We narrowed to 3,313 results for: phage
-
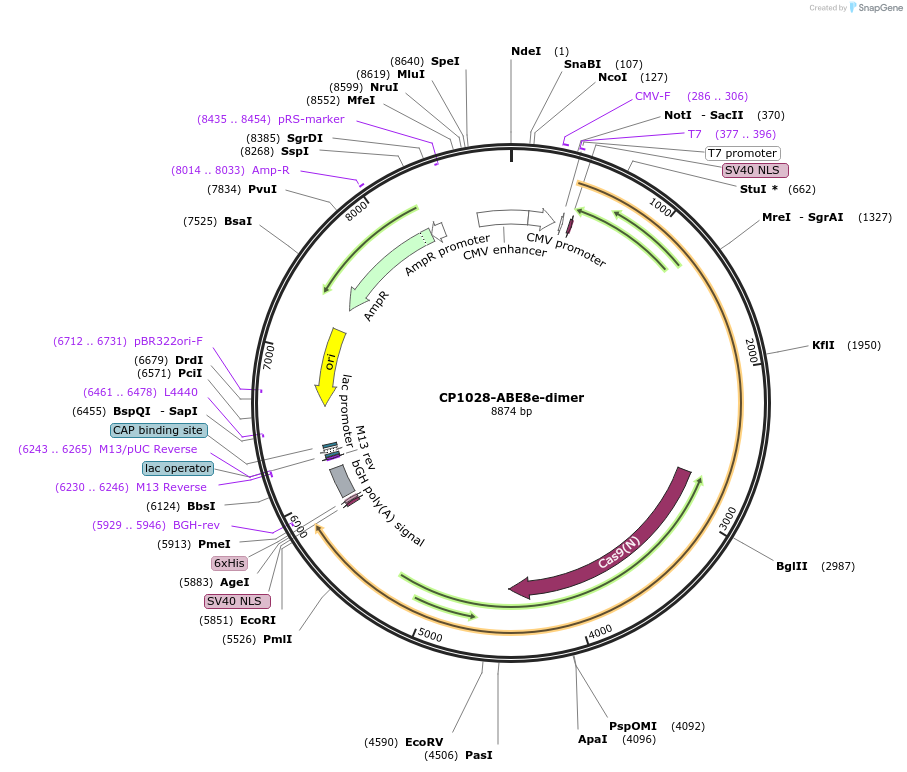
Plasmid#138498PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-nSpCas9 (CP1028)
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPSP6+t-gfp
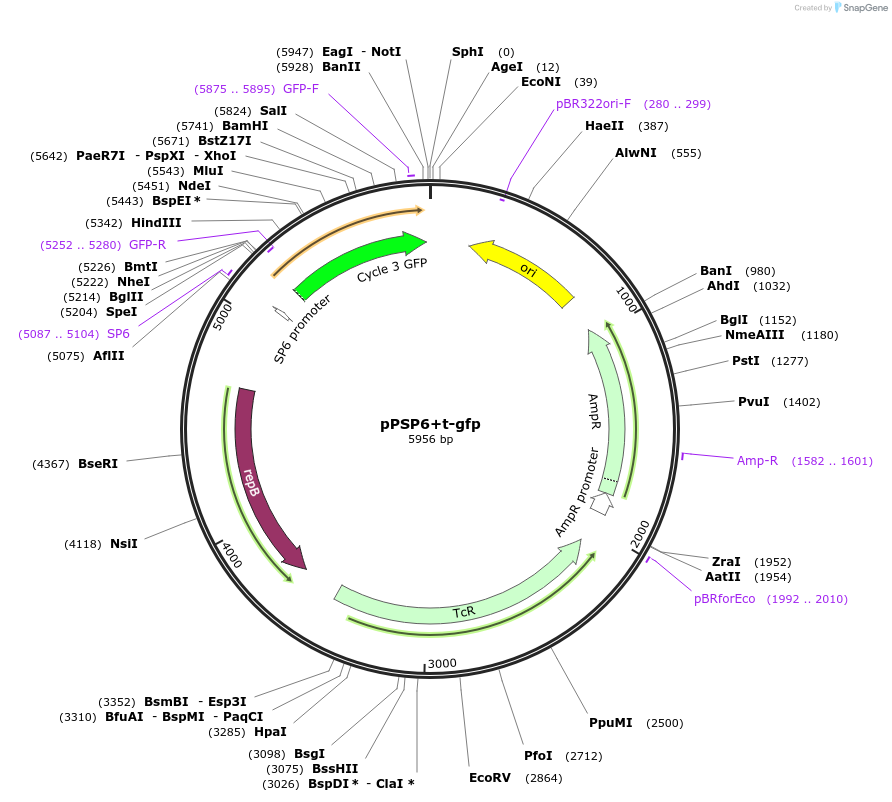
Plasmid#48130PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-SP6-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
SaKKH-ABE8e-dimer
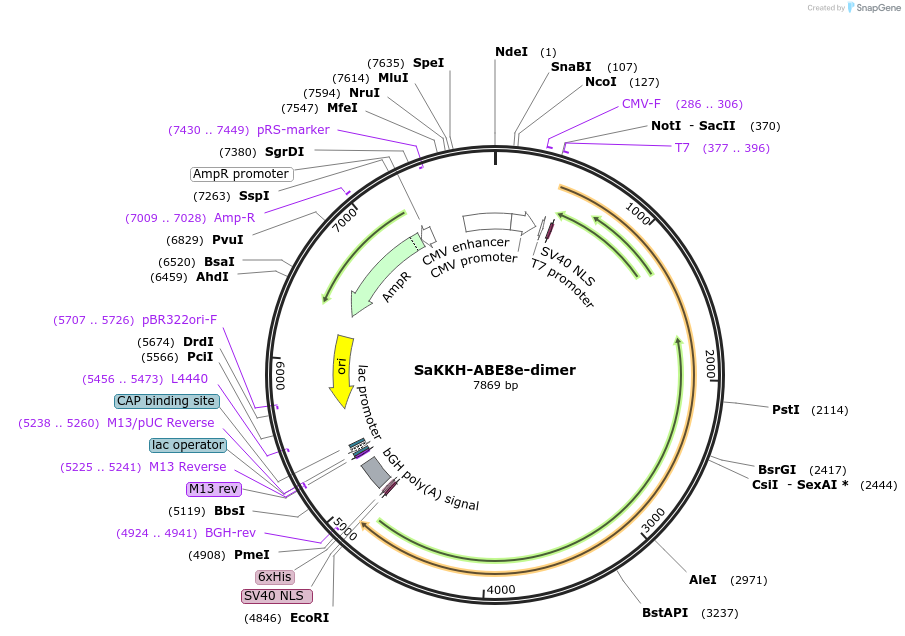
Plasmid#138503PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-nSaCas9 (KKH)
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
CP1041-ABE8e-dimer
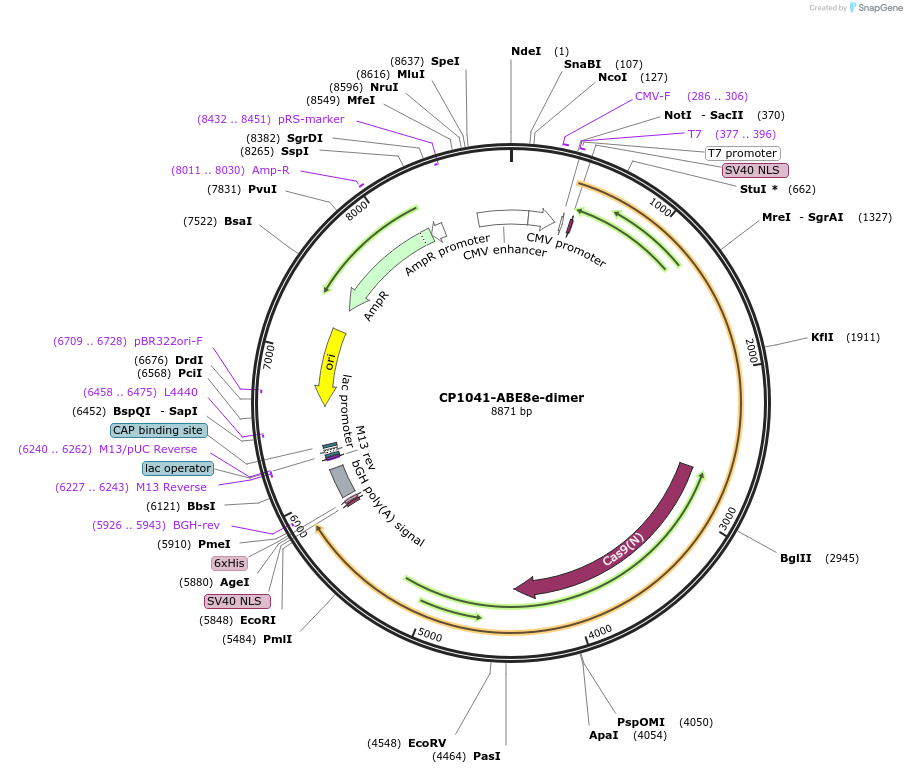
Plasmid#138499PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-nSpCas9 (CP1041)
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
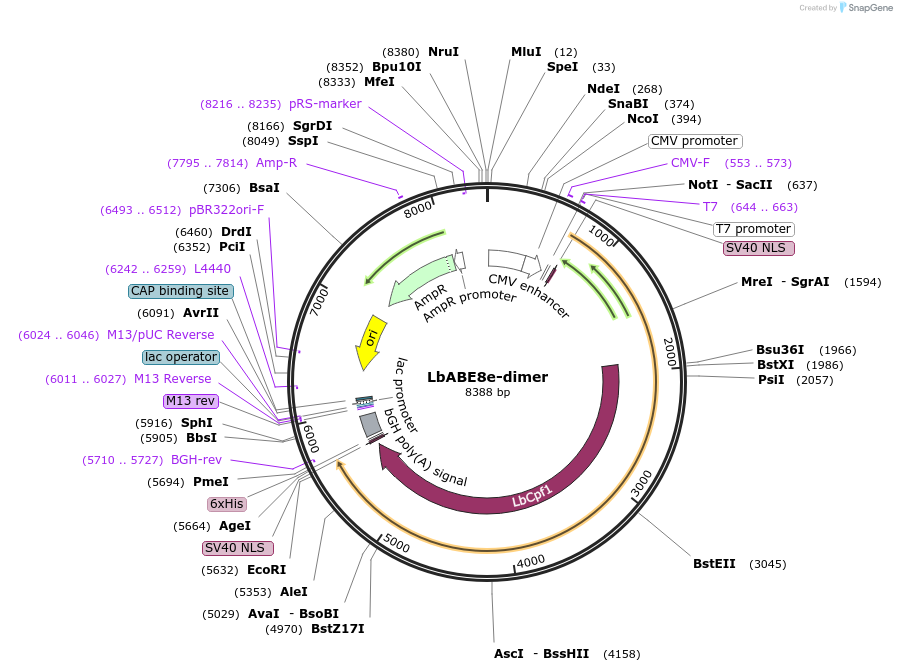
LbABE8e-dimer
Plasmid#138505PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-dLbCas12a
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
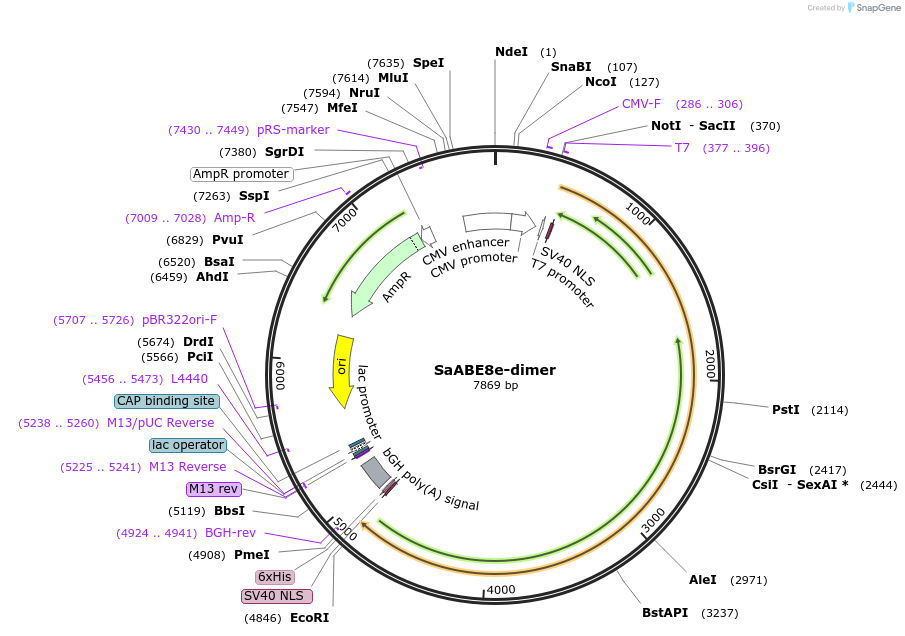
SaABE8e-dimer
Plasmid#138501PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-nSaCas9
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
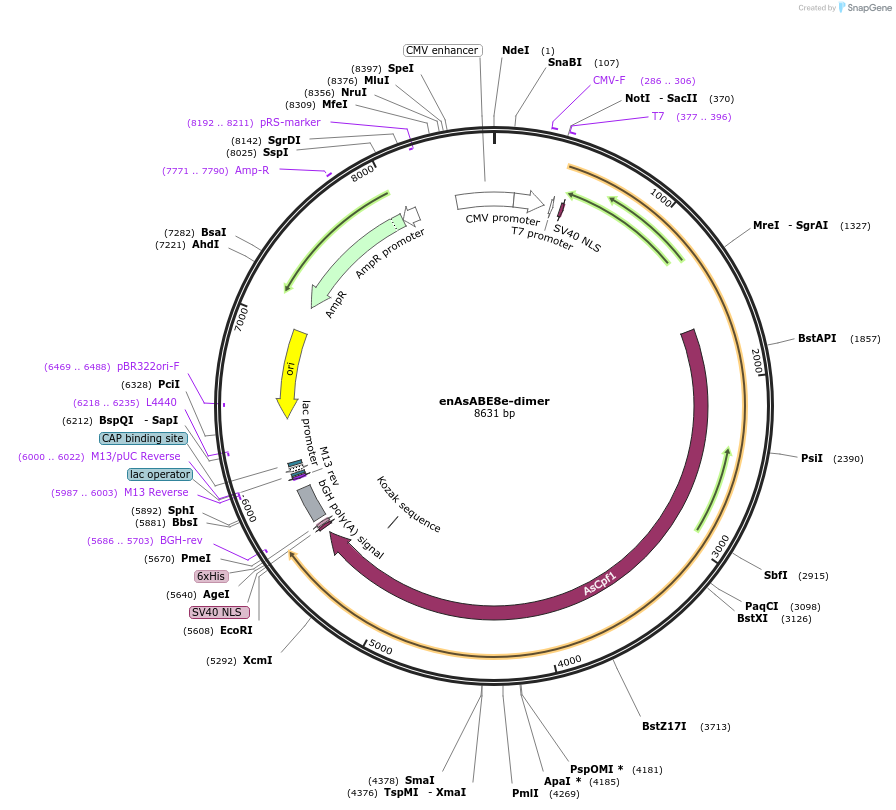
enAsABE8e-dimer
Plasmid#138507PurposeA-to-G base editorDepositorInsertecTadA(WT)-ecTadA(8e)-dAsCas12a (En)
ExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
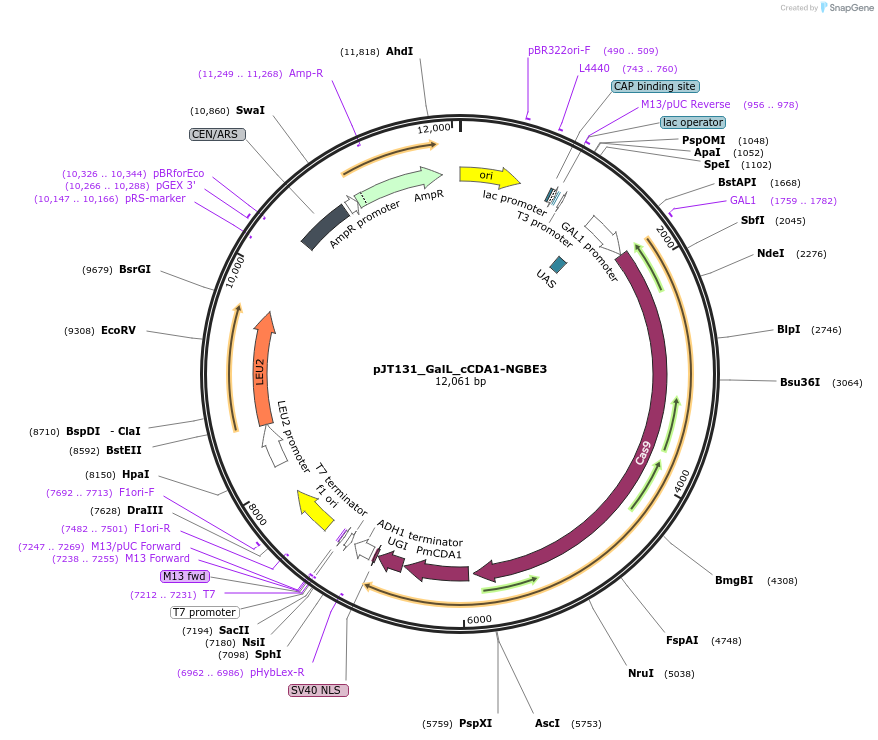
pJT131_GalL_cCDA1-NGBE3
Plasmid#145095PurposeExpressing base editor cCDA1-NGBE3 in yeast cellsDepositorInsertcCDA1-NGBE3
UseCRISPRExpressionYeastMutationspCas9(D10A/L1111R/D1135V/G1218R/E1219F/A1322R/R1…PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
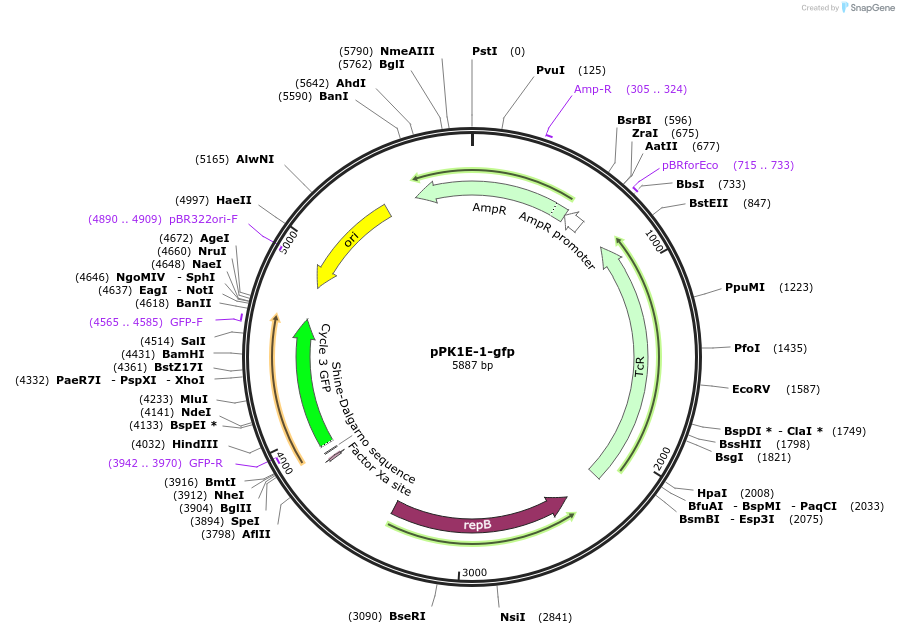
pPK1E-1-gfp
Plasmid#48133PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
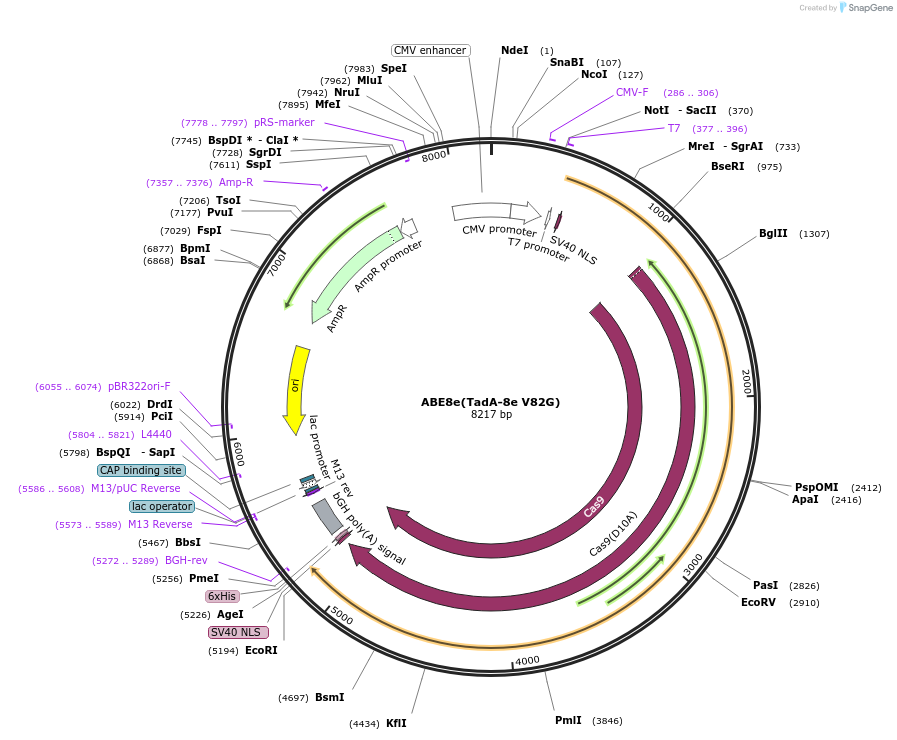
ABE8e(TadA-8e V82G)
Plasmid#138494PurposeA-to-G base editorDepositorInsertecTadA(8e V82G)-nSpCas9
ExpressionMammalianMutationTadA-8e V82GAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
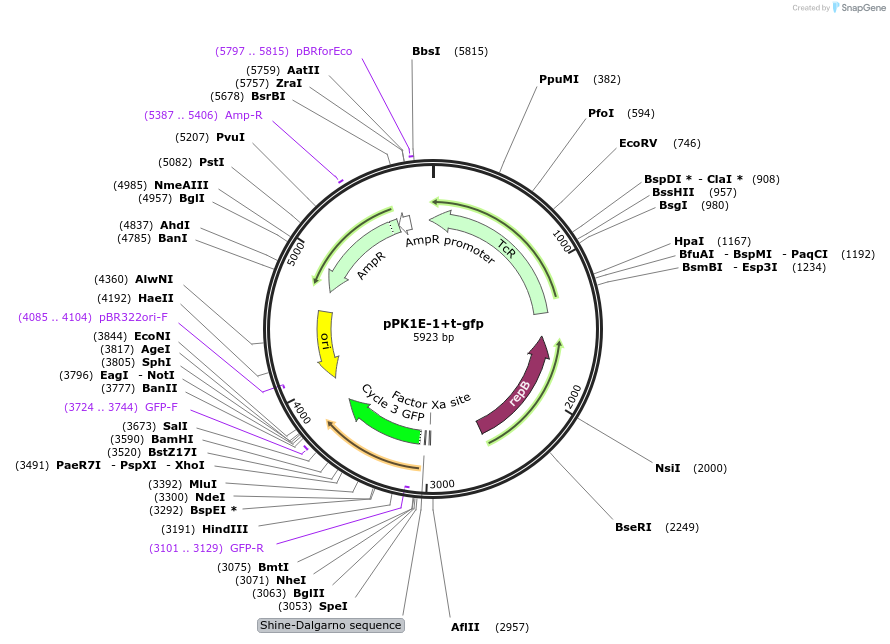
pPK1E-1+t-gfp
Plasmid#48131PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
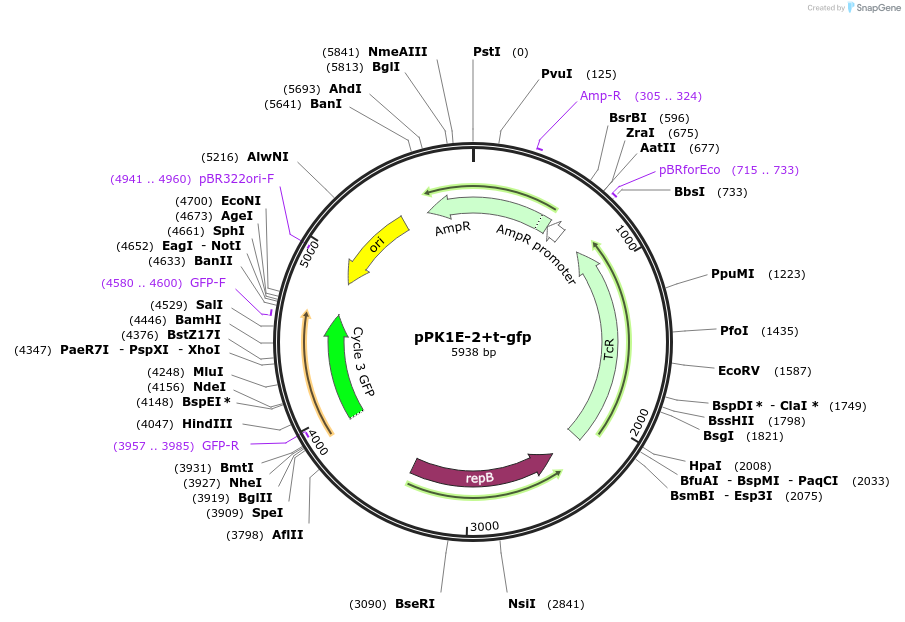
pPK1E-2+t-gfp
Plasmid#48132PurposeShuttle vector E. coli/B.meg.; gfp-expression under control of RNAP-K1E-inducible promoterDepositorInsertgfp
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
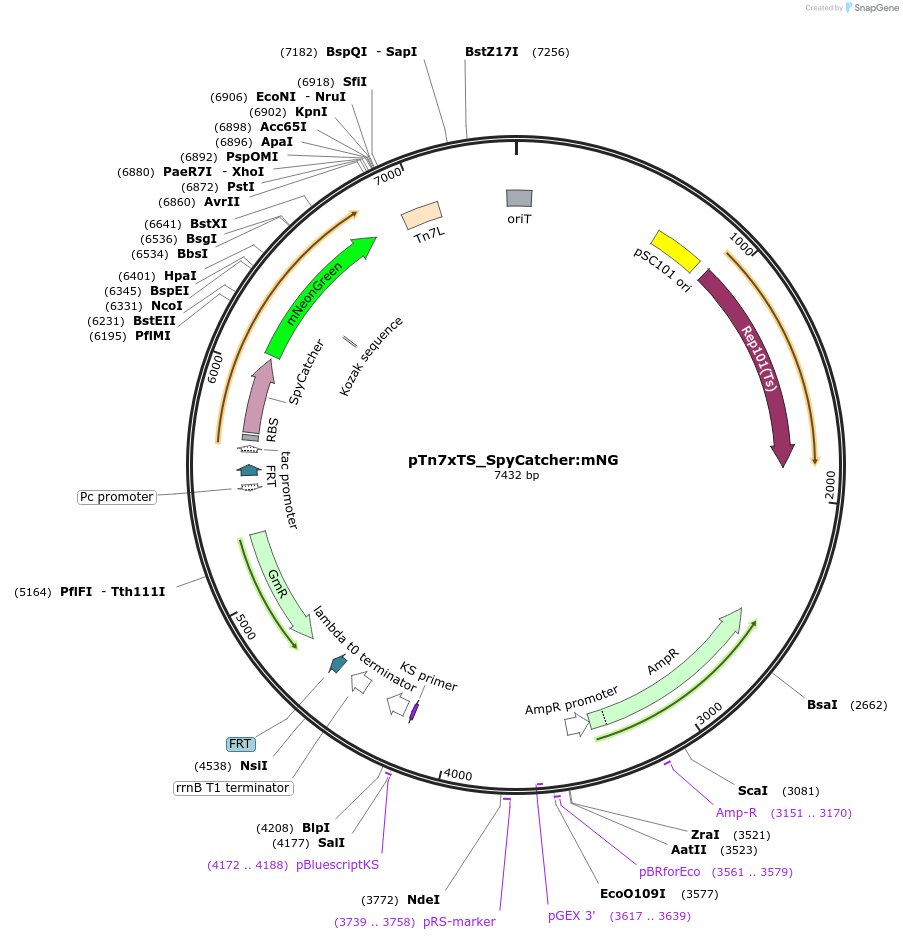
pTn7xTS_SpyCatcher:mNG
Plasmid#225185PurposeUsed for inserting SpyCatcher fused to mNeonGreen at the bacterial Tn7 siteDepositorInsertSpyCatcher:mNeonGreen
TagsSpyCatcherExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
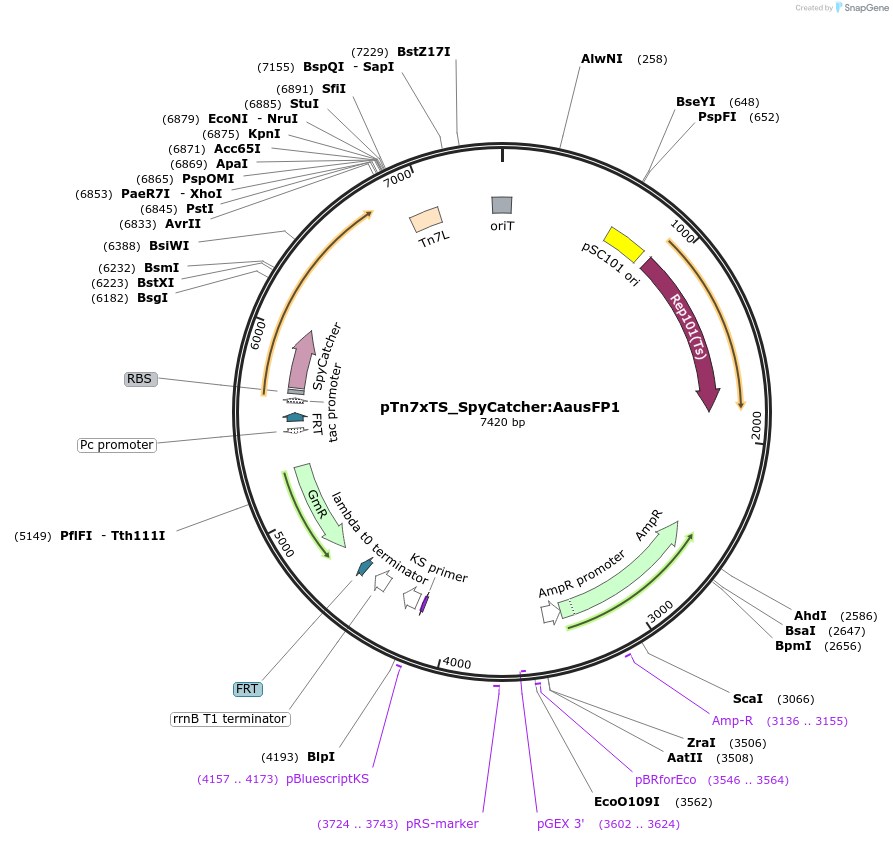
pTn7xTS_SpyCatcher:AausFP1
Plasmid#225186PurposeUsed for inserting SpyCatcher fused to AausFP1 at the bacterial Tn7 siteDepositorInsertSpyCatcher:AausFP1
TagsSpyCatcherExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
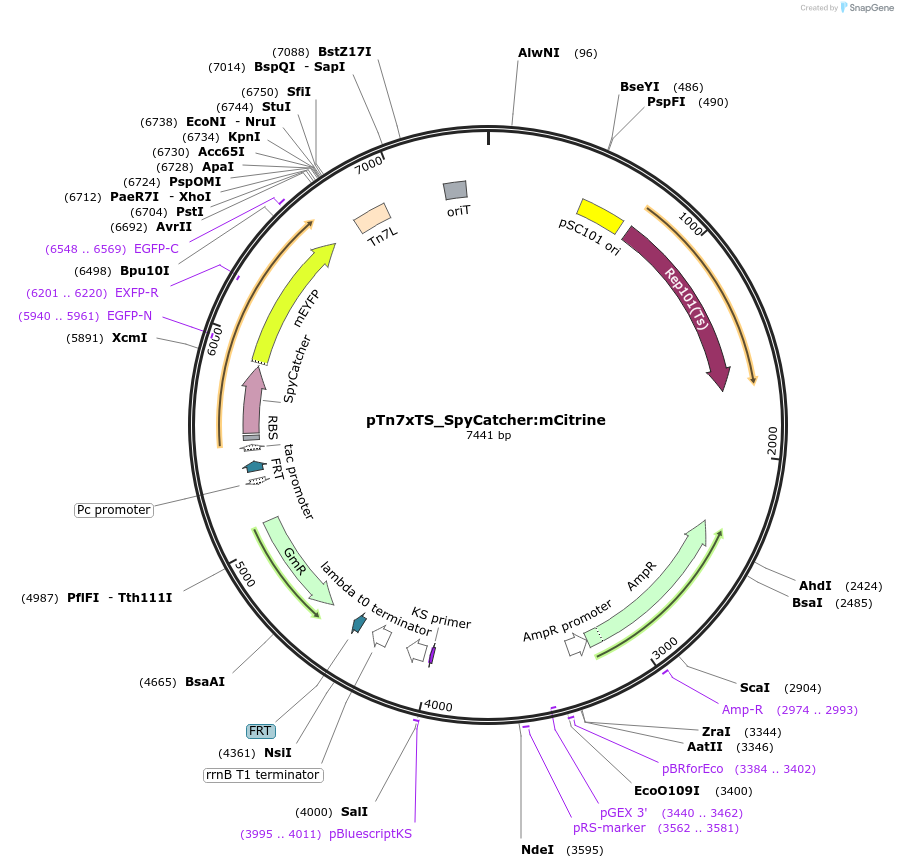
pTn7xTS_SpyCatcher:mCitrine
Plasmid#225187PurposeUsed for inserting SpyCatcher fused to mCitrine at the bacterial Tn7 siteDepositorInsertSpyCatcher:mCitrine
TagsSpyCatcherExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
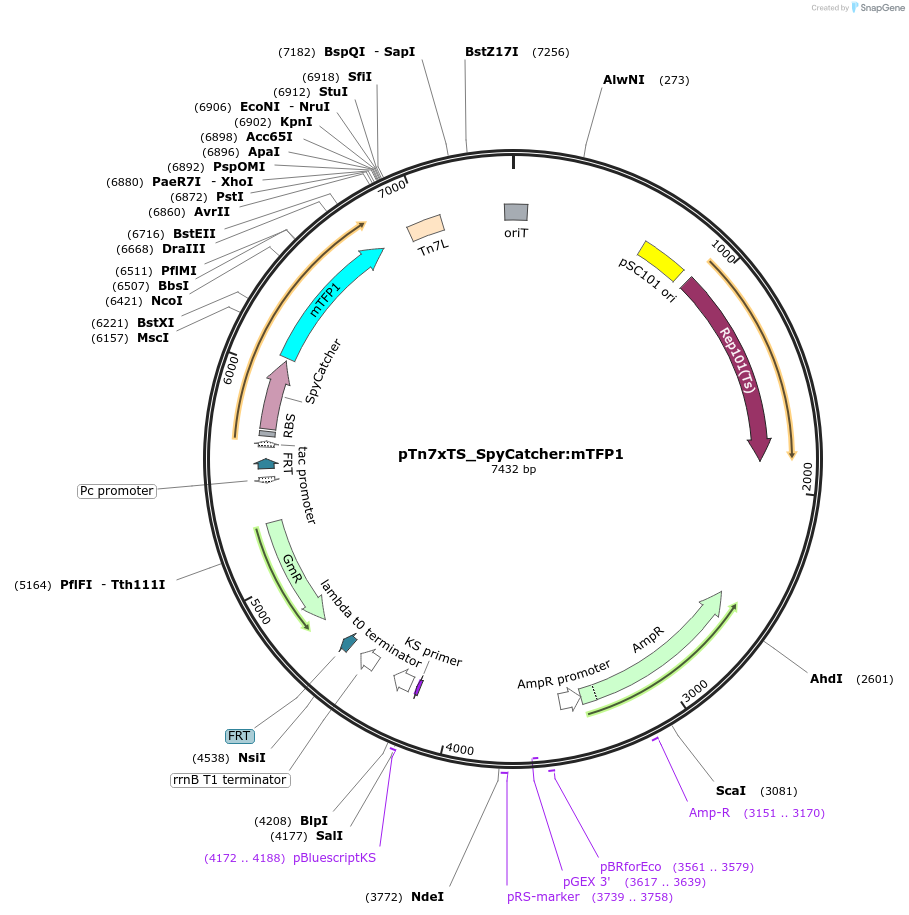
pTn7xTS_SpyCatcher:mTFP1
Plasmid#225189PurposeUsed for inserting SpyCatcher fused to mTFP1 at the bacterial Tn7 siteDepositorInsertSpyCatcher:mTFP1
TagsSpyCatcherExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
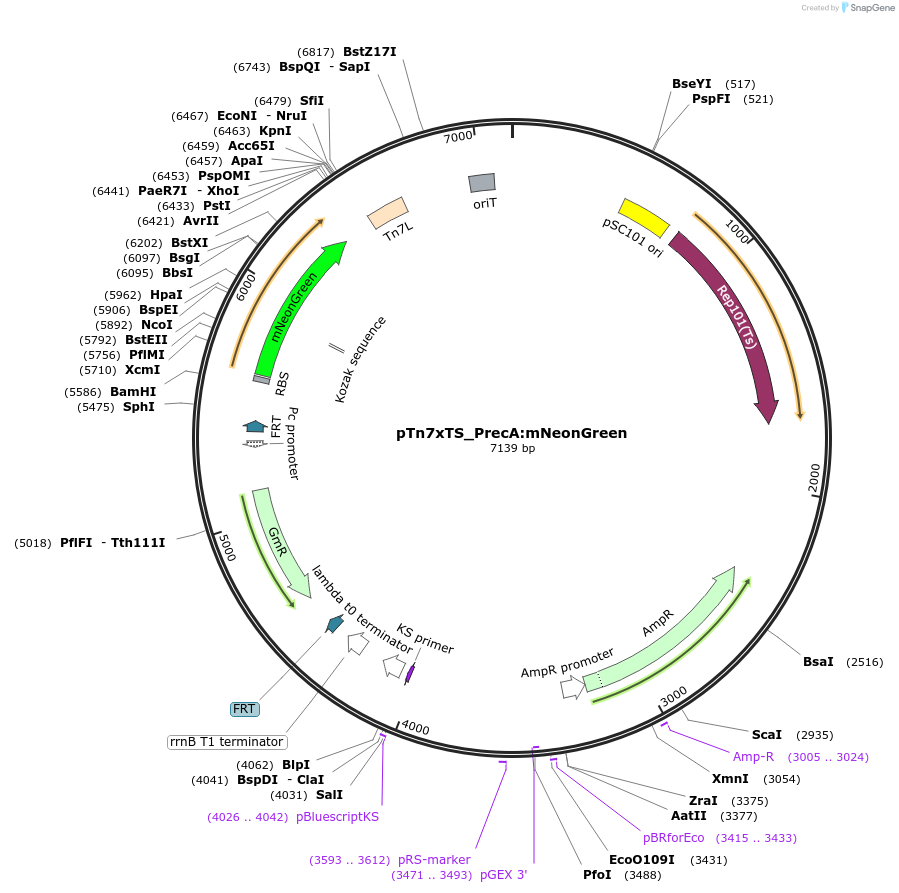
pTn7xTS_PrecA:mNeonGreen
Plasmid#225192PurposeUsed for inserting PrecA upstream of mNeonGreen at the bacterial Tn7 site for use as a RecA reporterDepositorInsertPrecA:mNeonGreen
ExpressionBacterialAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
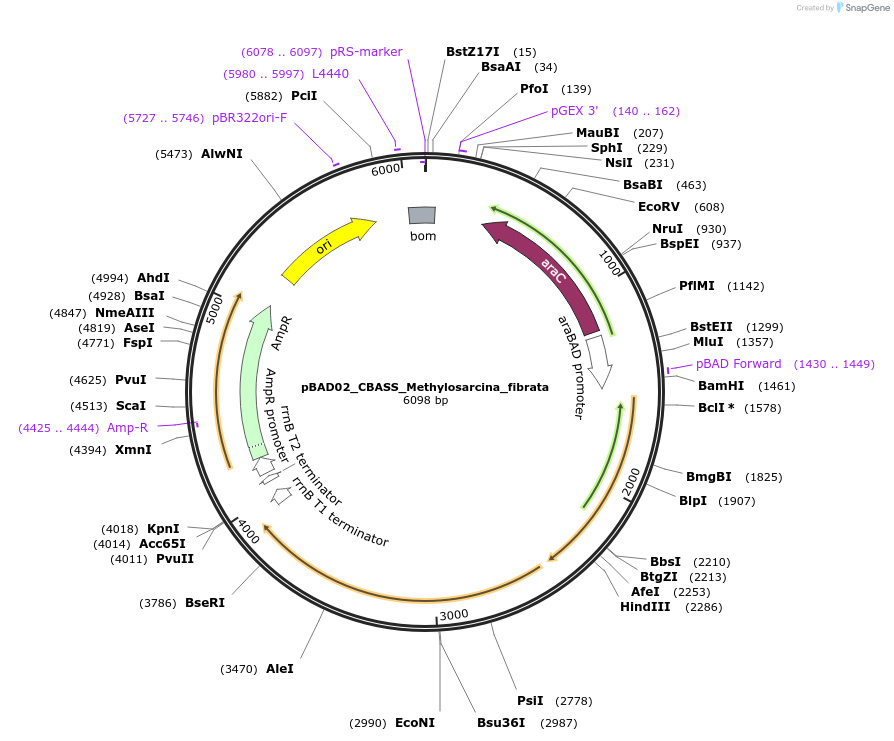
pBAD02_CBASS_Methylosarcina_fibrata
Plasmid#224406PurposeFor expression of the type I CBASS system encoded by Methylosarcina fibrataDepositorInsert4TM_MfCdnH_AGS-C
ExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
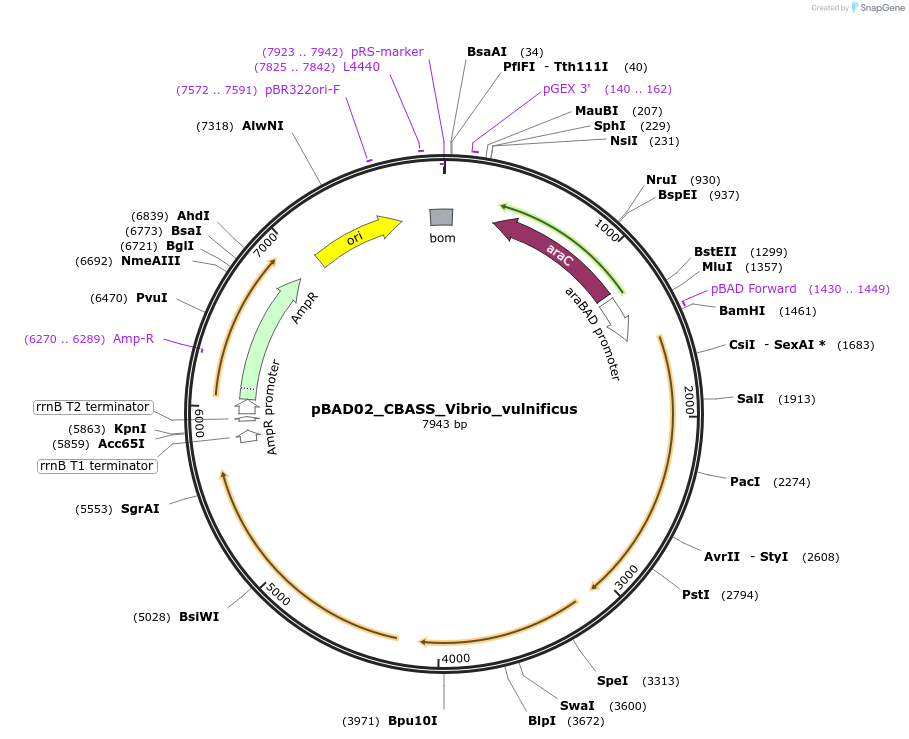
pBAD02_CBASS_Vibrio_vulnificus
Plasmid#224405PurposeFor expression of the type I CBASS system encoded by Vibrio vulnificusDepositorInsertEndonuclease_Phospholipase_VvCdnG_AGS-C
ExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
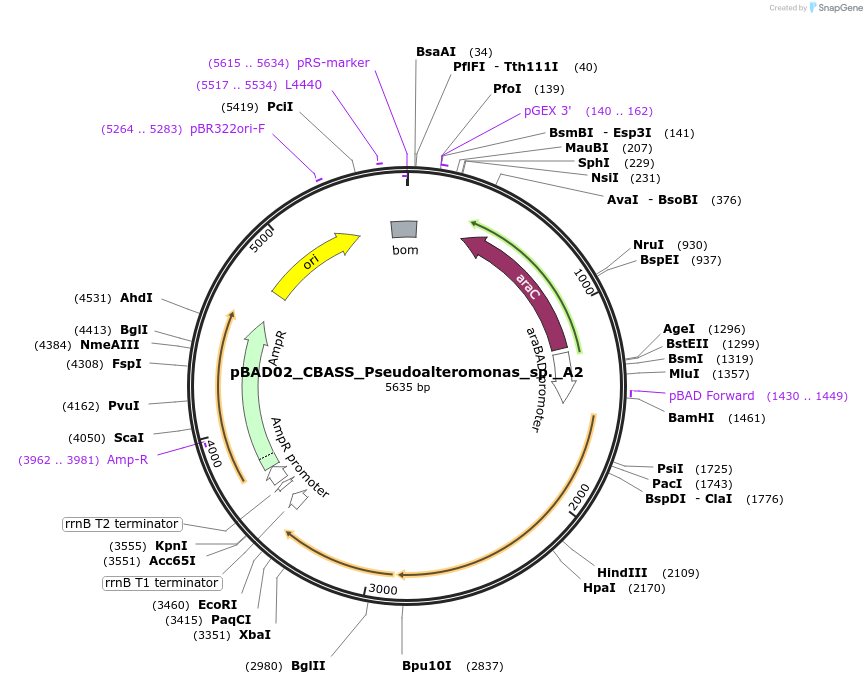
pBAD02_CBASS_Pseudoalteromonas_sp._A2
Plasmid#224401PurposeFor expression of the type I CBASS system encoded by Pseudoalteromonas sp. A2DepositorInsertPsp.CdnB_AGS-C_Cap15
ExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only