We narrowed to 1,917 results for: TRIM
-
Plasmid#40848PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–Bartel
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-EGFP-4xlet-7 (pAW-AS27)
Plasmid#40834PurposemiRNA sensor used in Drosophila cell linesDepositorInsertFour target sites perfectly complementary to let-7
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-GFP-2xmiR-34 (pAW-AS-25)
Plasmid#40833PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-34
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-EGFP-2xmiR-1 (pAW-AS-21)
Plasmid#40832PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-1
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-8 (seed1-8)
Plasmid#40843PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–seed1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-7 (mm12-21)
Plasmid#40842PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm12-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-6 (mm17-21)
Plasmid#40841PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm17-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-5 (mm18-21)
Plasmid#40840PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm18-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-4 (mm19-21)
Plasmid#40839PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm19-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-3 (mm20-21)
Plasmid#40838PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm20-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-16 (mm14-21)
Plasmid#40851PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm14-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-15 (mm15-21)
Plasmid#40850PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm15-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS14 (mm16-21)
Plasmid#40849PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm16-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-12 (mm1-8)
Plasmid#40847PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-11 (bulge 9-15)
Plasmid#40846PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge9-15
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-10 (bulge 9-13)
Plasmid#40845PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-13
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-9 (bulge 9-11)
Plasmid#40844PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-11
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
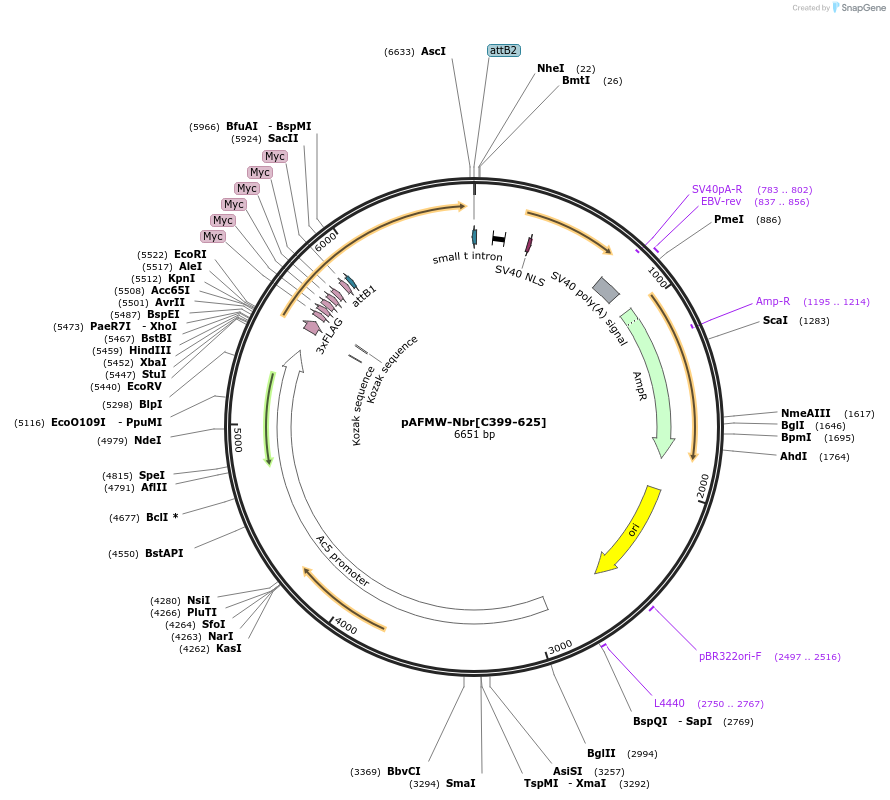
pAFMW-Nbr[C399-625]
Plasmid#188587PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorInsertNibbler C-terminal domain
TagsFLAG-MycExpressionInsectAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
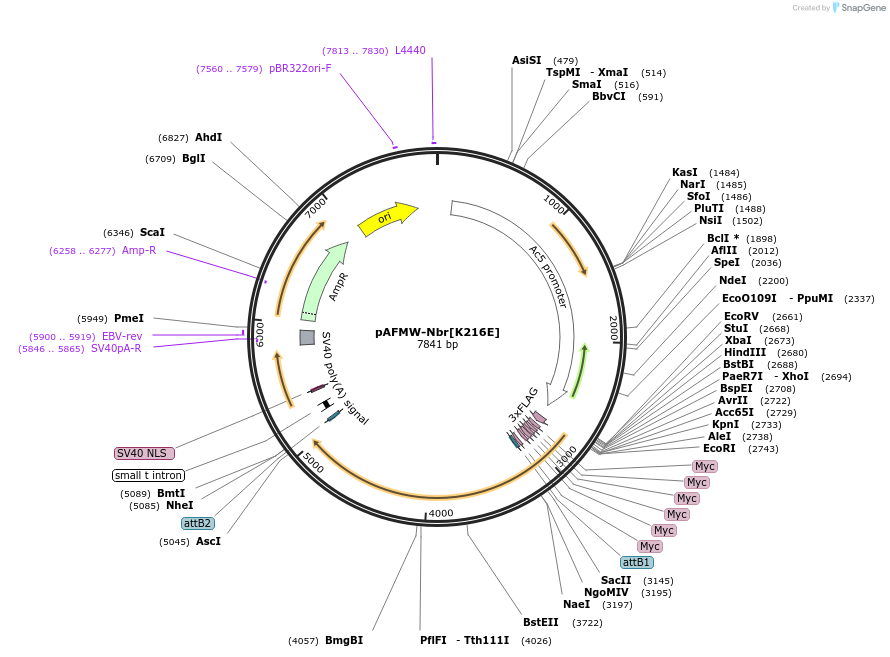
pAFMW-Nbr[K216E]
Plasmid#188600PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
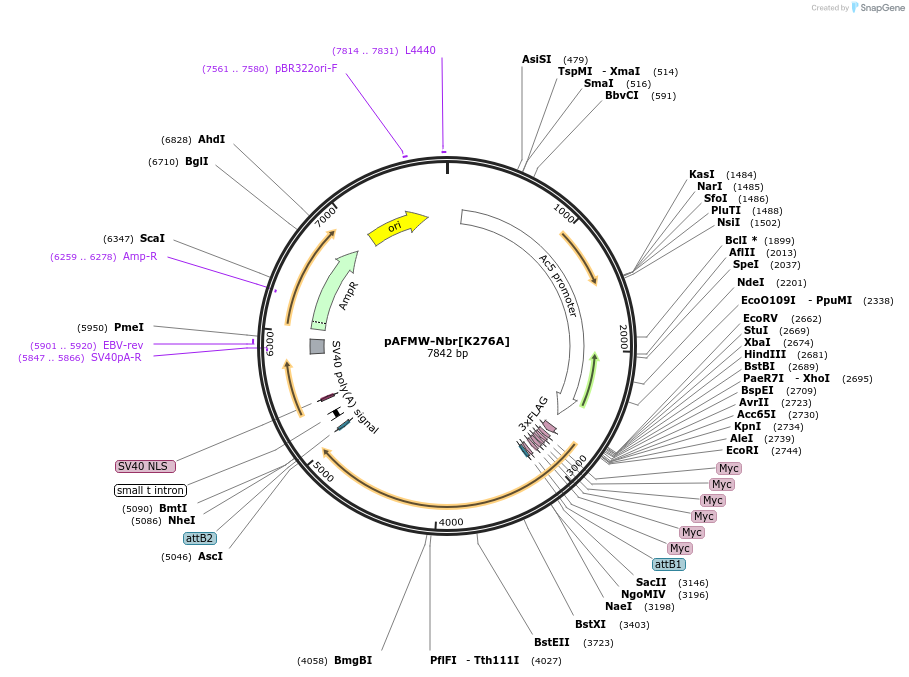
pAFMW-Nbr[K276A]
Plasmid#188597PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only