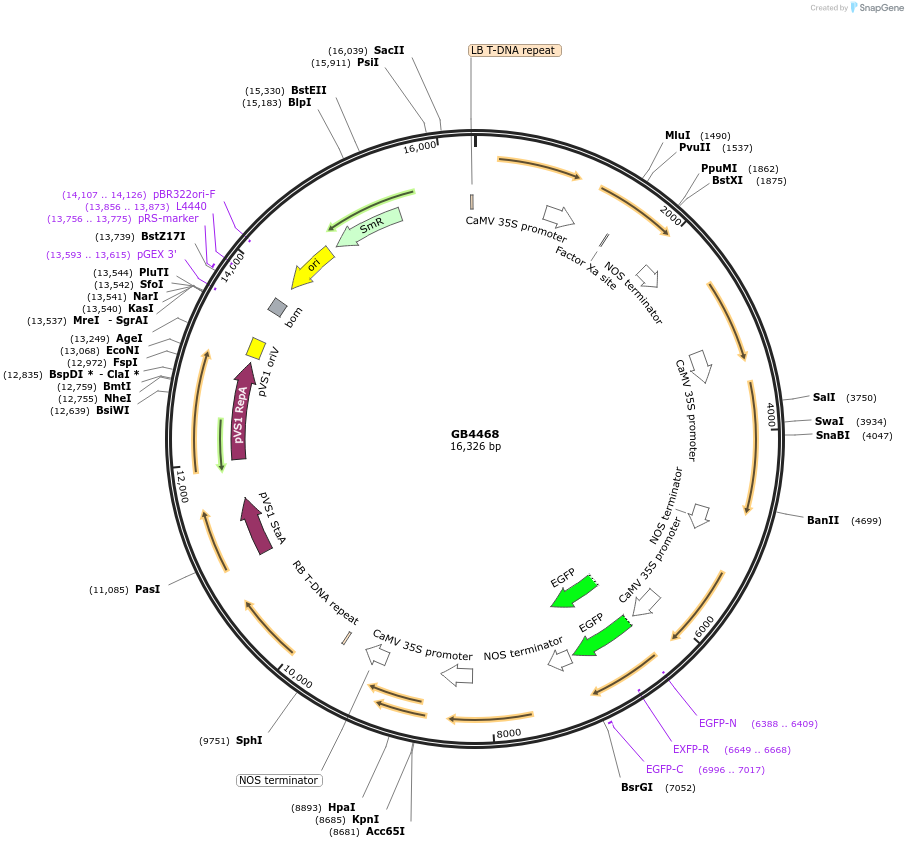
Plasmid#215257PurposeModule for the constitutive expression of the N. nambi LUZ, H3H genes, EGFP and p19 fused to a SF in omega2.DepositorInsertSF-LUZ-H3H-EGFP-p19
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJuly 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
GB4665
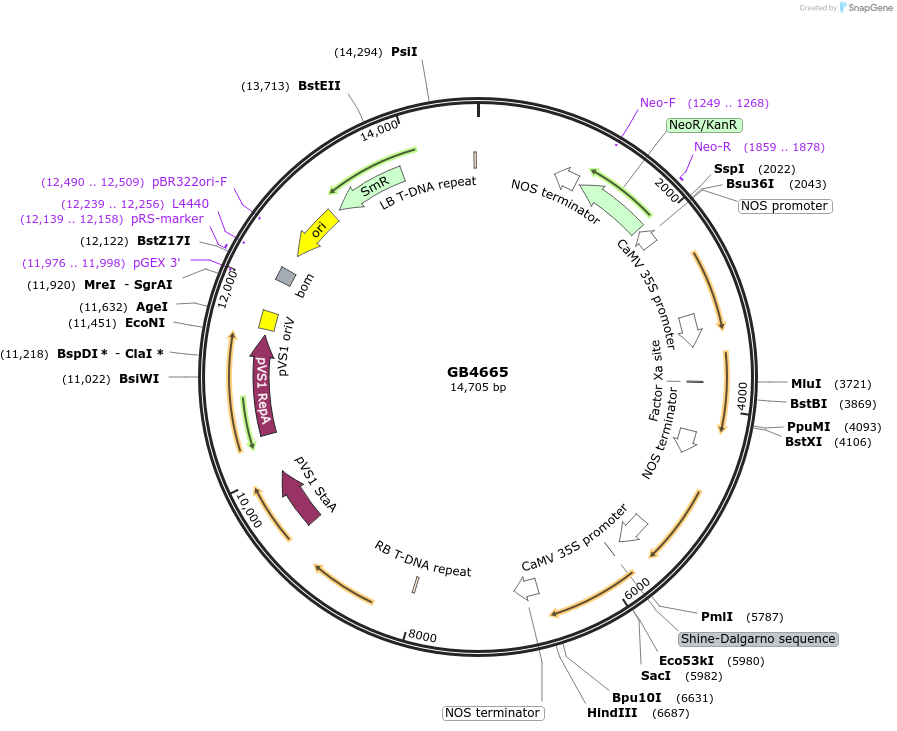
Plasmid#215270PurposeModule for the expression of nptII and the N. nambi LUZ and CPH genes flanked by insulators on the sides of the construction.DepositorInsertMAR10-SF-nptII-LUZ-CPH-MAR10-SF
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJuly 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
GB4666
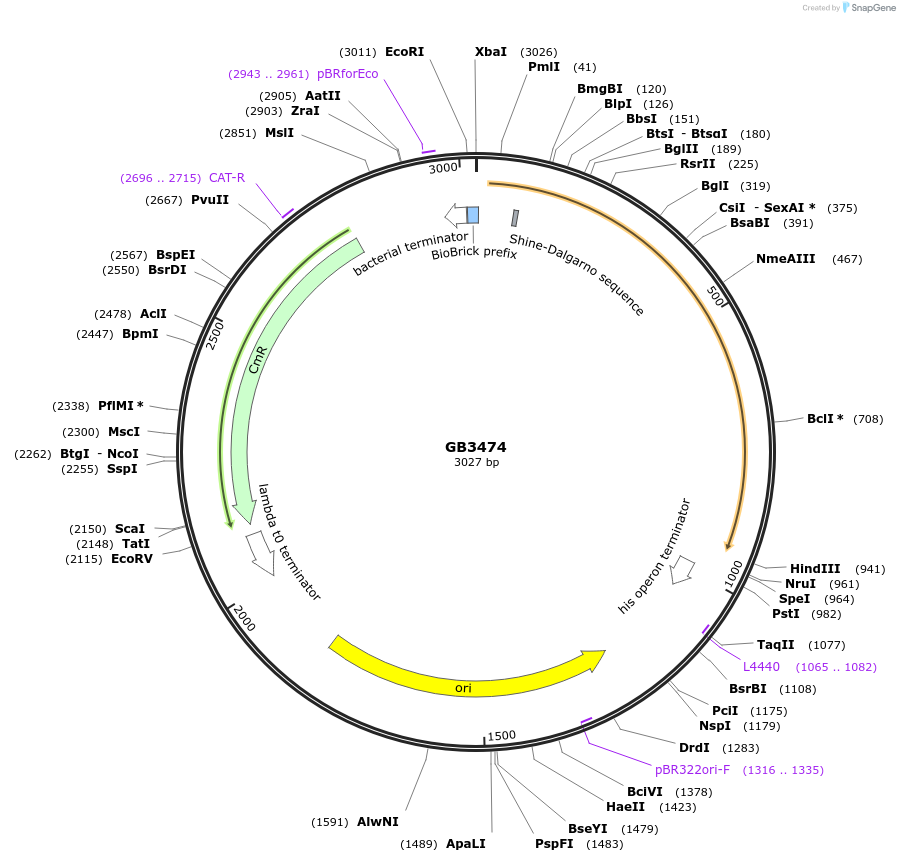
Plasmid#215271PurposepUPD2 for the Arabidopsis thaliana ABA-responsive MAPKKK18 promoter.DepositorInsertpromMAPKKK18
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
GB3474
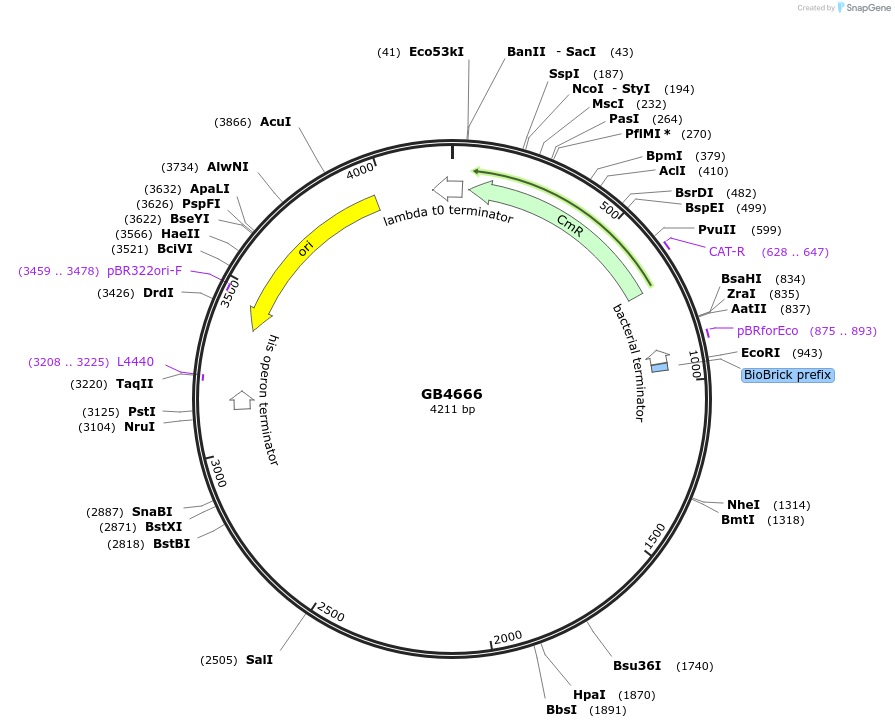
Plasmid#215370PurposeCDS of CPH from N. nambi codon optimized for N.benthamiana. This gene is the final step and recycling of the bioluminiscente pathwayDepositorInsertCPH
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
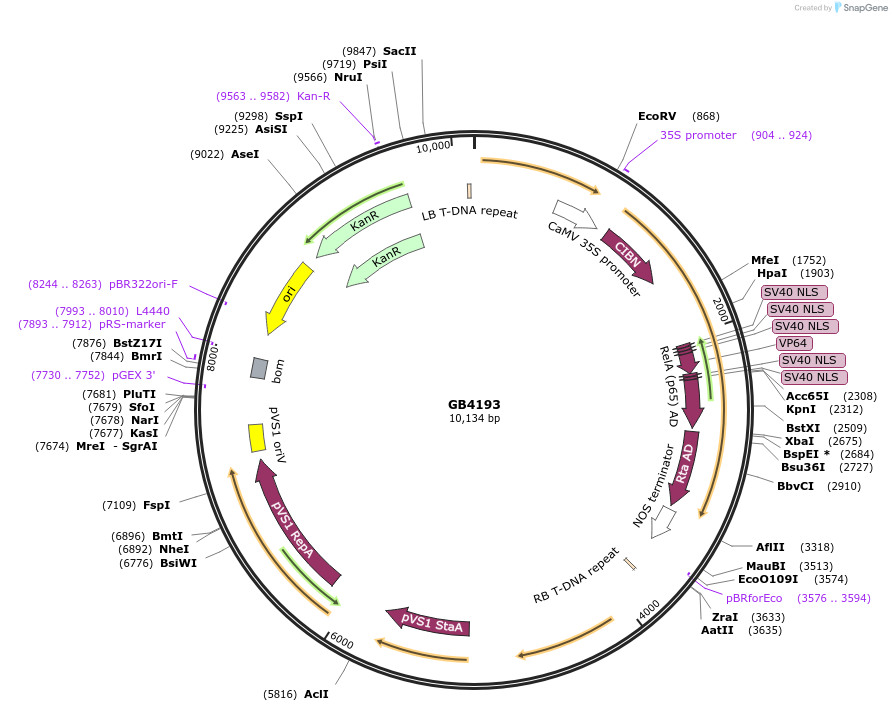
GB4193
Plasmid#215251PurposeTU for the expression of the CIB1 gene fused to the VPR activation domain under the P35S promoter and TNOS terminator in alpha1DepositorInsertP35S:CIB1:VPR:TNOS
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
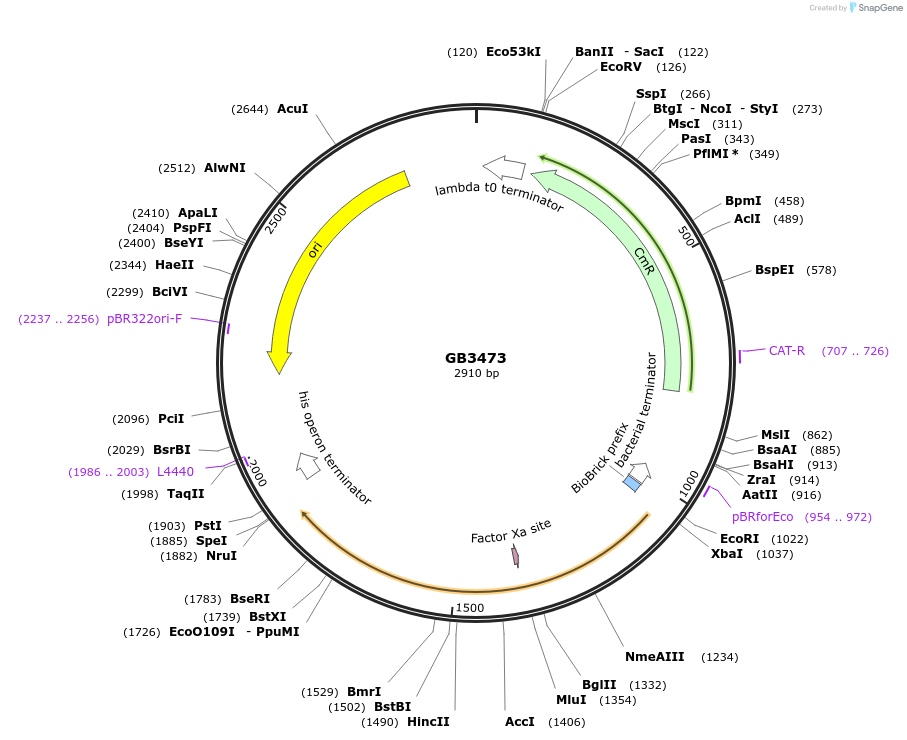
GB3473
Plasmid#215246PurposeCDS of Luz gene from N. nambi codon optimized for N.benthamiana. This gene generate the luminiscence of the bioluminiscente pathwayDepositorInsertLuz
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
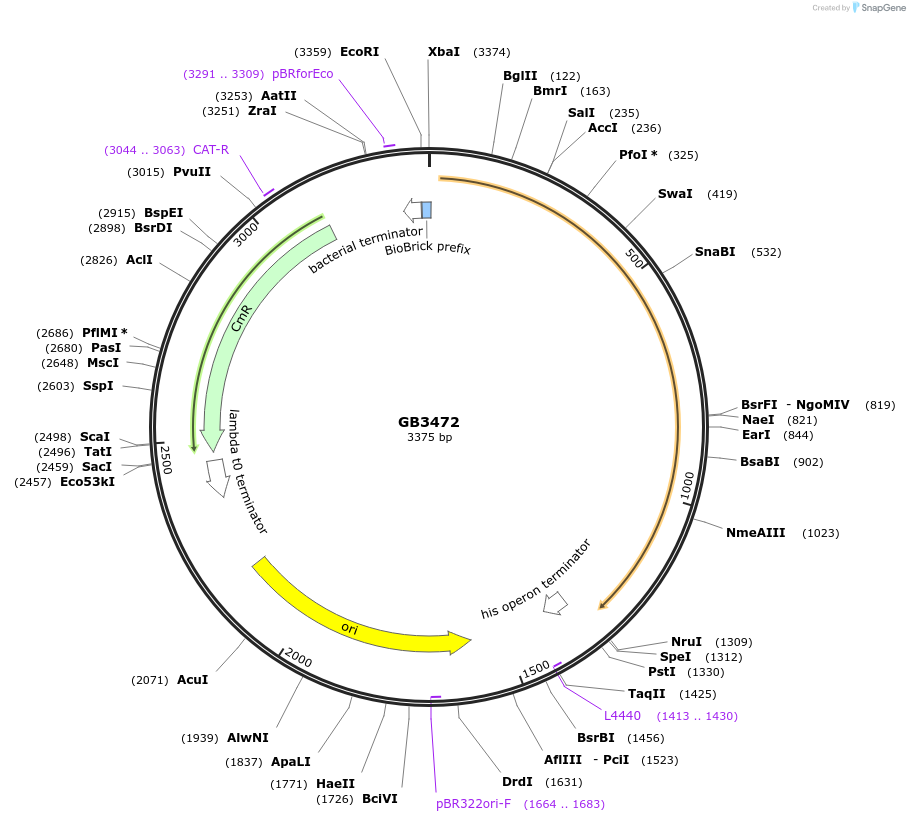
GB3472
Plasmid#215245PurposeCDS of H3H from N. nambi codon optimized for N.benthamiana. This gene is the second step of the bioluminiscente pathwayDepositorInsertH3H
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
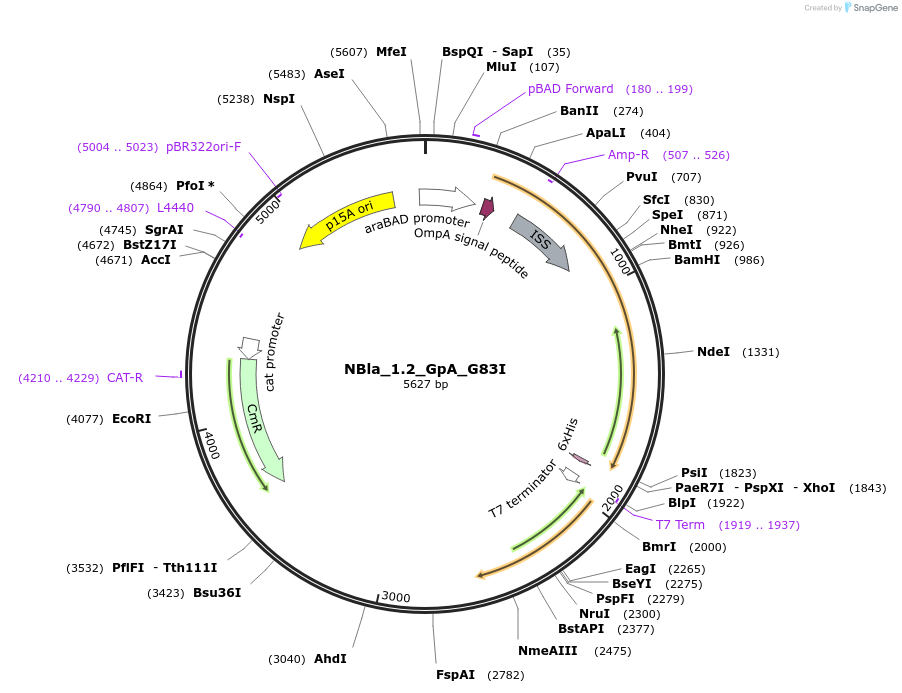
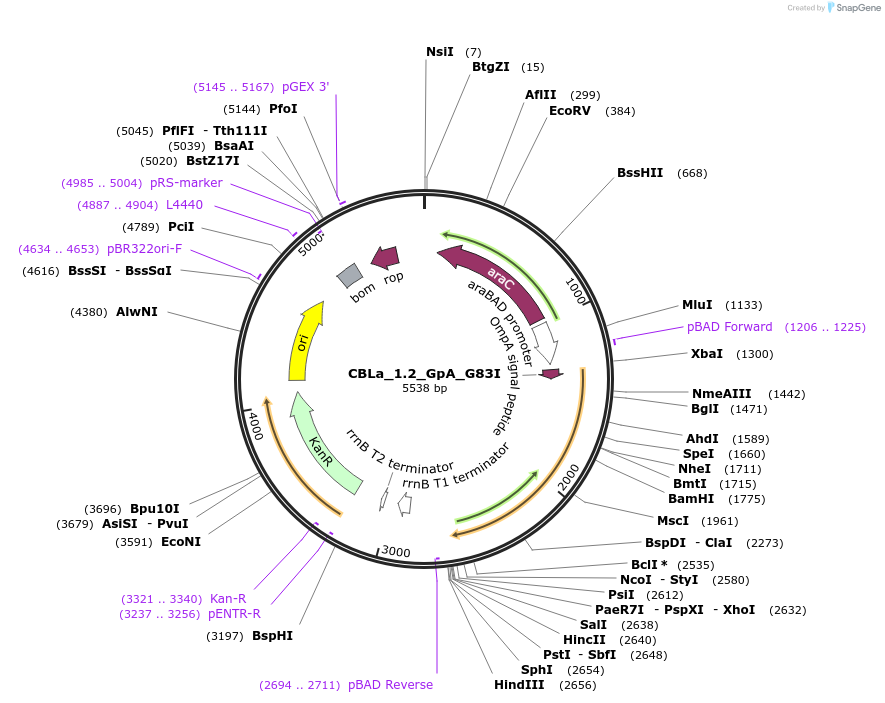
NBla_1.2_GpA_G83I
Plasmid#207844PurposeBLaTM-System GpA G83I negative control in NBLa 1.2 vectorDepositorInsertN-BLa 1.2 fusion protein
TagsFlagExpressionBacterialAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
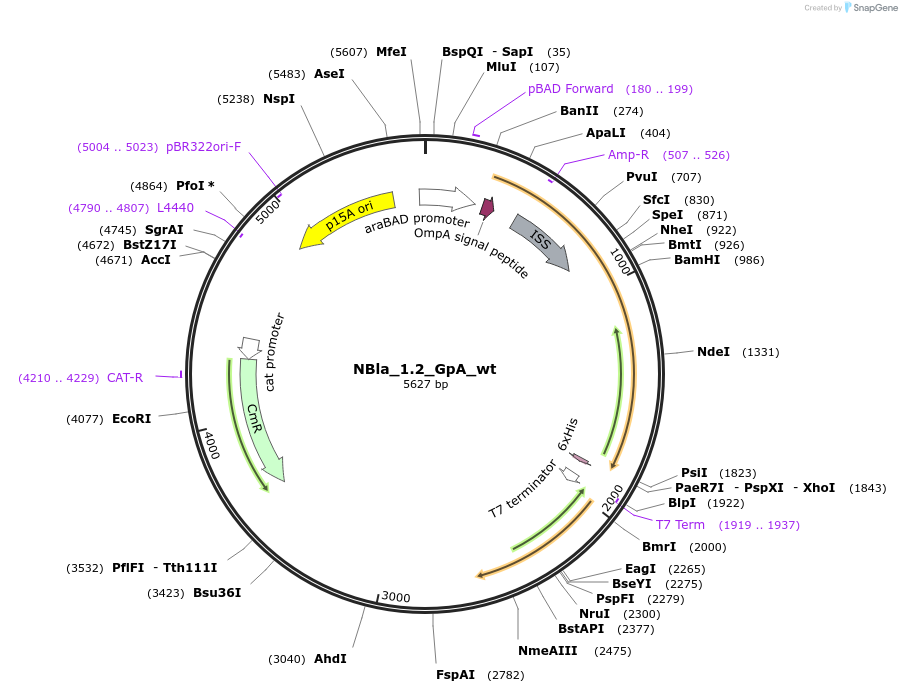
NBla_1.2_GpA_wt
Plasmid#207842PurposeBLaTM-System GpA wt positive control in NBLa 1.2 vectorDepositorInsertN-BLa 1.2 fusion protein
TagsFlagExpressionBacterialAvailable SinceDec. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
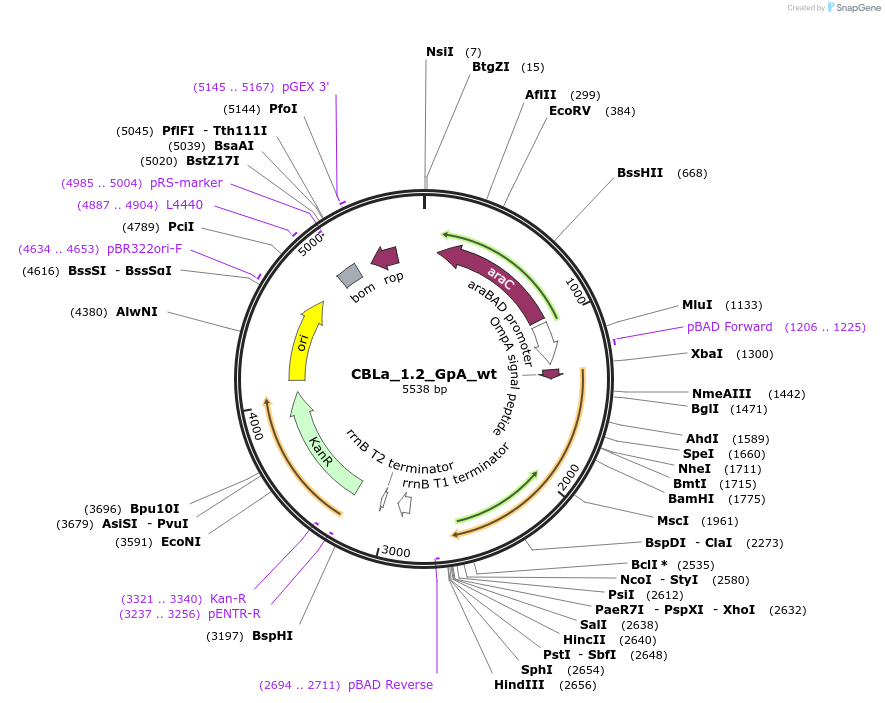
CBLa_1.2_GpA_wt
Plasmid#207843PurposeBLaTM-System GpA wt positive control in CBLa 1.2 vectorDepositorInsertN-BLa 1.2 fusion protein
TagsFlagExpressionBacterialAvailable SinceDec. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
CBLa_1.2_GpA_G83I
Plasmid#207845PurposeBLaTM-System GpA G83I negative control in CBLa 1.2 vectorDepositorInsertN-BLa 1.2 fusion protein
TagsFlagExpressionBacterialAvailable SinceDec. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
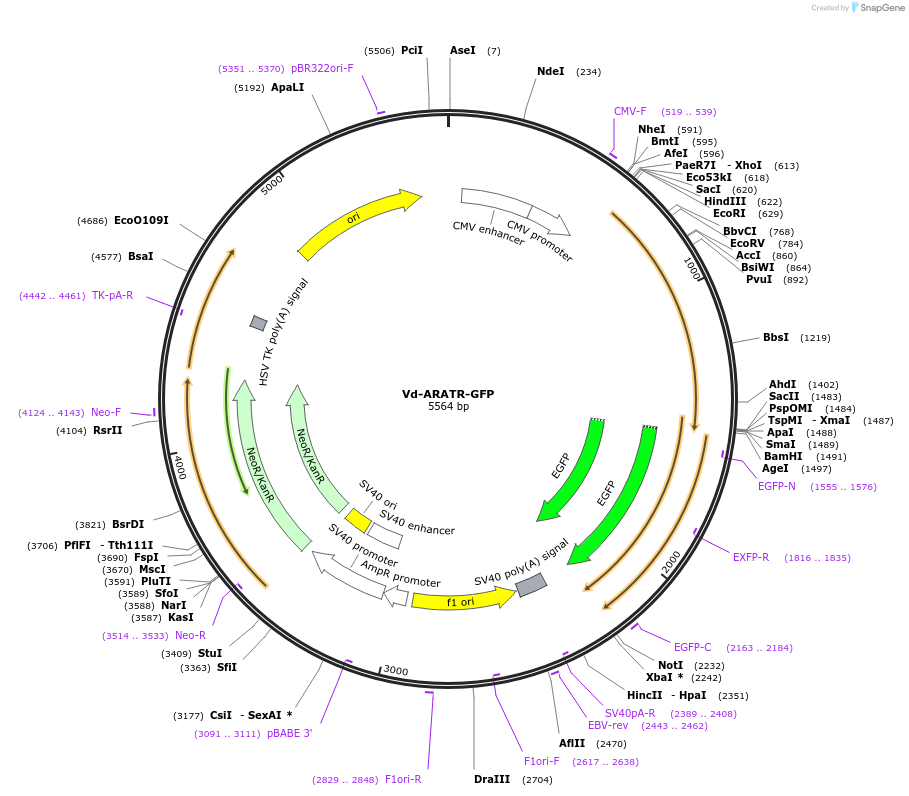
Vd-ARATR-GFP
Plasmid#208903Purposeencodes the arachnotocin receptor (oxytocin/vasopressin-like) from the mite Varroa destructor, tagged with GFPDepositorInsertARATR
TagsEGFPExpressionMammalianAvailable SinceNov. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
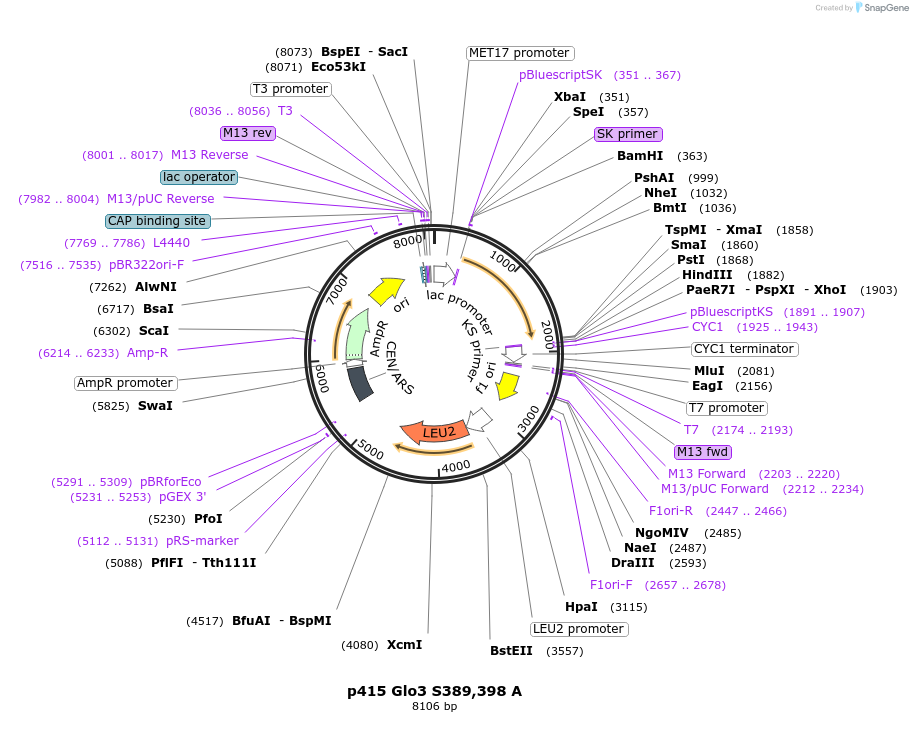
p415 Glo3 S389,398 A
Plasmid#129485PurposeExpression in S. cerevisiaeDepositorInsertGlo3 S389,398 A (GLO3 Budding Yeast)
ExpressionYeastMutationMutation of serines 389 and 398 to alanineAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
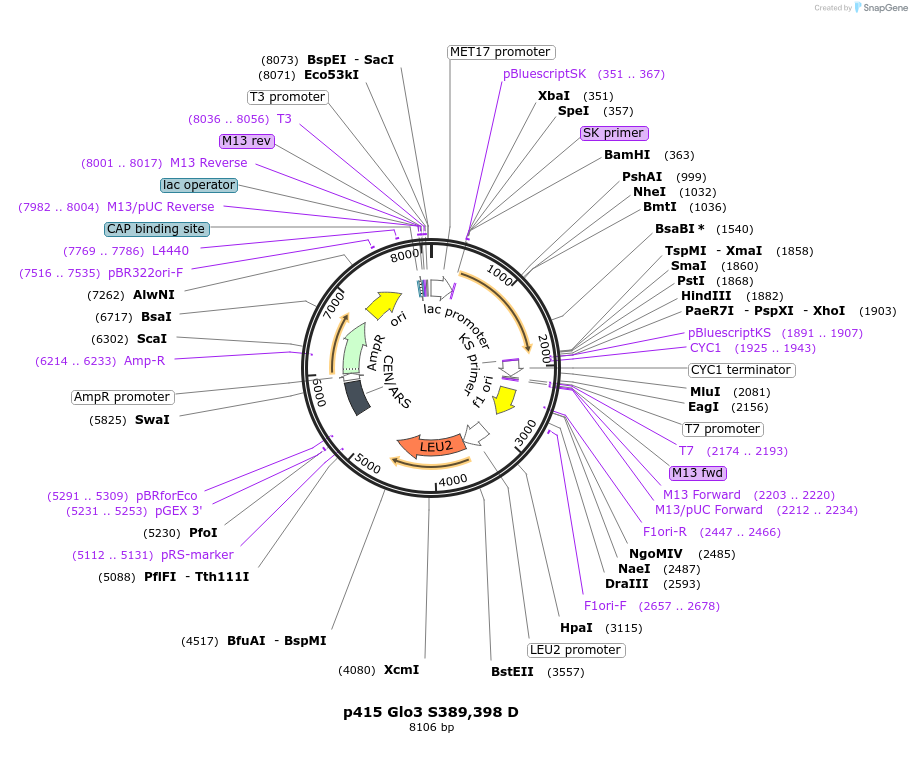
p415 Glo3 S389,398 D
Plasmid#129486PurposeExpression in S. cerevisiaeDepositorInsertGlo3 S389,398 D (GLO3 Budding Yeast)
ExpressionYeastMutationMutation of serines 389 and 398 to AspAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
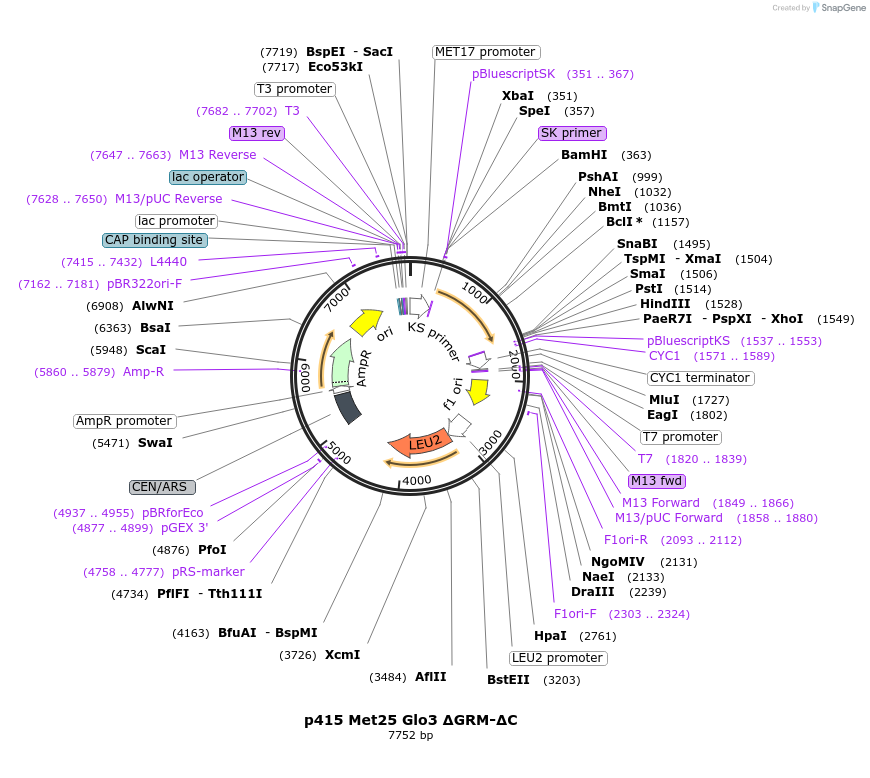
p415 Met25 Glo3 ΔGRM-ΔC
Plasmid#129487PurposeExpression in S. cerevisiaeDepositorInsertGlo3 ΔGRM ΔC (GLO3 Budding Yeast)
ExpressionYeastMutationΔ376-493 (deletion of Glo3 motif & C-terminal…Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
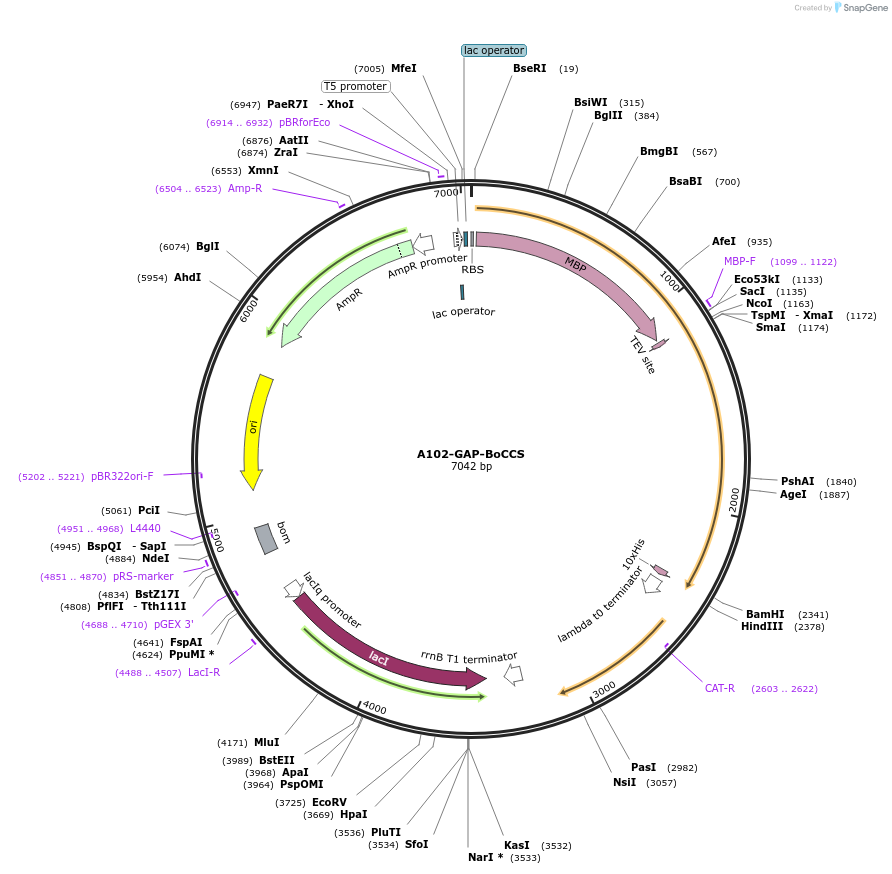
A102-GAP-BoCCS
Plasmid#123287PurposeExpression of truncated Glo3 (1-375) in E.ColiDepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
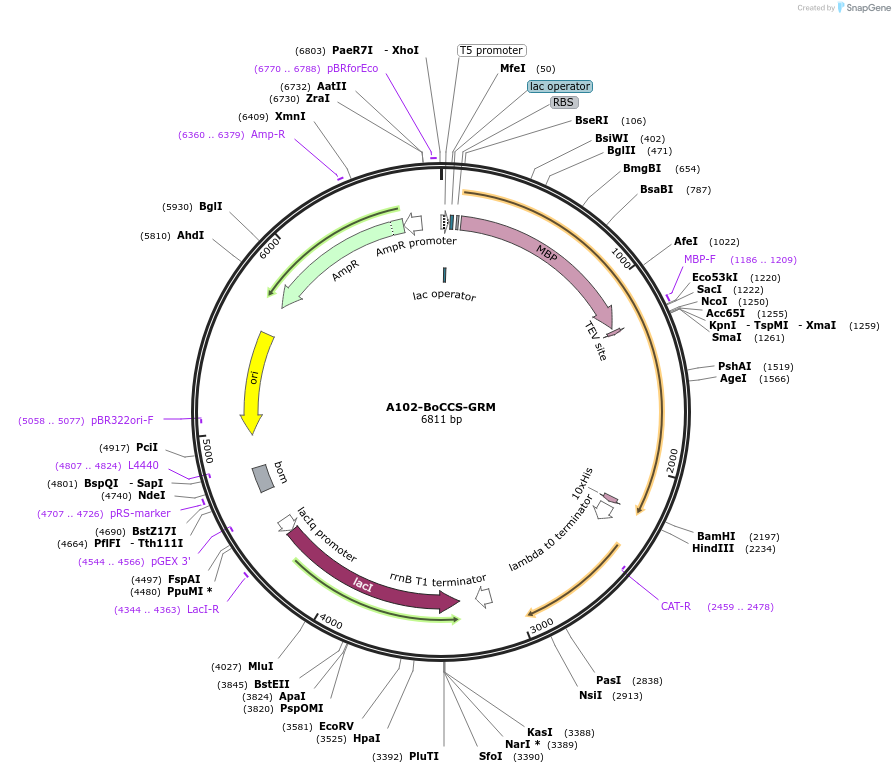
A102-BoCCS-GRM
Plasmid#123288PurposeExpression of truncated Glo3 (137-459) in E.ColiDepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
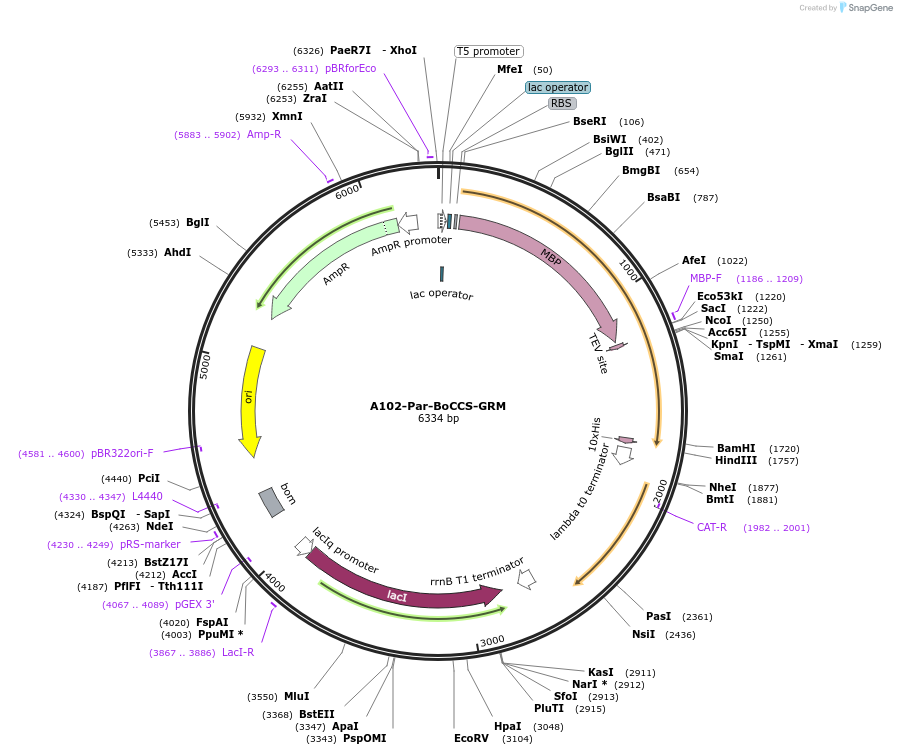
A102-Par-BoCCS-GRM
Plasmid#123289PurposeExpression of truncated Glo3 (296-459) in E.ColiDepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
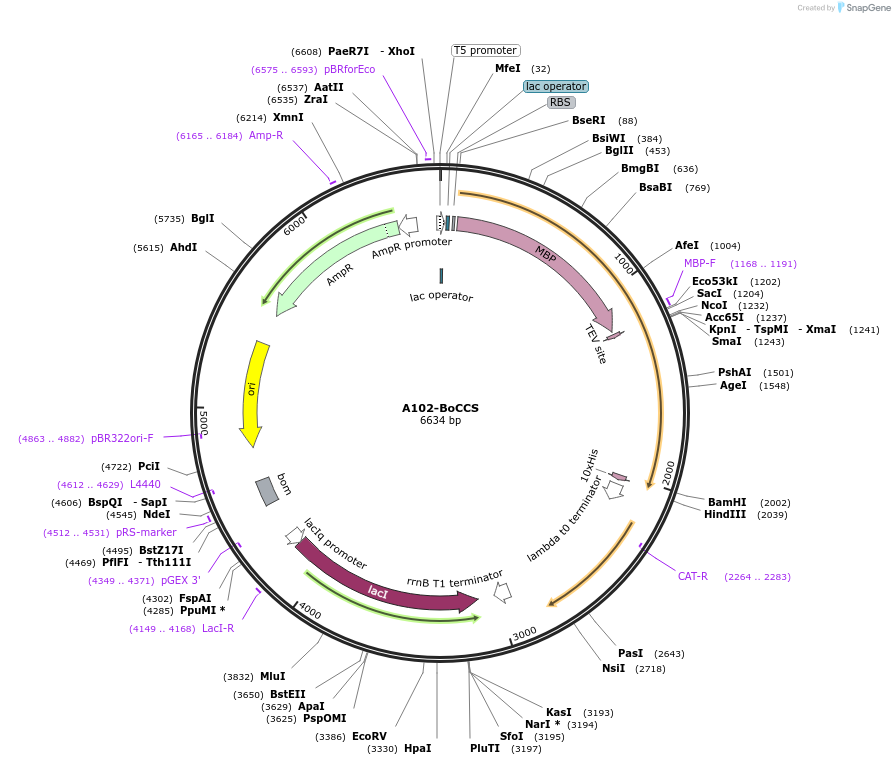
A102-BoCCS
Plasmid#123290PurposeExpression of truncated Glo3 (137-375) in E.ColiDepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
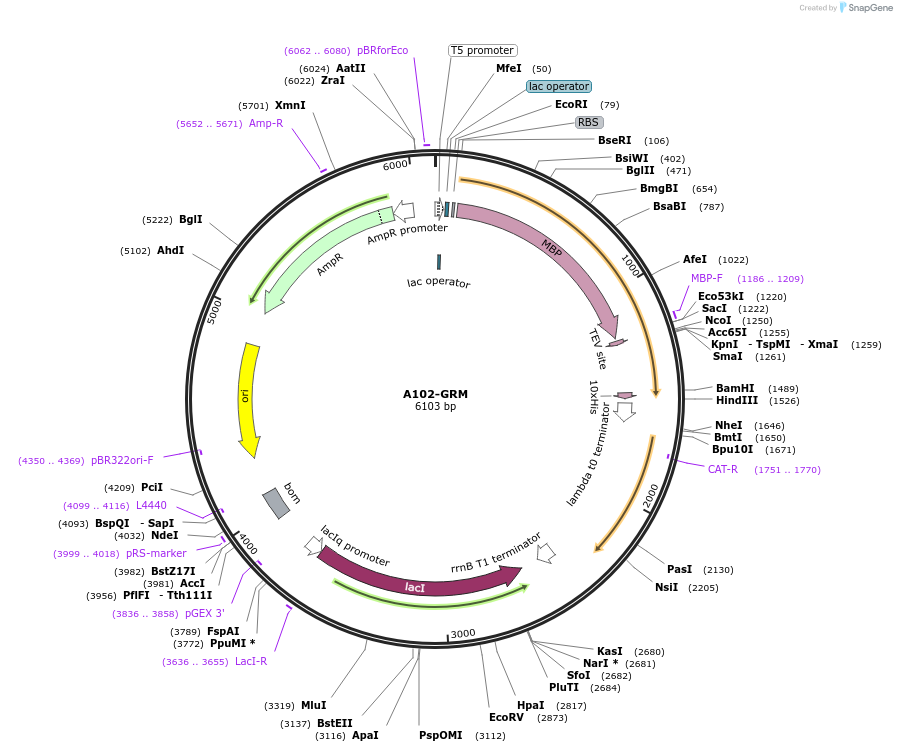
A102-GRM
Plasmid#123291PurposeExpression of truncated Glo3 (373-459) in E.ColiDepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only