We narrowed to 4,424 results for: ARA-2
-
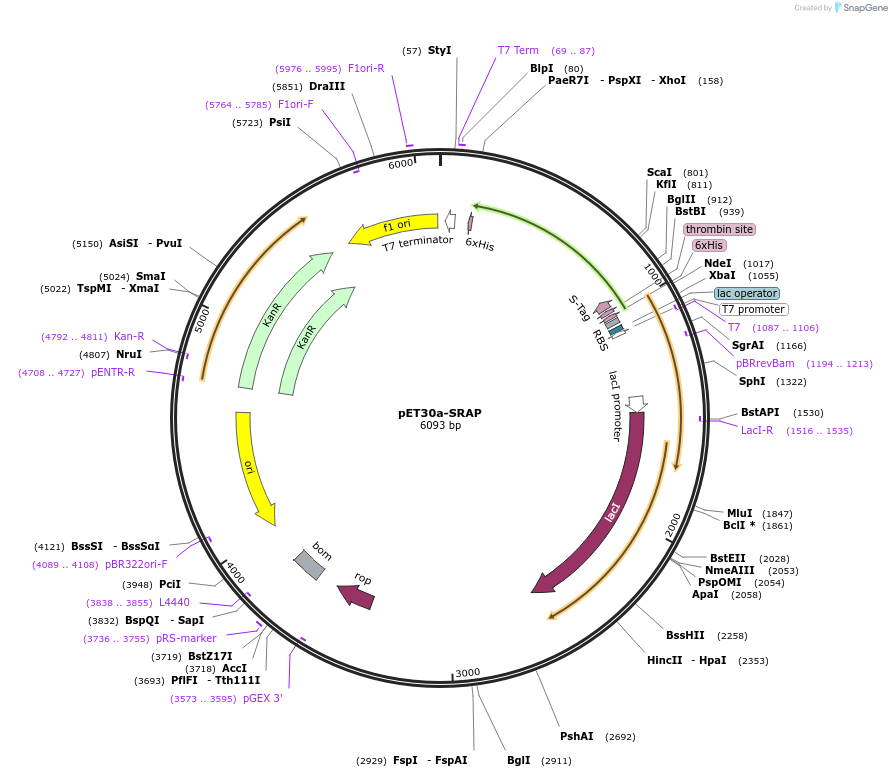
Plasmid#84524PurposeExpresses murine SRAP domain (250 aa) in bacteriaDepositorAvailable SinceNov. 18, 2016AvailabilityAcademic Institutions and Nonprofits only
-
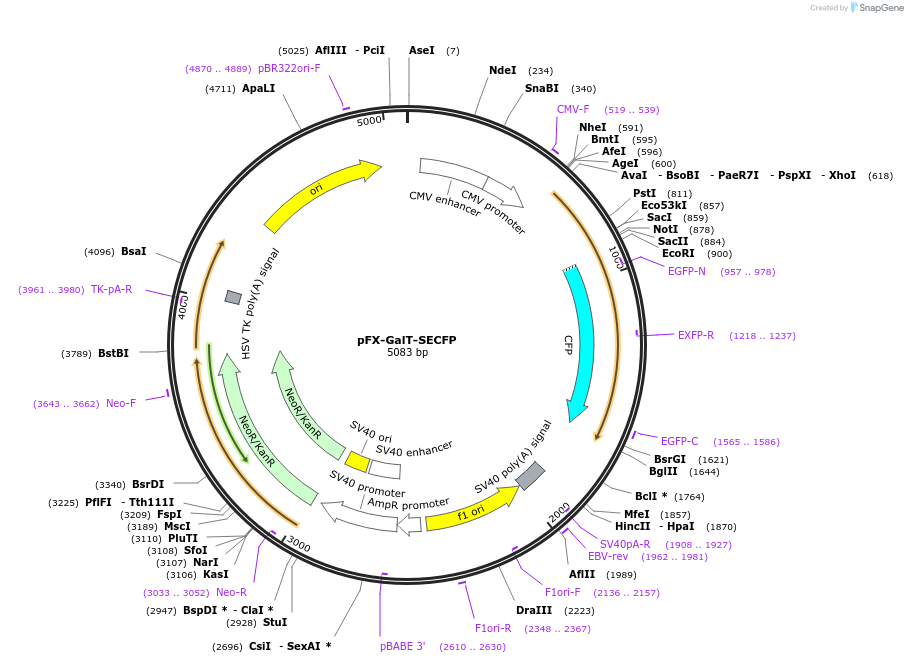
pFX-GalT-SECFP
Plasmid#174470PurposeExpress SECFP with golgi apparatus-targeting sequence in mammalian cellsDepositorInsertSECFP with golgi apparatus-targeting sequence
Tagsβ1,4-galactose transferaseExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
pFX-GalT-iRFP
Plasmid#174474PurposeExpress iRFP with golgi apparatus-targeting sequence in mammalian cellsDepositorInsertiRFP with golgi apparatus-targeting sequence
Tagsβ1,4-galactose transferaseExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
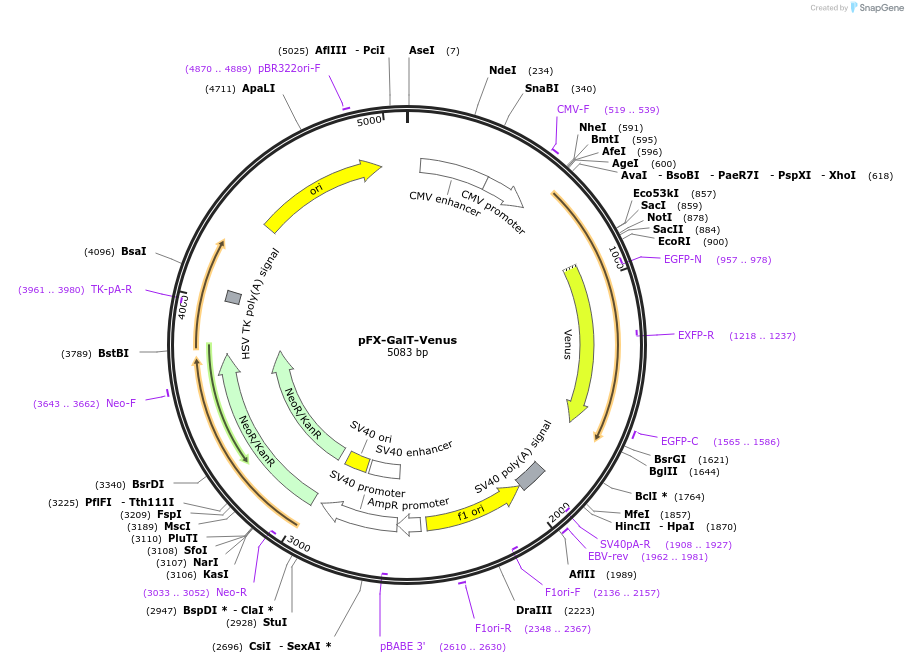
pFX-GalT-Venus
Plasmid#174472PurposeExpress Venus with golgi apparatus-targeting sequence in mammalian cellsDepositorInsertVenus with golgi apparatus-targeting sequence
Tagsβ1,4-galactose transferaseExpressionMammalianPromoterCMVAvailable SinceFeb. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
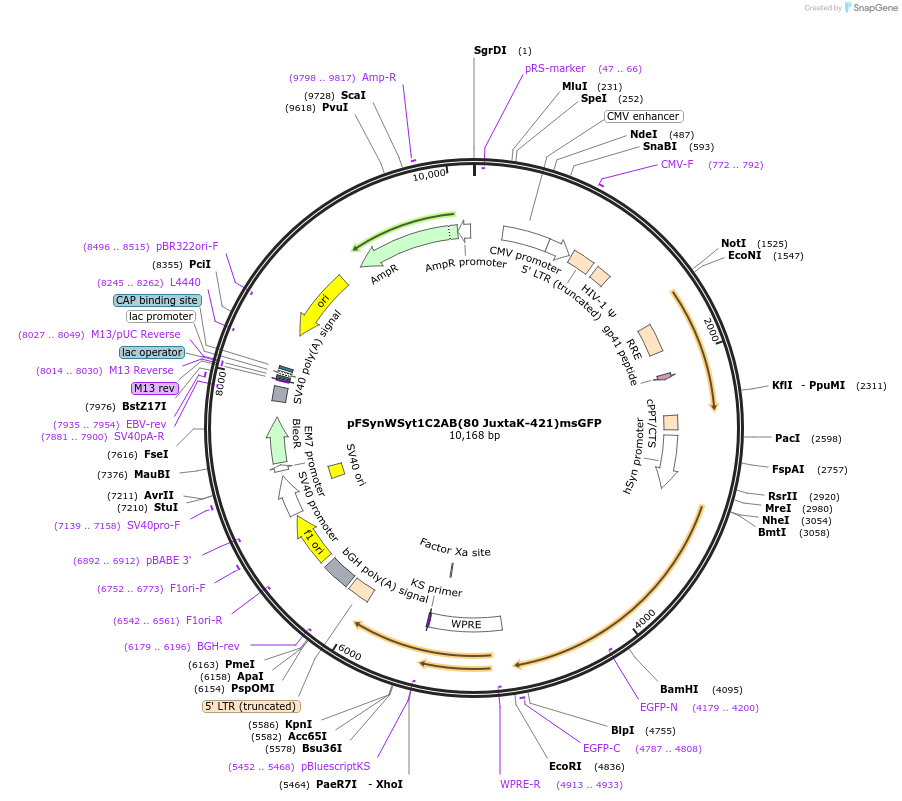
pFSynWSyt1C2AB(80 JuxtaK-421)msGFP
Plasmid#209016PurposeOverexpression of truncated syt1 with Juxta K mutations and tagged with GFP in mammalian cellsDepositorInsertsyt1 (Syt1 Rat)
UseLentiviralTagsmsfGFPExpressionMammalianMutationaa 80-421 only, Juxta K mutant (80KKCLFKKKNKKKGKE…Available SinceNov. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
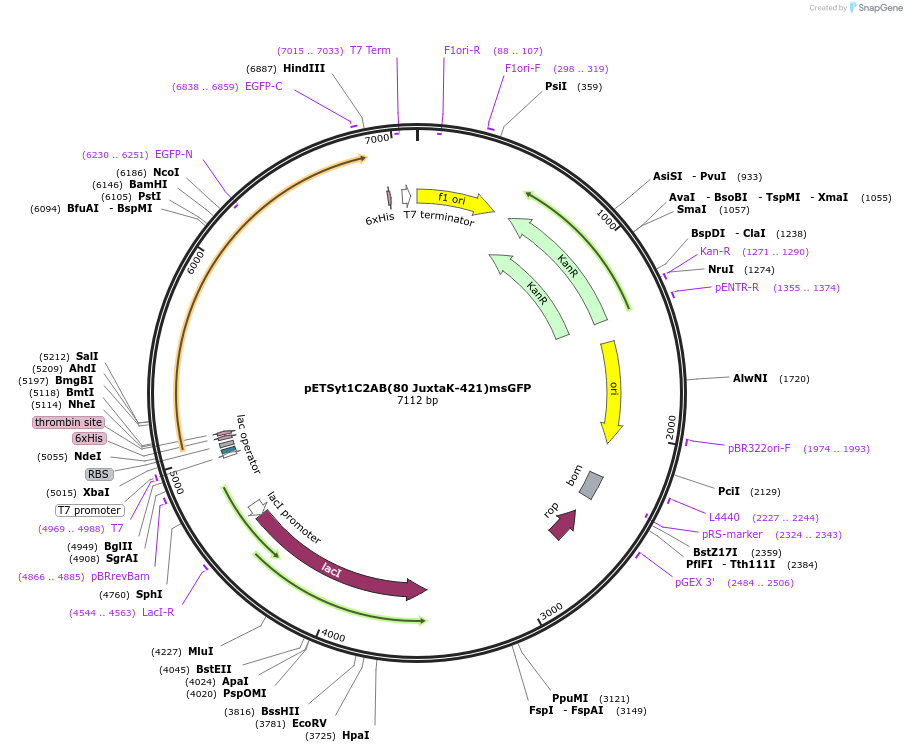
pETSyt1C2AB(80 JuxtaK-421)msGFP
Plasmid#209011PurposeRecombinant protein expression of truncated syt1 construct with Juxta K mutations and tagged with GFPDepositorInsertsyt1 (Syt1 Rat)
Tags6xHis and msfGFPExpressionBacterialMutationaa 80-421 only, Juxta K mutant (80KKCLFKKKNKKKGKE…Available SinceNov. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
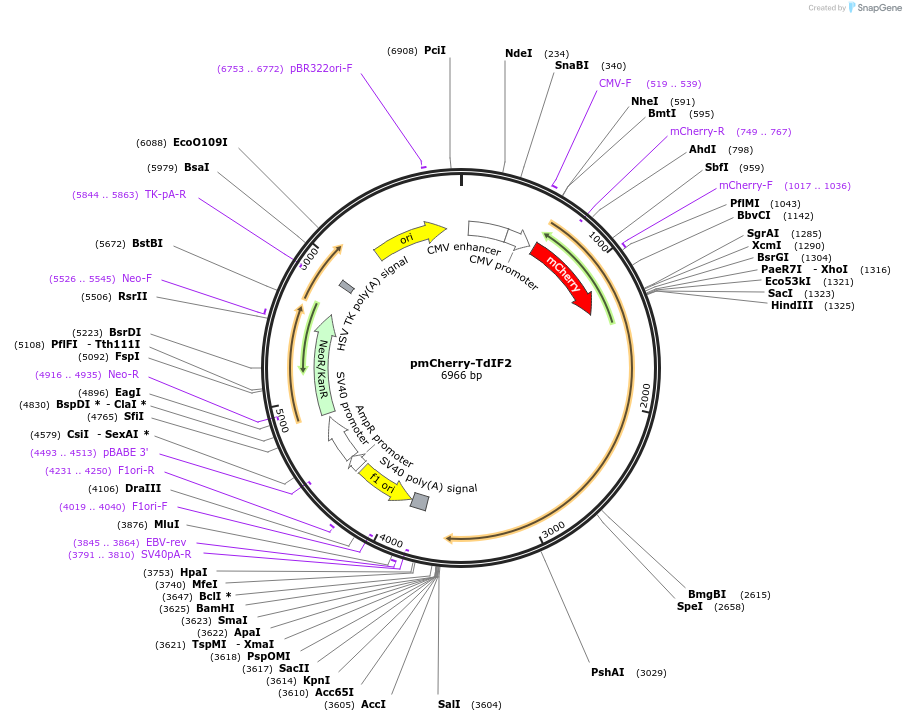
pmCherry-TdIF2
Plasmid#196906Purposevisualization of TdIF2/ERBP as a fusion to mCherryDepositorInsertTdIF2
TagsmCherryExpressionMammalianPromoterCMVAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
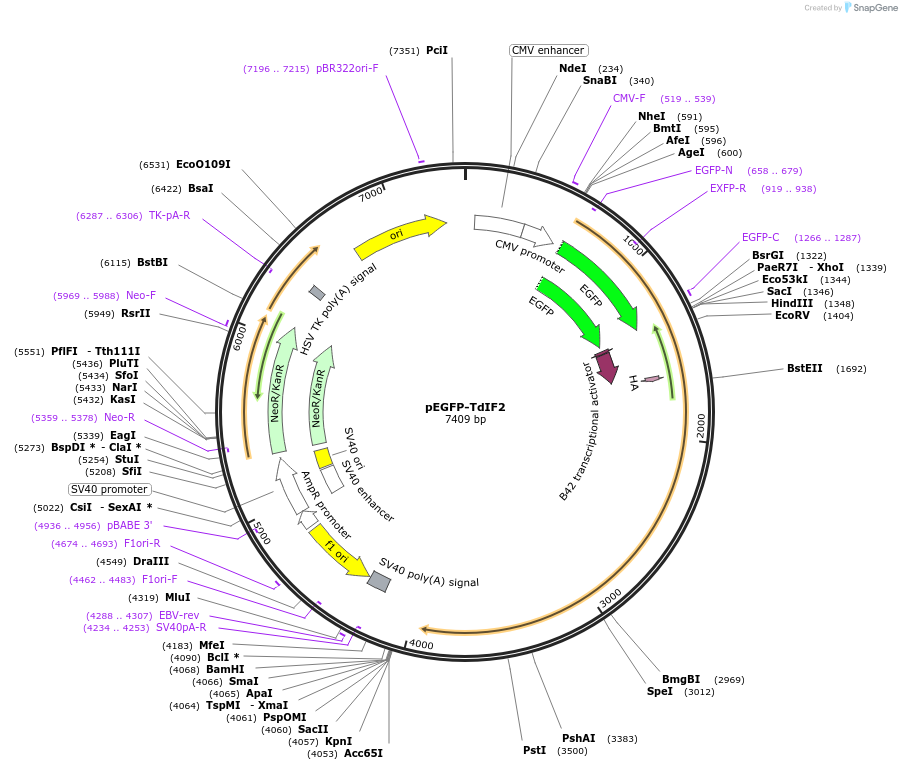
pEGFP-TdIF2
Plasmid#199699Purposevisualization of nucleolus in living mammalian cellsDepositorInsertTdIF2
TagsEGFPExpressionMammalianPromoterCMVAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
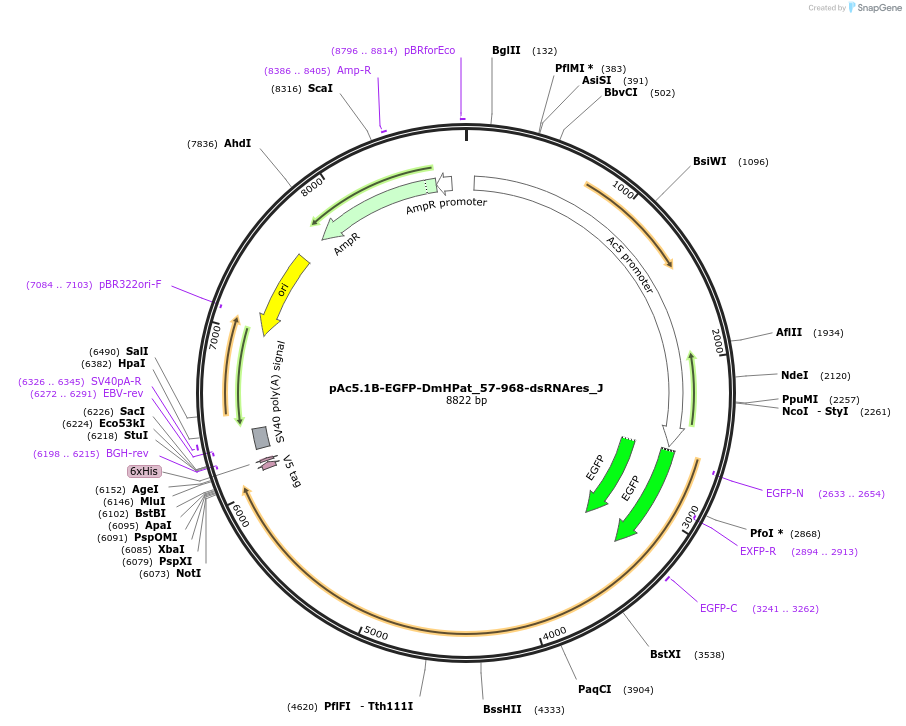
pAc5.1B-EGFP-DmHPat_57-968-dsRNAres_J
Plasmid#146690PurposeInsect Expression of DmHPat_57-968-dsRNAresDepositorInsertDmHPat_57-968-dsRNAres (Patr-1 Fly)
ExpressionInsectMutationone silent mutation A1758G and one non silent G56…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
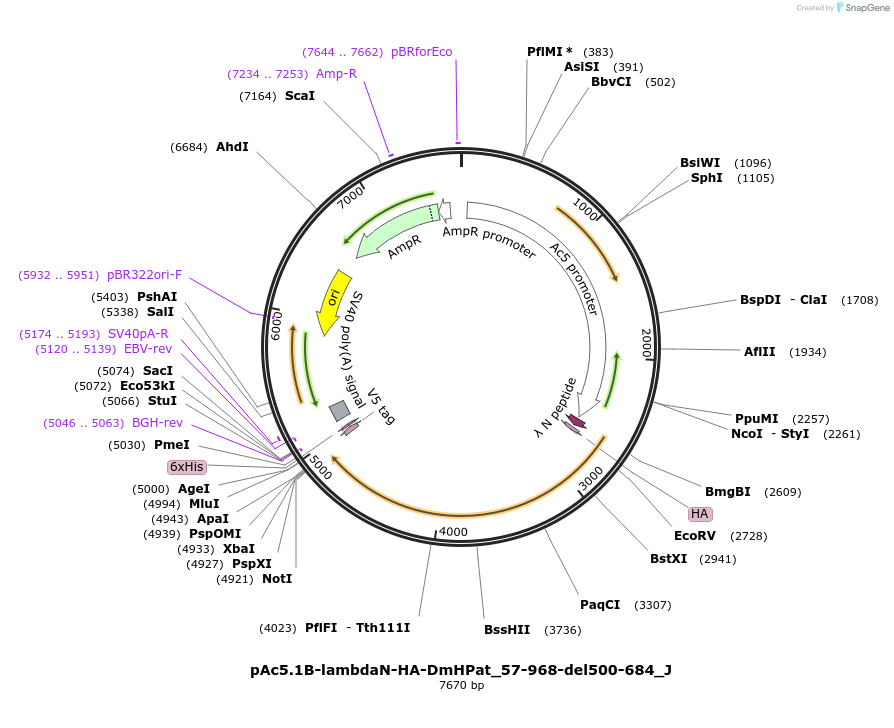
pAc5.1B-lambdaN-HA-DmHPat_57-968-del500-684_J
Plasmid#146695PurposeInsect Expression of DmHPat_57-968-del500-684DepositorInsertDmHPat_57-968-del500-684 (Patr-1 Fly)
ExpressionInsectMutationtwo silent mutations compared to the sequence giv…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
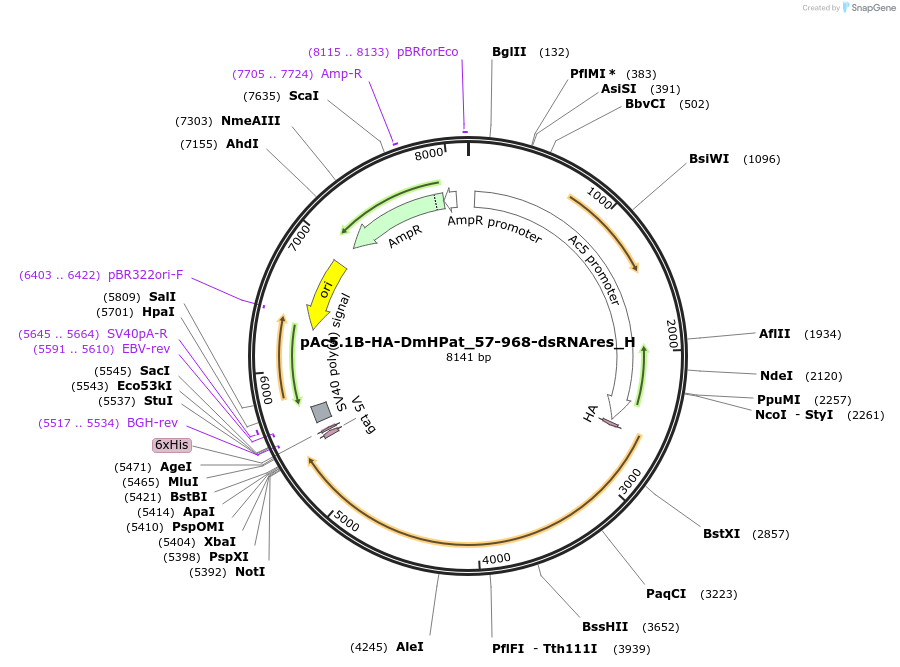
pAc5.1B-HA-DmHPat_57-968-dsRNAres_H
Plasmid#146464PurposeInsect Expression of DmHPat_57-968-dsRNAresDepositorInsertDmHPat_57-968-dsRNAres (Patr-1 Fly)
ExpressionInsectMutationone silent mutation T477C and one non silent muta…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
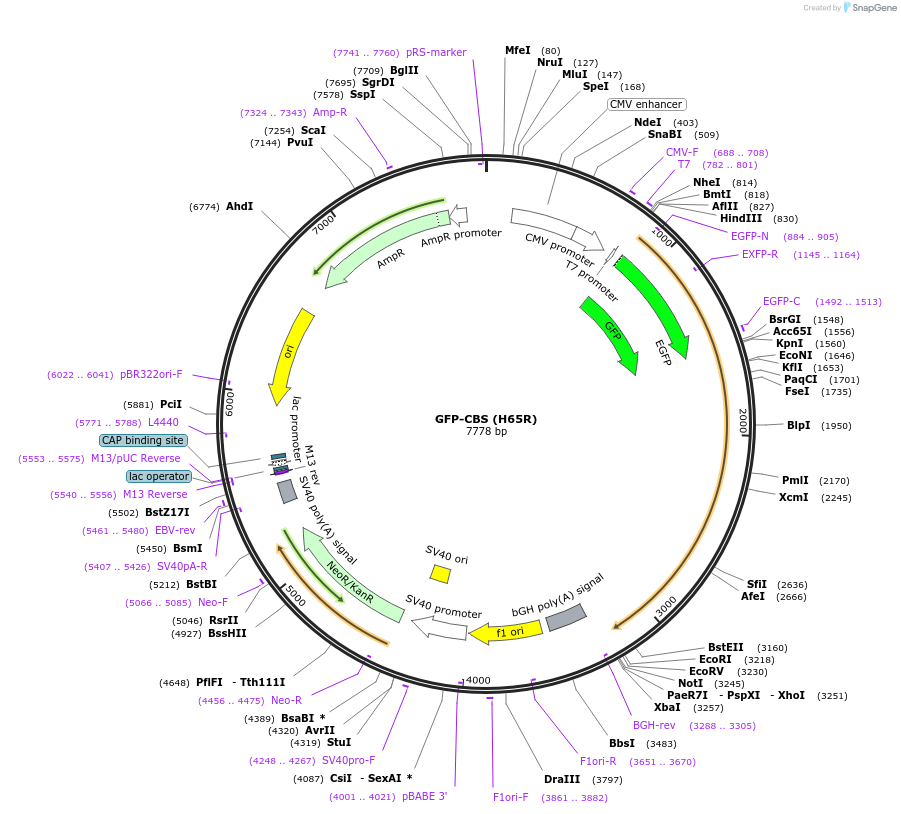
GFP-CBS (H65R)
Plasmid#182924PurposeMammalian expression plasmid for human CBS mutant H65RDepositorInsertCystathionine Beta-Synthase (CBS Human)
TagseGFPExpressionMammalianMutationchanged histidine 65 to argininePromoterCMVAvailable SinceJan. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
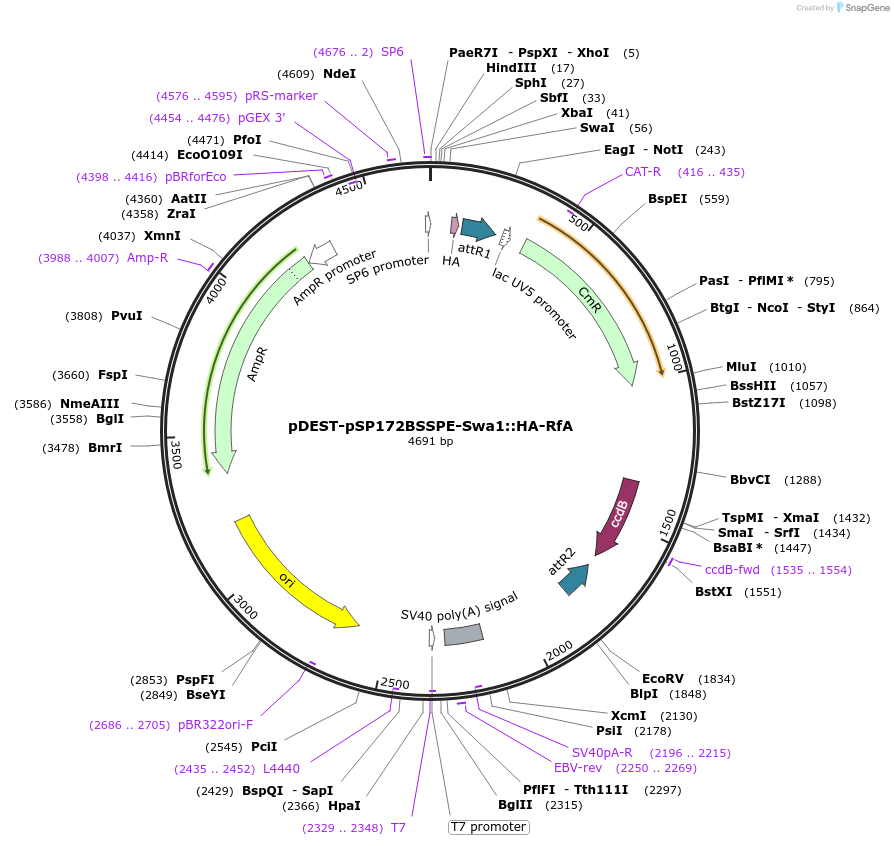
pDEST-pSP172BSSPE-Swa1::HA-RfA
Plasmid#186404PurposeDestination vector for ascidian transgenesis by electroporation of a Gateway-cloned fusion of N-ter HA tag and a gene of interest under control of a reg. sequence in the Swa1 restriction site.DepositorTypeEmpty backboneUseGateway destination vectorAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
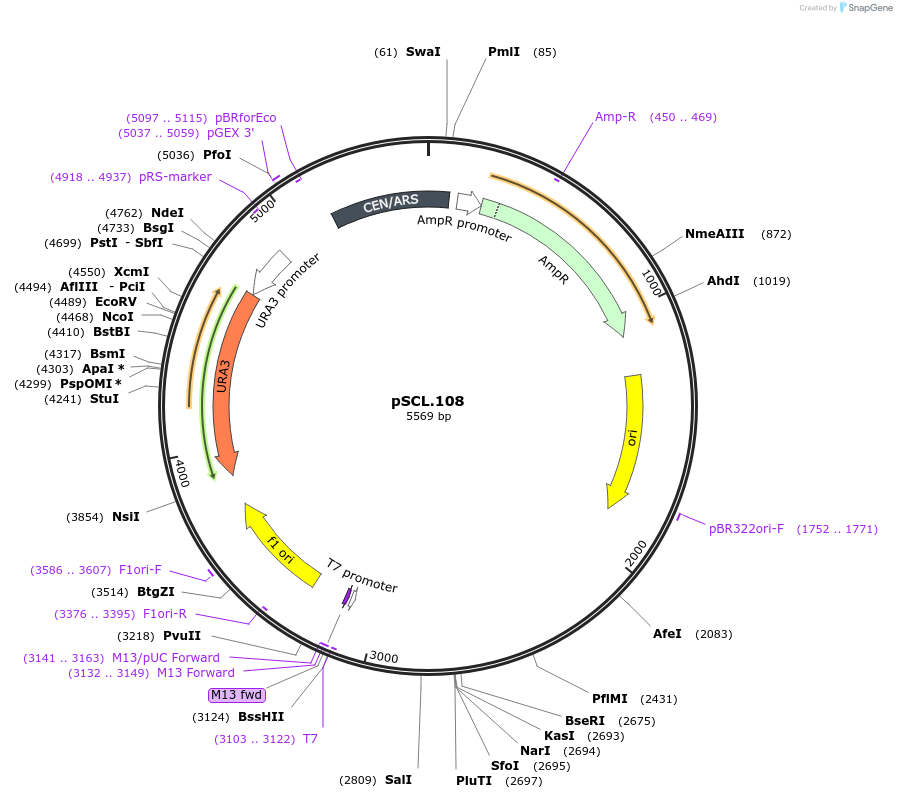
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
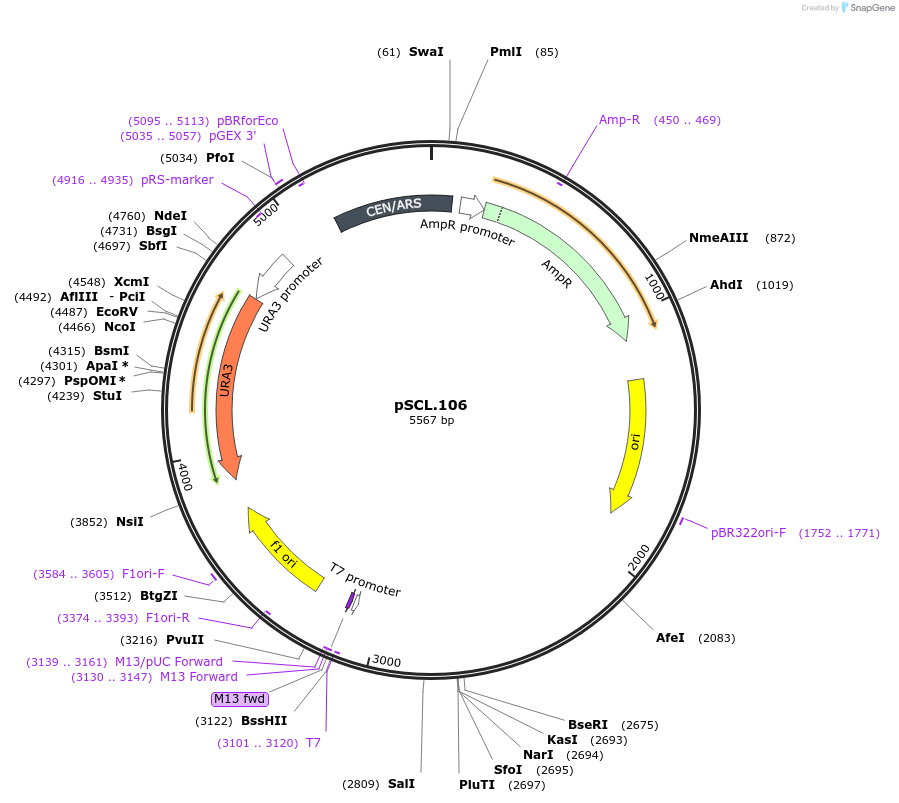
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
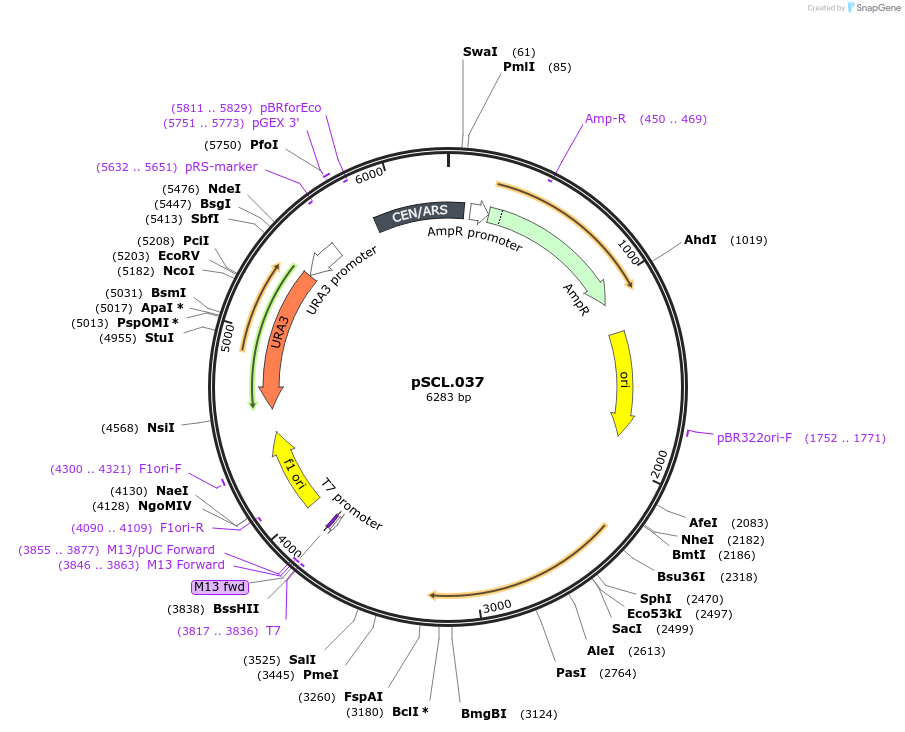
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
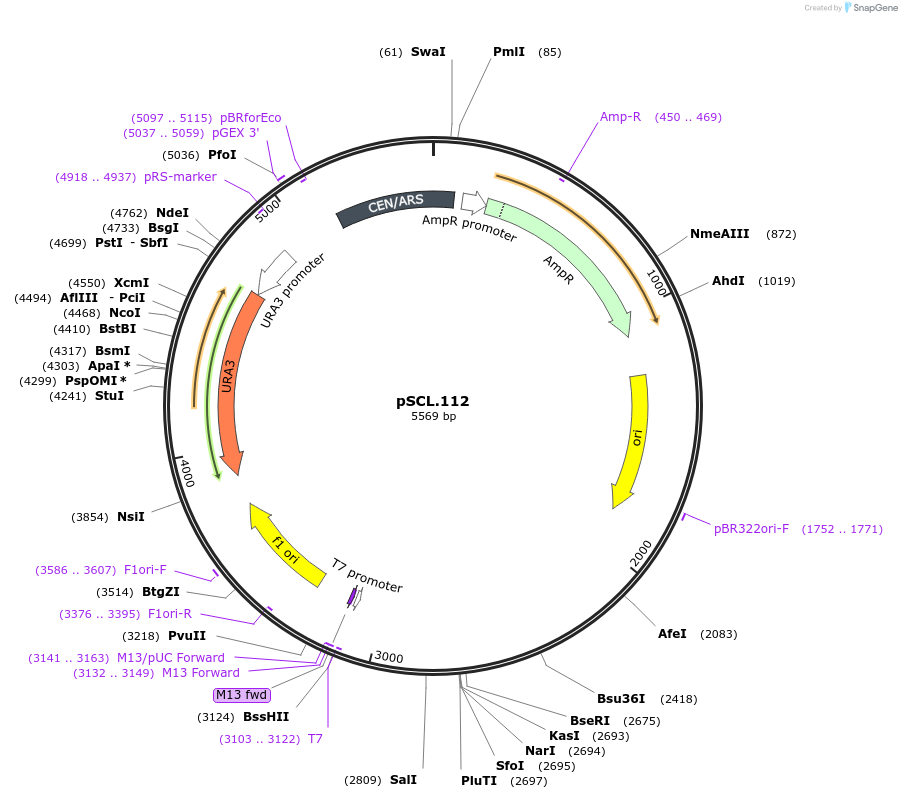
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
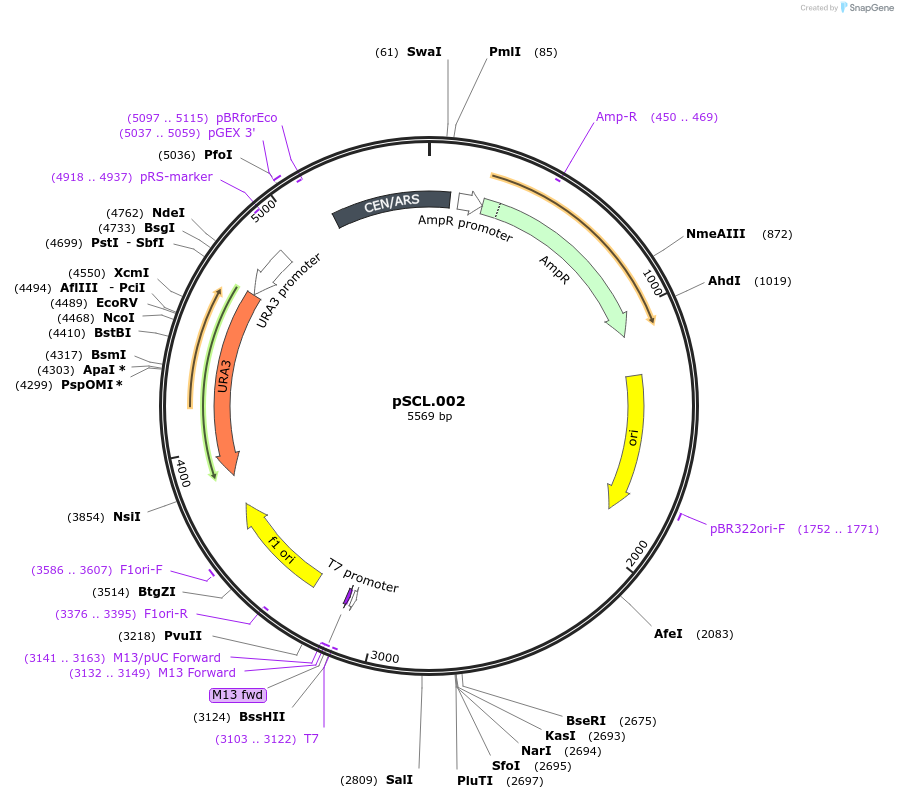
pSCL.002
Plasmid#184972PurposeExpress -Eco1 ADE2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 12
ExpressionYeastMutationADE2 donor P272stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
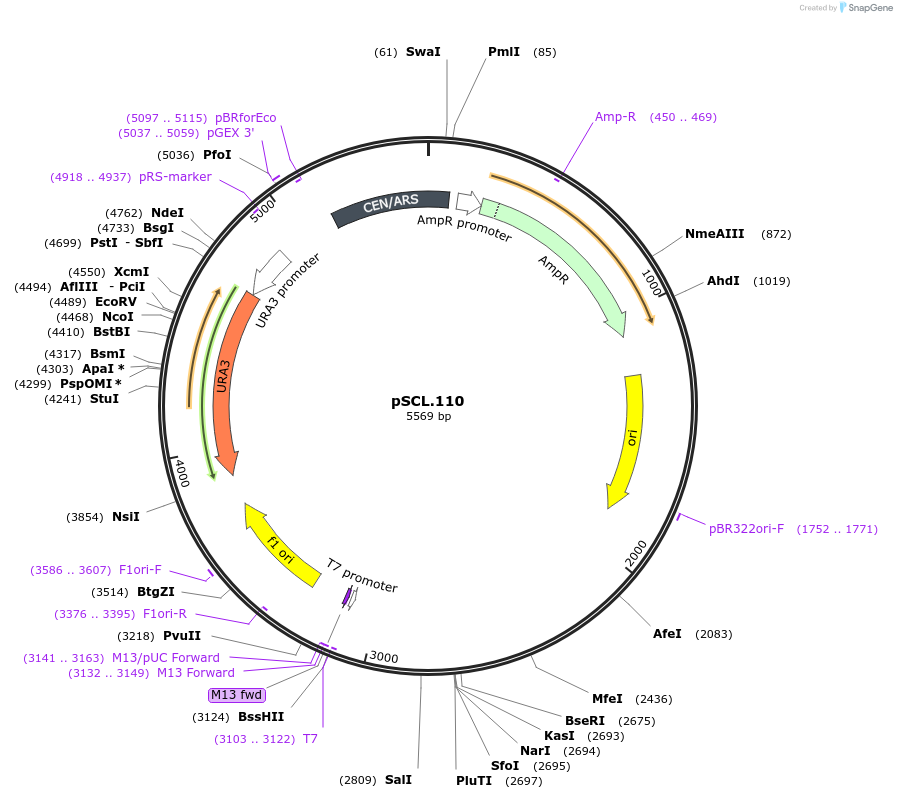
pSCL.110
Plasmid#184979PurposeExpress -Eco1 TRP2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 12
ExpressionYeastMutationTRP2 donor E64stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
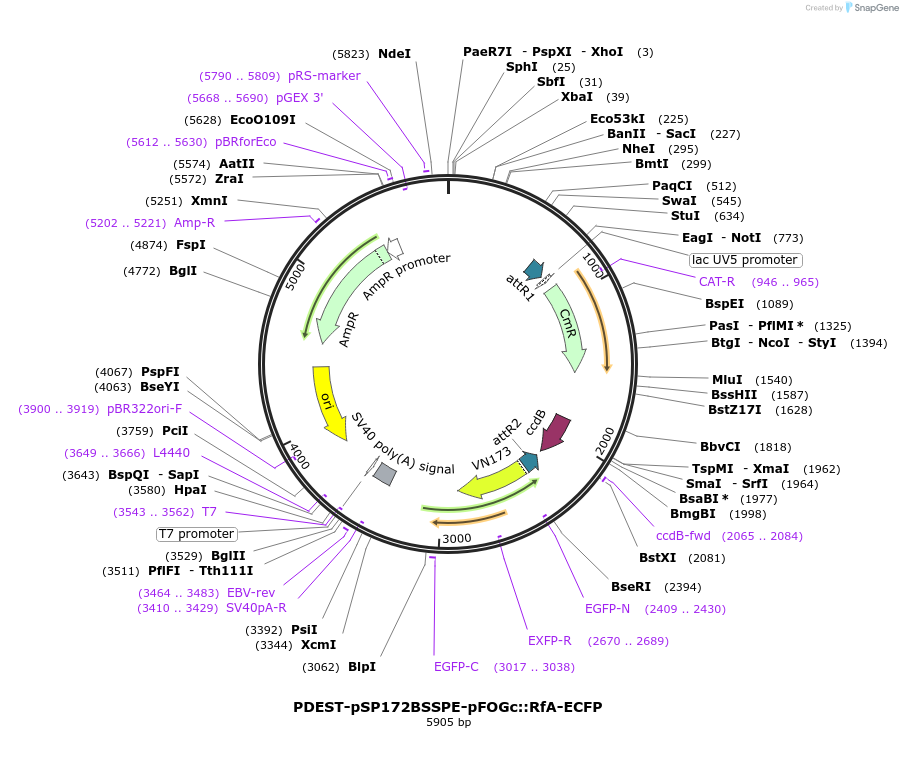
PDEST-pSP172BSSPE-pFOGc::RfA-ECFP
Plasmid#186403PurposeDestination vector for ascidian transgenesis by electroporation of a Gateway-cloned fusion of a gene of interest with a C-ter ECFP under control of the pFOG ectodermal regulatory sequence.DepositorTypeEmpty backboneUseGateway destination vectorAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only