We narrowed to 15,478 results for: pET
-
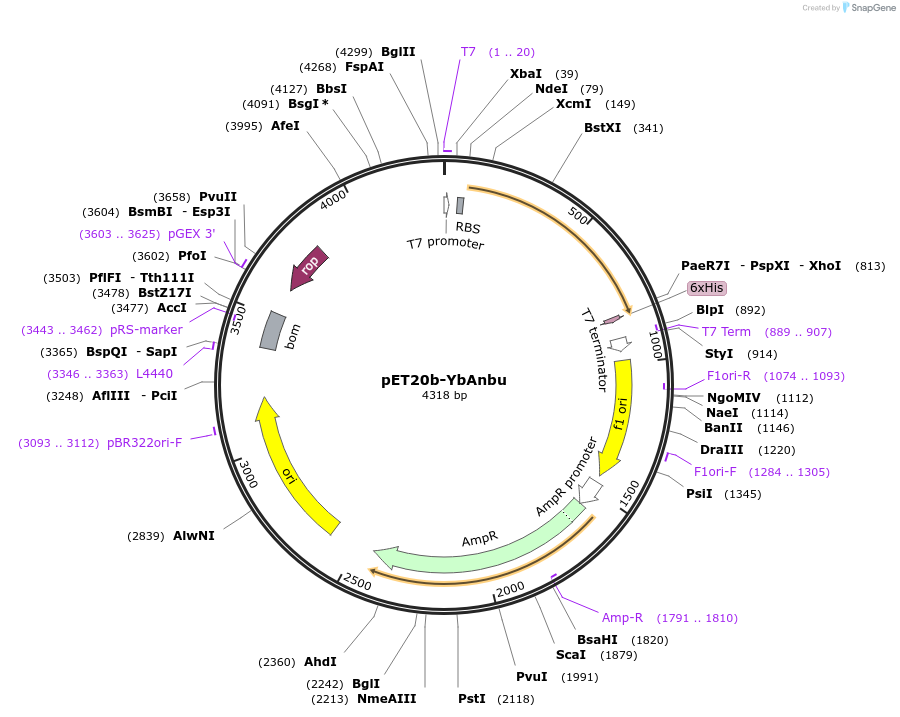
Plasmid#129637PurposeExpression plasmid of Anbu from Y. bercovieri (UNIPROT A0A0T9RXH3 variant with V14A and G107H mutations)DepositorInsertYbAnbu
TagsHis6-tagExpressionBacterialMutationV14A and G107HPromoterT7Available SinceNov. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET20b-YbAnbuT1A
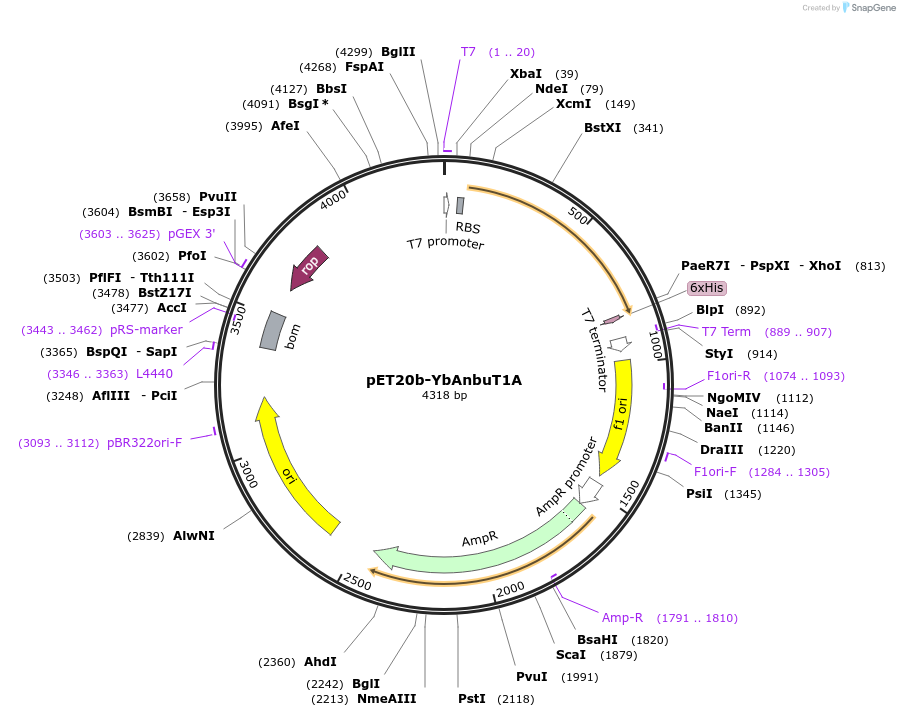
Plasmid#129638PurposeExpression plasmid of Anbu from Y. bercovieri (inactive mutant; (UNIPROT A0A0T9RXH3 variant with T1A, V14A and G107H mutations)DepositorInsertYbAnbuT1A
TagsHis6-tagExpressionBacterialMutationT1A, V14A and G107HPromoterT7Available SinceNov. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
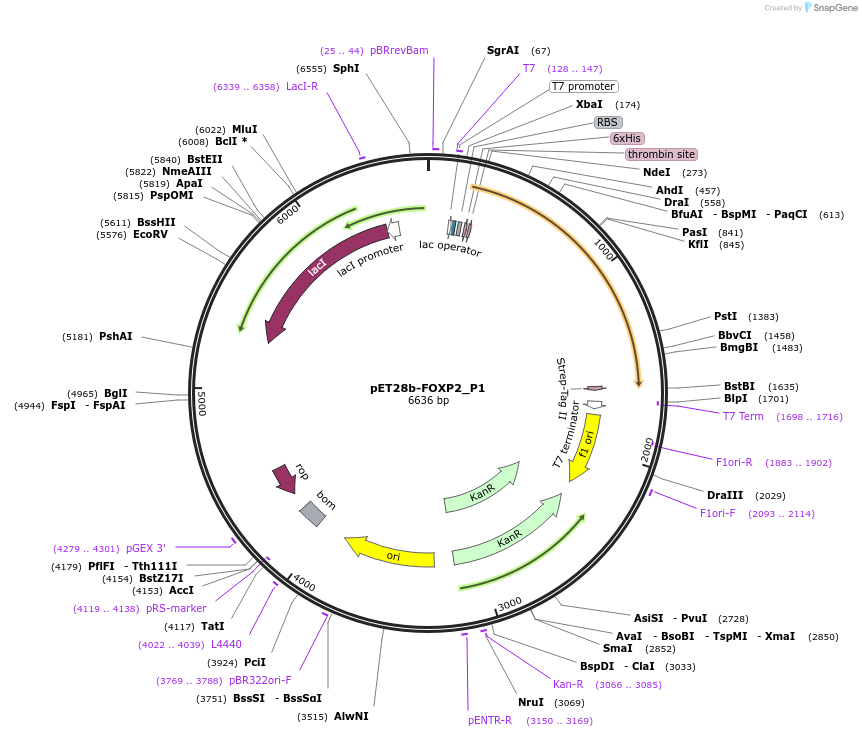
pET28b-FOXP2_P1
Plasmid#122645PurposeExpresses truncated variant (aa271-aa714) of human FOXP2 in bacterial cellsDepositorInsertHomo sapiens forkhead box P2 (FOXP2 Human)
TagsHis-tag and Strep-tag IIExpressionBacterialPromoterT7Available SinceSept. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
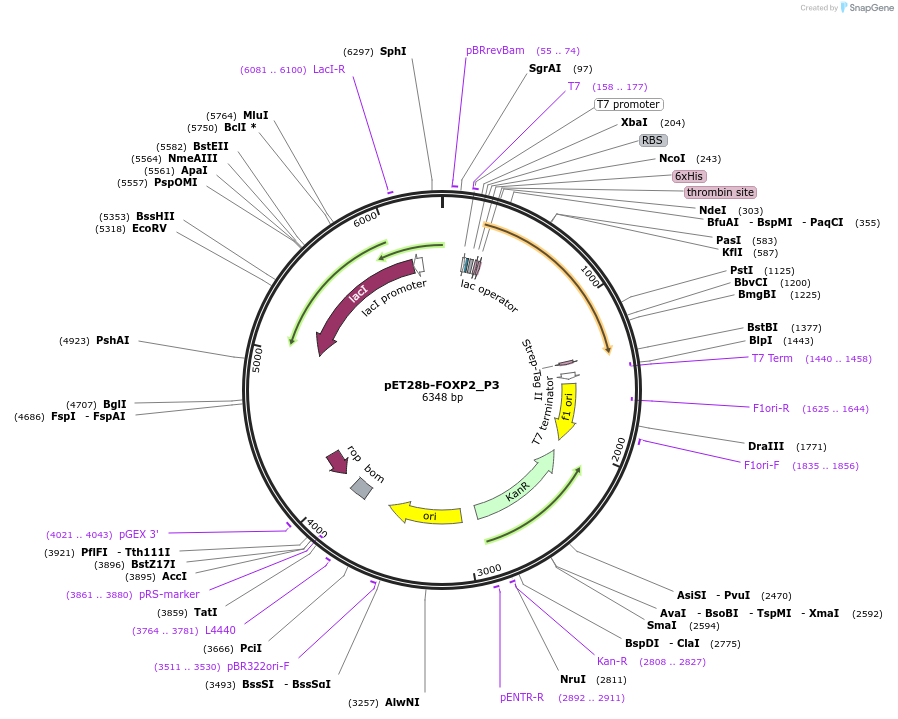
pET28b-FOXP2_P3
Plasmid#122647PurposeExpresses truncated variant (aa367-aa714) of human FOXP2 in bacterial cellsDepositorInsertHomo sapiens forkhead box P2 (FOXP2 Human)
TagsHis-tag and Strep-tag IIExpressionBacterialPromoterT7Available SinceSept. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
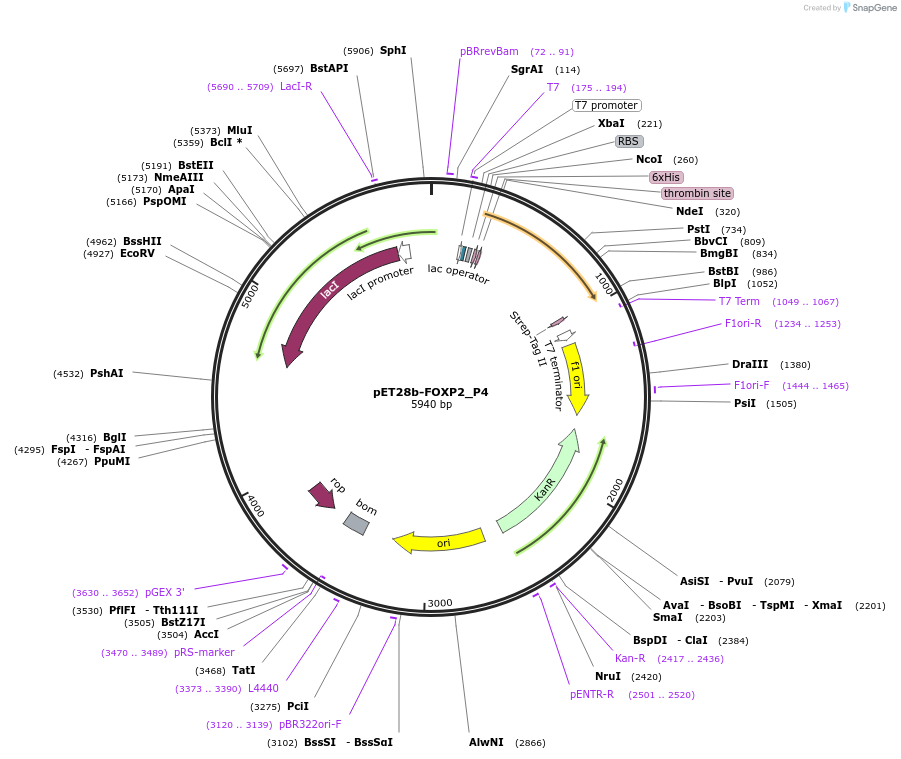
pET28b-FOXP2_P4
Plasmid#122648PurposeExpresses truncated variant (aa503-aa714) of human FOXP2 in bacterial cellsDepositorInsertHomo sapiens forkhead box P2 (FOXP2 Human)
TagsHis-tag and Strep-tag IIExpressionBacterialPromoterT7Available SinceSept. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
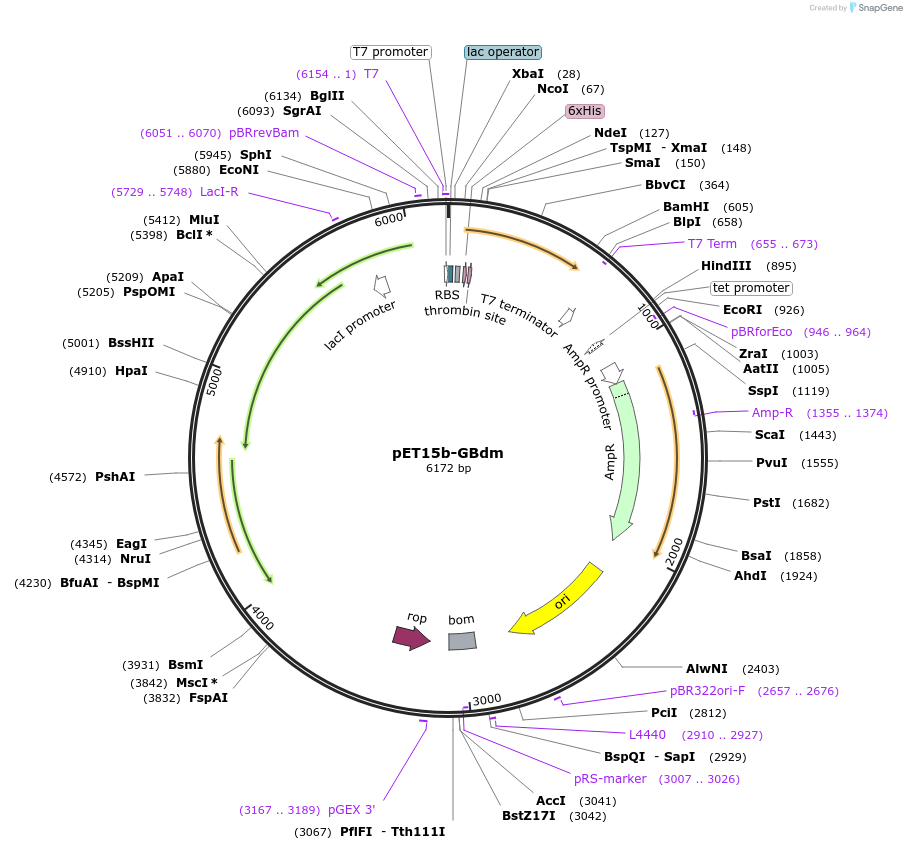
pET15b-GBdm
Plasmid#129690Purposeexpress E. coli GreB E82C/C68SDepositorAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
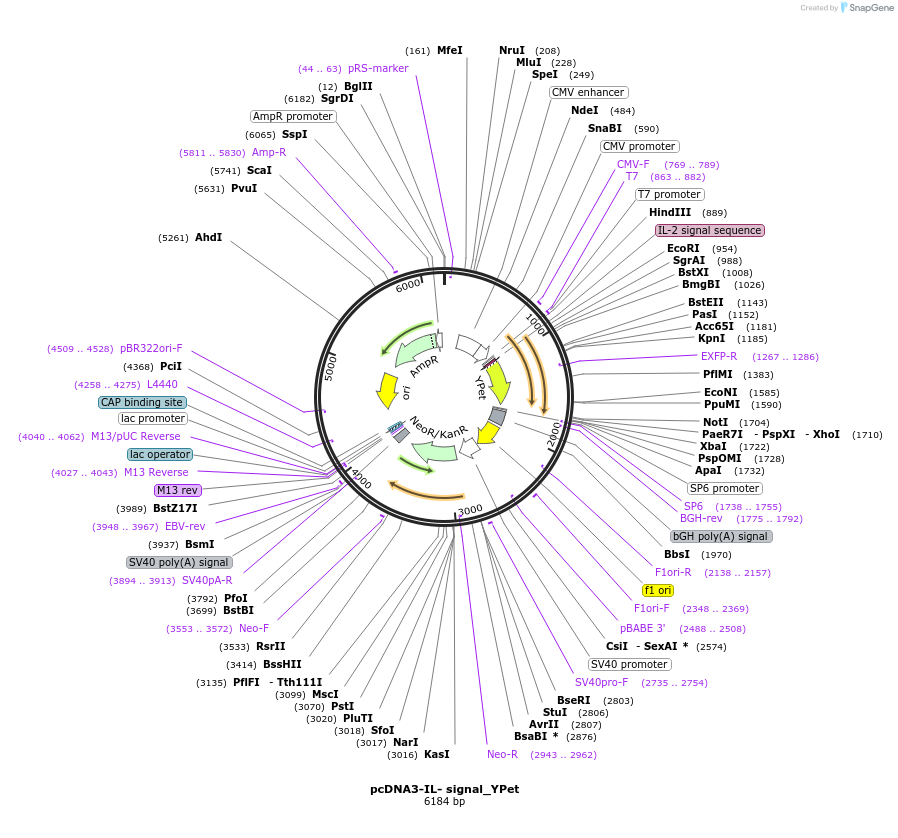
pcDNA3-IL- signal_YPet
Plasmid#128030PurposeMammalian expression vector with N terminal IL-2 signal peptide and YPetDepositorTypeEmpty backboneTagsIL-2 signal peptide and YPetExpressionMammalianAvailable SinceAug. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
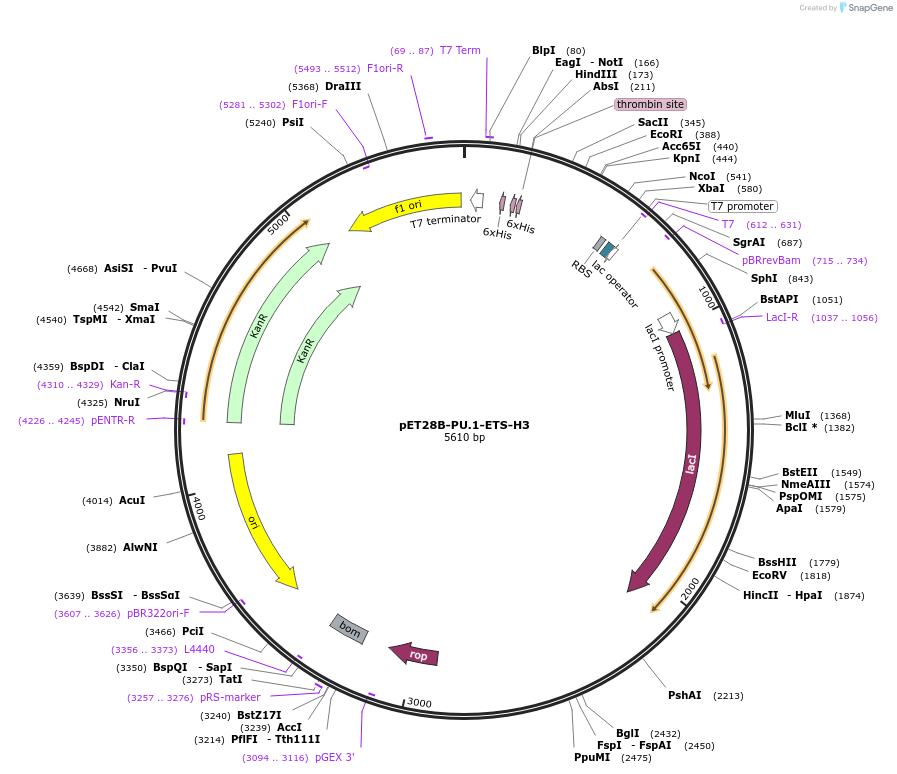
pET28B-PU.1-ETS-H3
Plasmid#124435PurposeChimeric ETS domain: H3 of Ets-1 in PU.1 scaffoldDepositorTags6xHis TagExpressionBacterialMutationResidues 225 to 239 replaced with corresponding s…Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
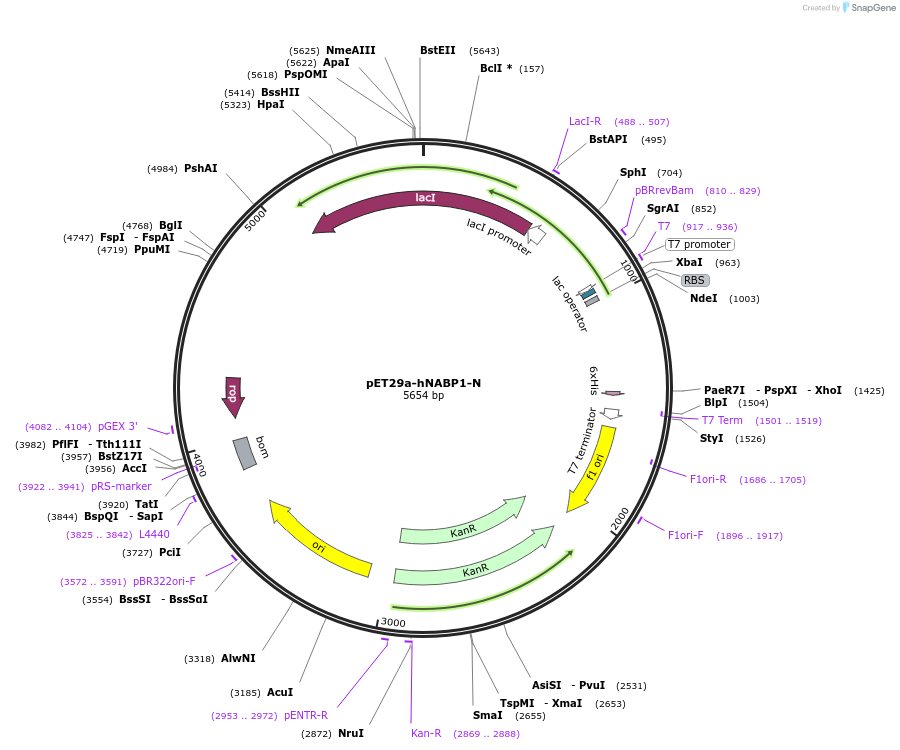
pET29a-hNABP1-N
Plasmid#128310PurposeExpress hNABP1 N-terminus (1–141 aa) in E. coli with a C-terminal His-tagDepositorInserthNABP1 N-terminal domain (OB fold, 1–141 aa) (NABP1 Human)
TagsHisExpressionBacterialMutationN-terminal domain (OB fold, 1–140 aa)PromoterT7Available SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
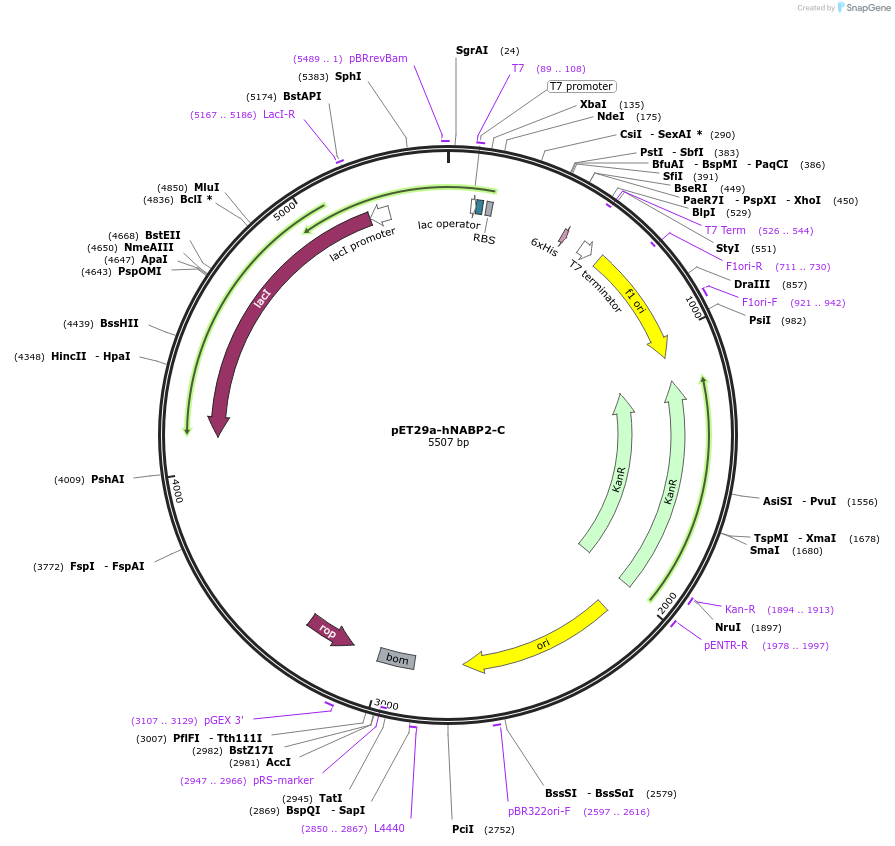
pET29a-hNABP2-C
Plasmid#128309PurposeExpress hNABP2 C-terminus(122–211 aa) in E. coli with a C-terminal His-tagDepositorInserthNABP2 C-terminal domain (122–211 aa) (NABP2 Human)
TagsHisExpressionBacterialMutationC-terminal domain (122–211 aa)PromoterT7Available SinceAug. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
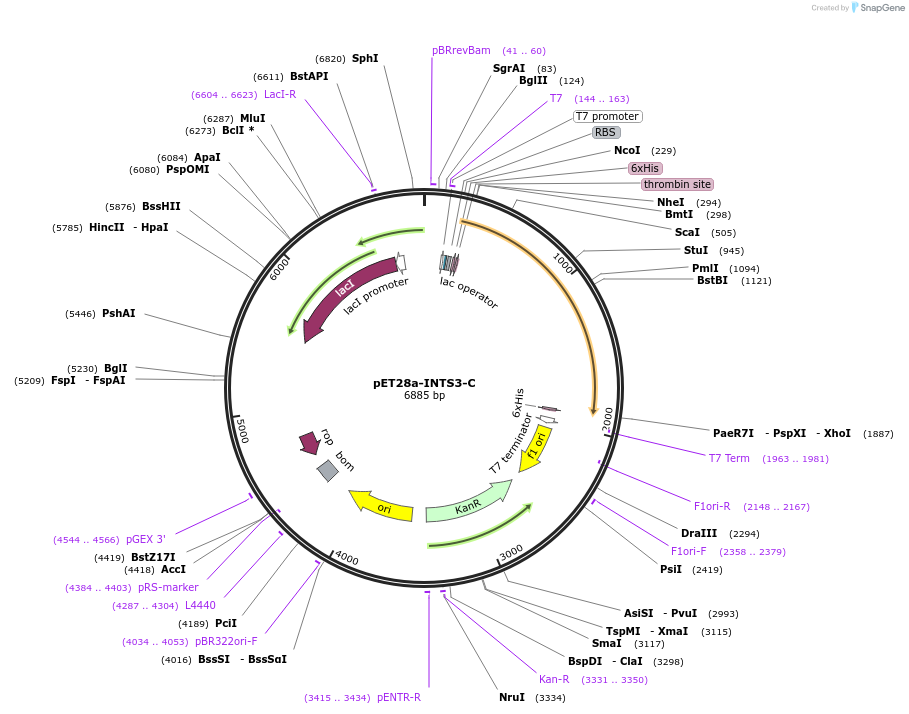
pET28a-INTS3-C
Plasmid#128642PurposeExpress INTS3 C-termini (514-1042 aa) in E. coli with N-terminal and C-terminal His tagDepositorInsertINTS3 C-termini (INTS3 Human)
TagsHis tag on N and C-terminalExpressionBacterialMutationINTS3 C-termini (514–1042 aa)PromoterT7Available SinceAug. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
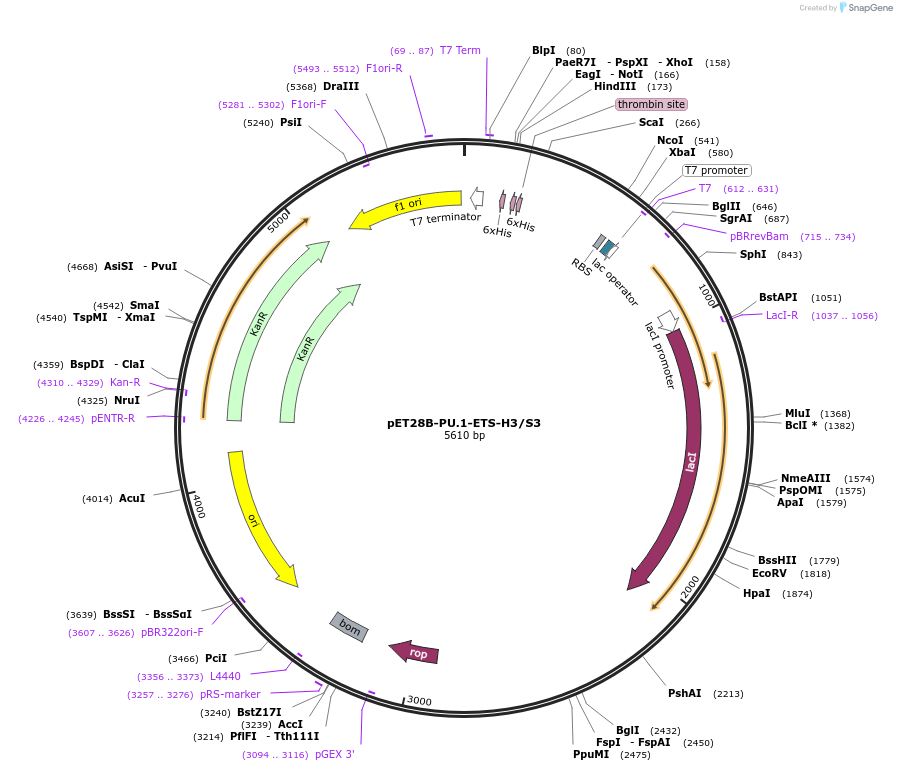
pET28B-PU.1-ETS-H3/S3
Plasmid#124627PurposeChimeric ETS domain: H3/S3 of Ets-1 in PU.1 scaffoldDepositorTags6xHis tagExpressionBacterialMutationResidues 237 to 241 replaced with corresponding s…Available SinceApril 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
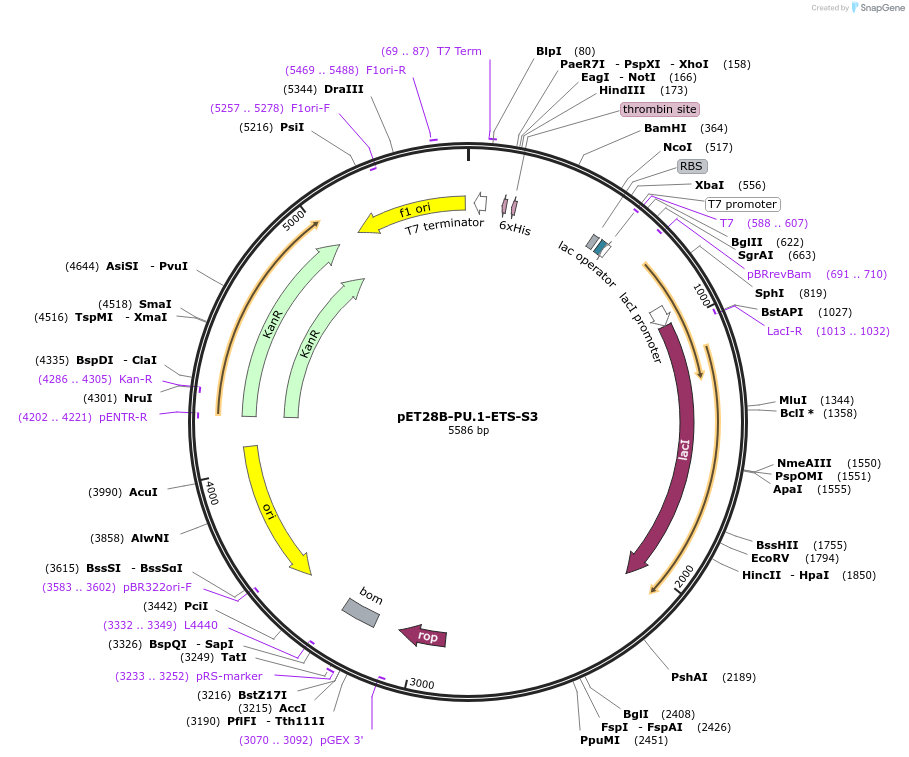
pET28B-PU.1-ETS-S3
Plasmid#124550PurposeChimeric ETS domain: S3 of Ets-1 in PU.1 scaffoldDepositorTags6xHis tagExpressionBacterialMutationResidues 242 to 246 replaced with corresponding s…Available SinceApril 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
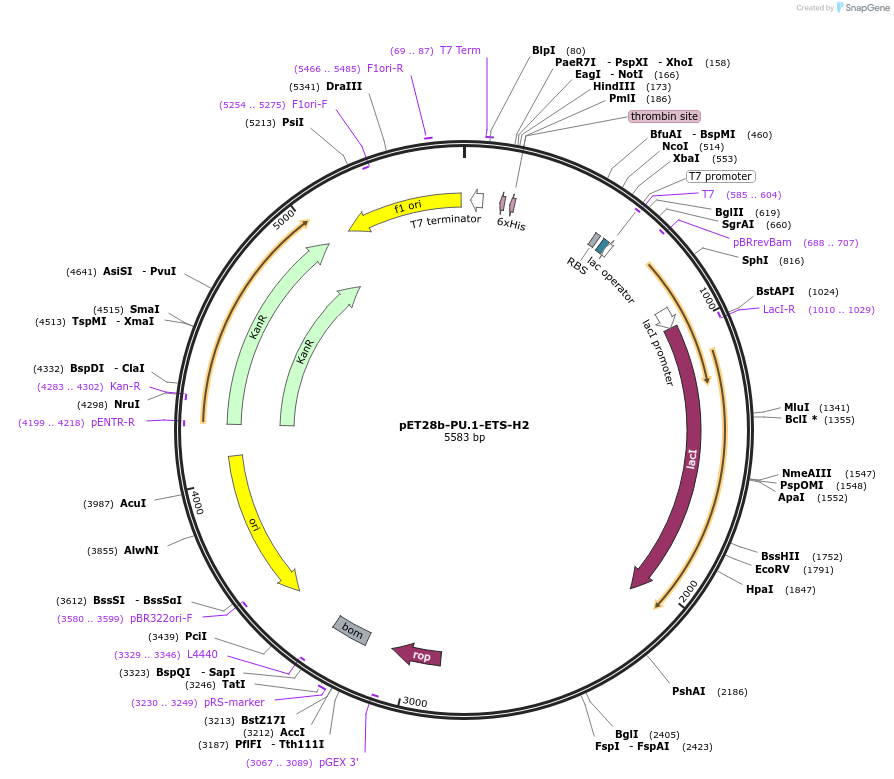
pET28b-PU.1-ETS-H2
Plasmid#123358PurposeChimeric ETS domain: H2 of Ets-1 in PU.1 scaffoldDepositorTags6xHis tagExpressionBacterialMutationResidues 207 to 215 replaced with corresponding s…Available SinceApril 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
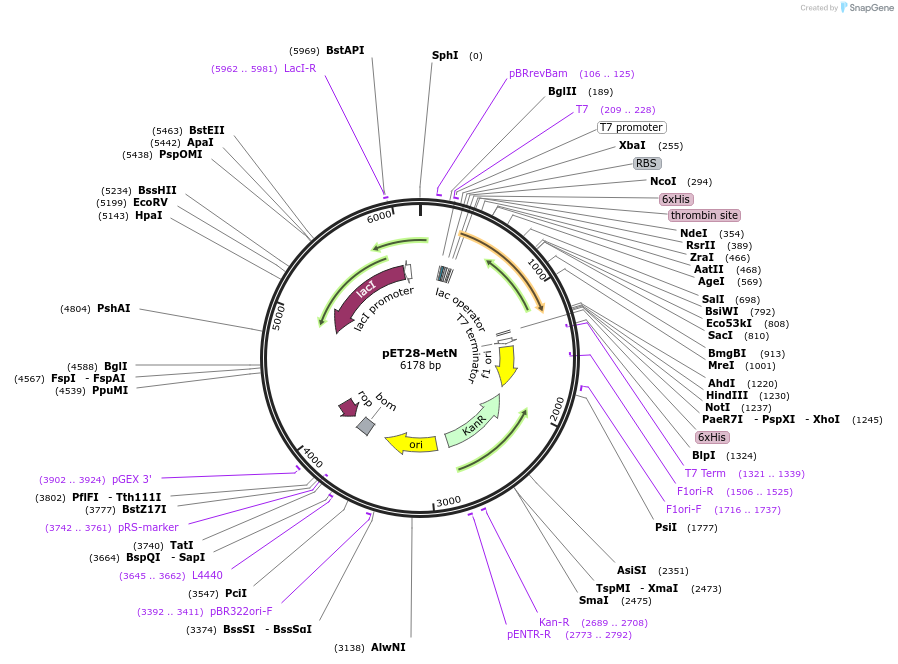
pET28-MetN
Plasmid#114458PurposeExpression of hexahistidine-tagged AceS methylase in E. coliDepositorInsertaceS
TagsHexahistidineExpressionBacterialPromoterT7Available SinceFeb. 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
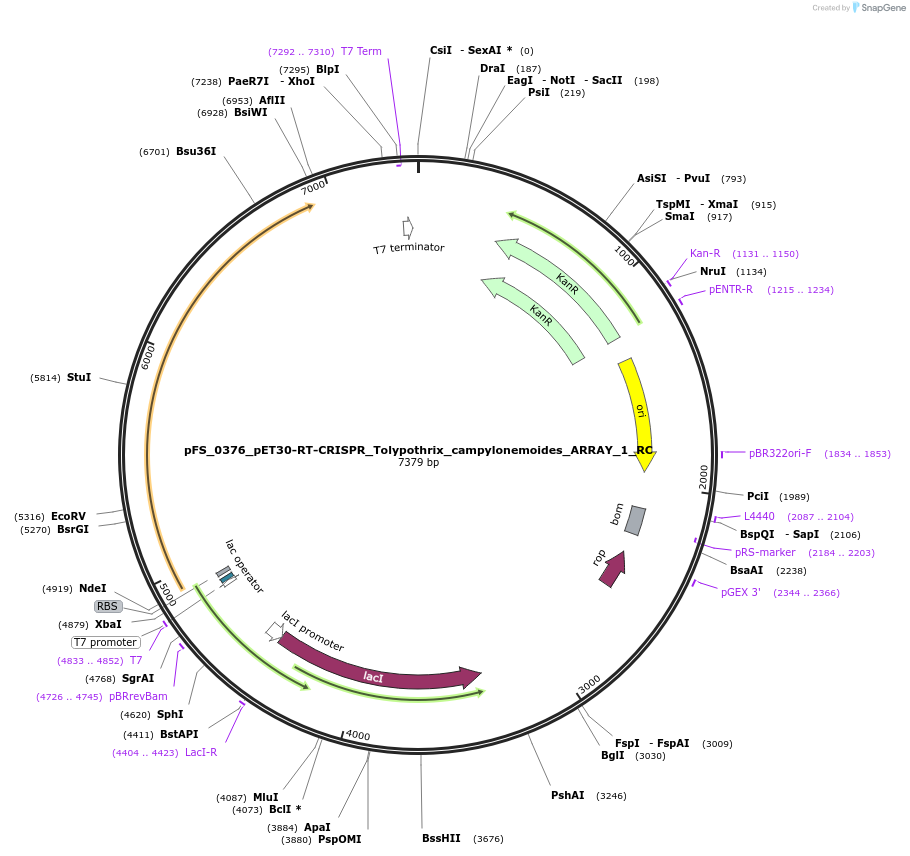
pFS_0376_pET30-RT-CRISPR_Tolypothrix_campylonemoides_ARRAY_1_RC
Plasmid#116988PurposeExpresses TcRT-Cas1-Cas2 under pT7lac promoter, encodes TcCRISPR array 1 RC, compatible with SENECA acquisition readoutDepositorInsertTolypothrix campylonemoides RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceDec. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
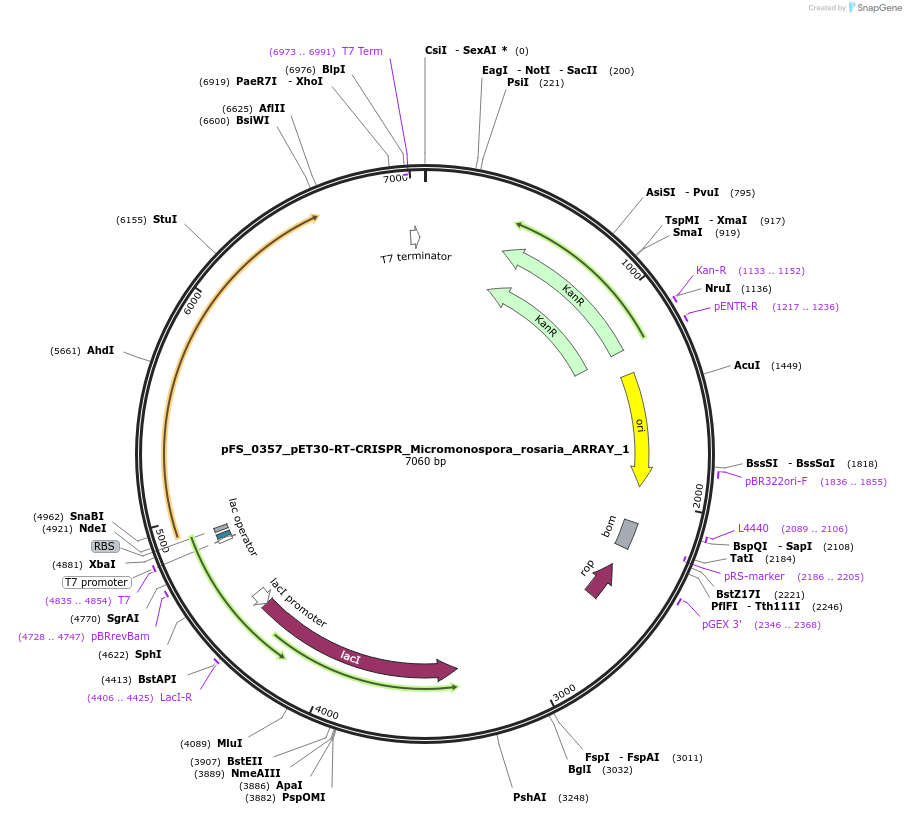
pFS_0357_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_1
Plasmid#116969PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 1, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
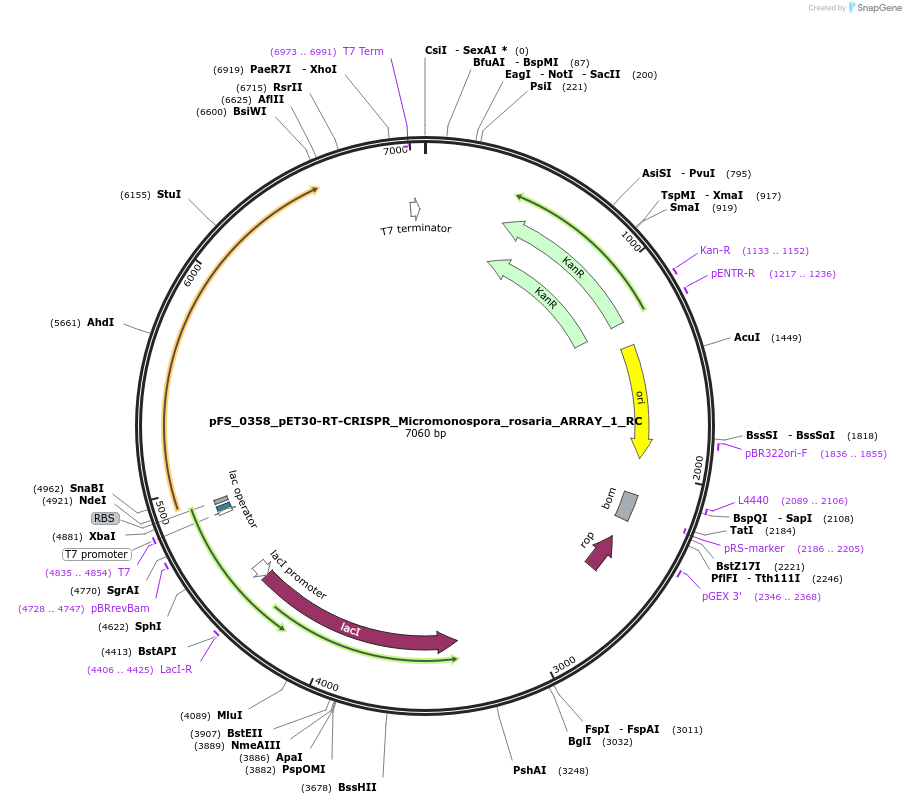
pFS_0358_pET30-RT-CRISPR_Micromonospora_rosaria_ARRAY_1_RC
Plasmid#116970PurposeExpresses MrRT-Cas1-Cas2 under pT7lac promoter, encodes MrCRISPR array 1 RC, compatible with SENECA acquisition readoutDepositorInsertMicromonospora rosaria RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
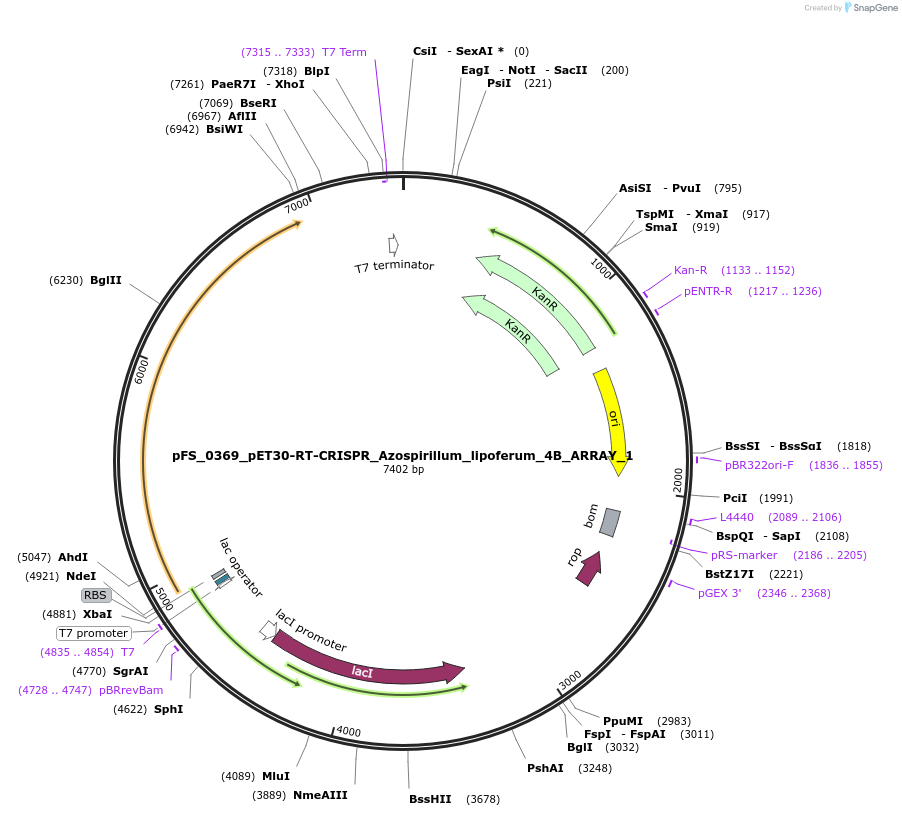
pFS_0369_pET30-RT-CRISPR_Azospirillum_lipoferum_4B_ARRAY_1
Plasmid#116981PurposeExpresses AlRT-Cas1-Cas2 under pT7lac promoter, encodes AlCRISPR array 1, compatible with SENECA acquisition readoutDepositorInsertAzospirillum lipoferum 4B RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
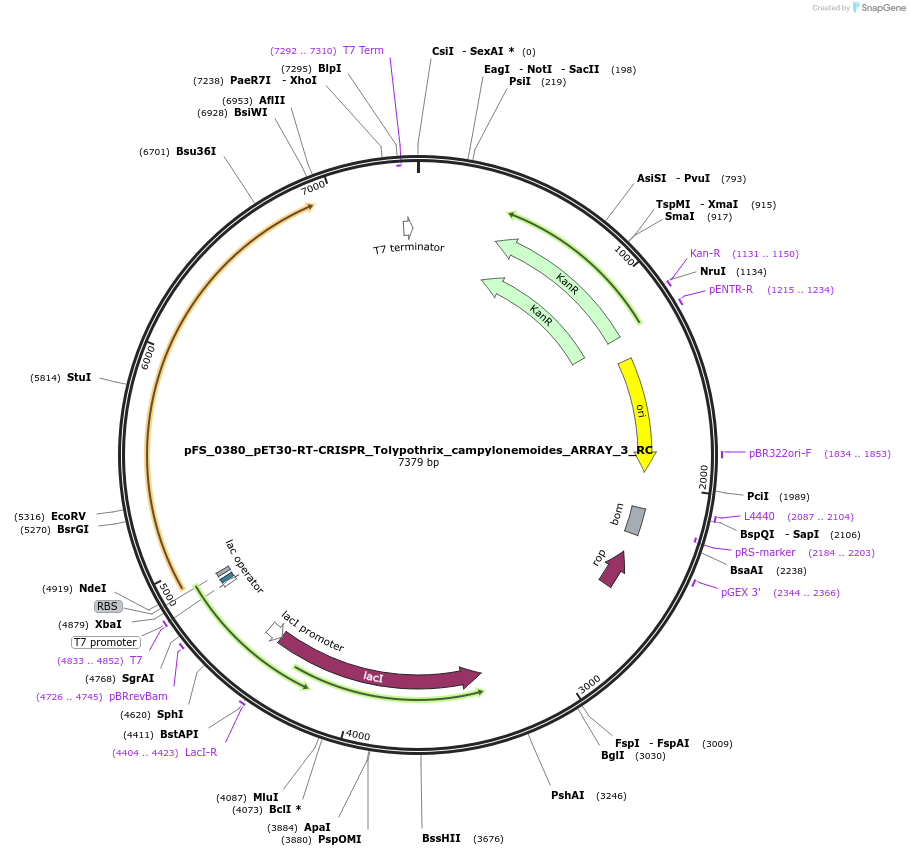
pFS_0380_pET30-RT-CRISPR_Tolypothrix_campylonemoides_ARRAY_3_RC
Plasmid#116992PurposeExpresses TcRT-Cas1-Cas2 under pT7lac promoter, encodes TcCRISPR array 3 RC, compatible with SENECA acquisition readoutDepositorInsertTolypothrix campylonemoides RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 29, 2018AvailabilityAcademic Institutions and Nonprofits only