We narrowed to 9,520 results for: BLI;
-
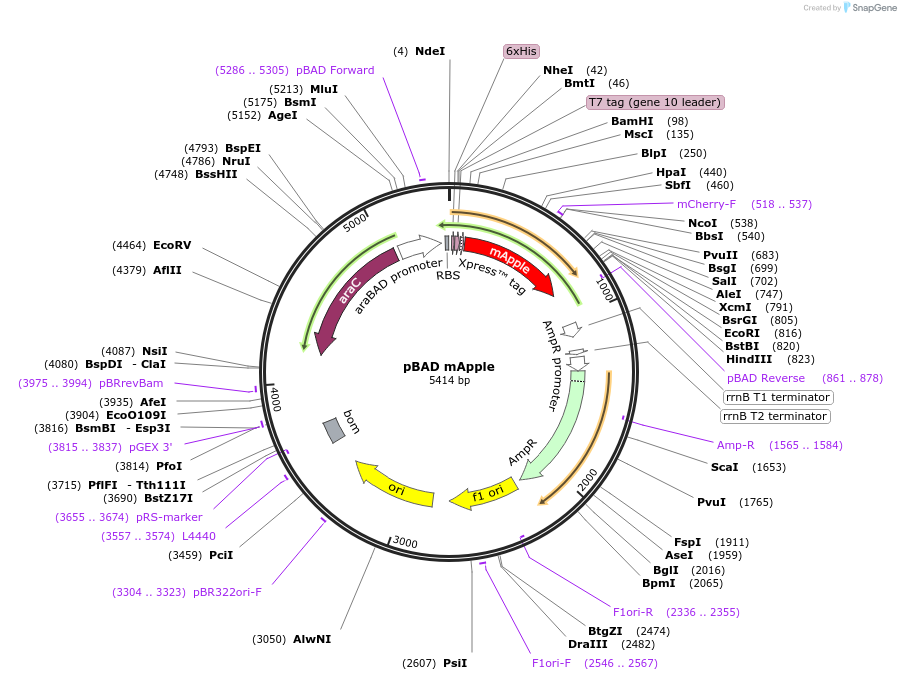
Plasmid#91772PurposeBacterial expression of a fluorescent protein, monomeric Apple (mApple). Note: Addgene identified several mutations compared to the final published sequence.DepositorInsertmonomeric Apple
TagsHexa-Histidine tag, Xpress epitope for detection …ExpressionBacterialMutationR97K, E152S, A180S, V207T compared to mApple (FP …PromoteraraBADAvailable SinceJune 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
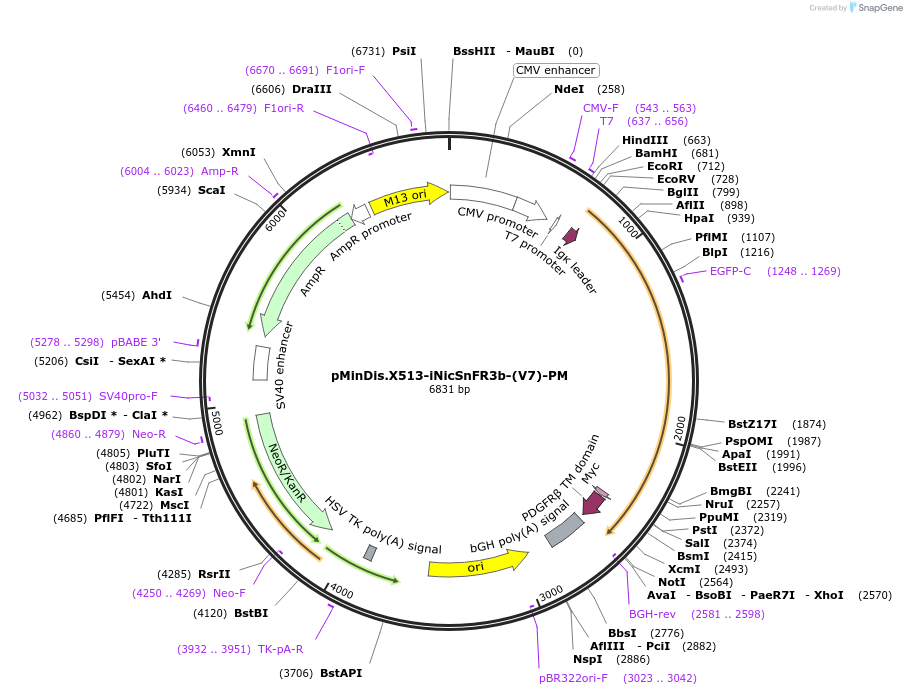
pMinDis.X513-iNicSnFR3b-(V7)-PM
Plasmid#125124PurposeDevelop a family of genetically encoded fluorescent biosensors for nicotine, termed iNicSnFRs thus enabling optical subcellular pharmacokinetics for nicotineDepositorInsertiNicSnFR3b-(V7)-PM
TagsIg-K-Chain and membrane anchoring protein-PDGFRExpressionMammalianPromoterT7Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
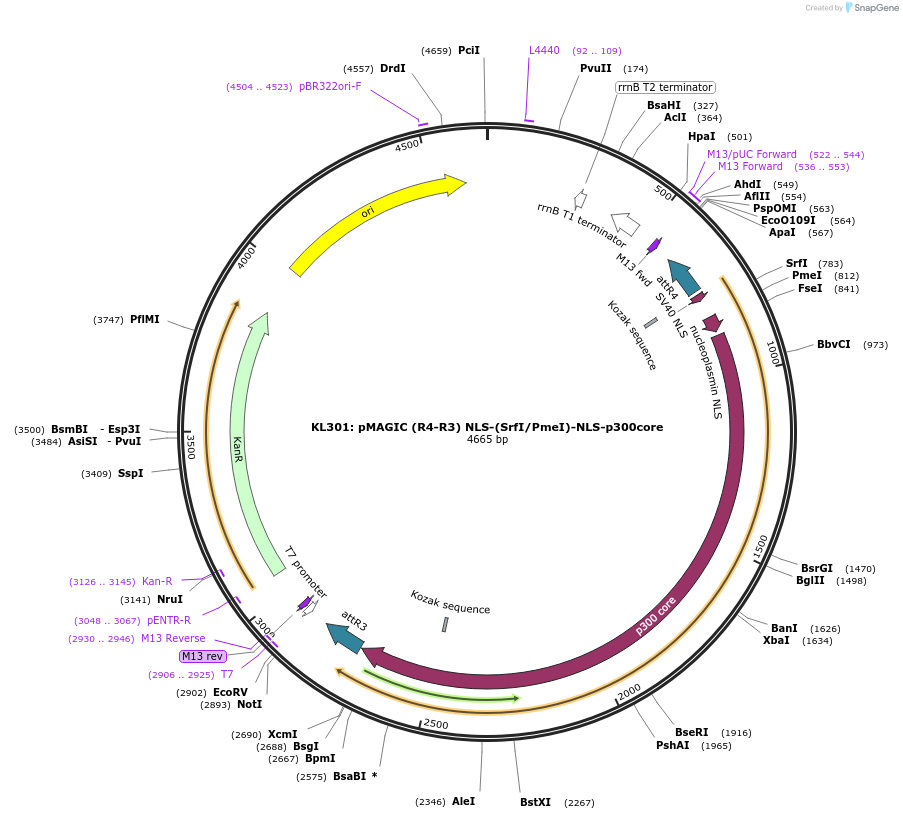
KL301: pMAGIC (R4-R3) NLS-(SrfI/PmeI)-NLS-p300core
Plasmid#121839PurposepMAGIC R4-R3 entry plasmid, contains 2x NLS/p300core scaffold enabling addition of dCas9 mutants into pMAGIC. dCas9 can be inserted via unique SrfI/PmeI restriction sitesDepositorTypeEmpty backboneUseSynthetic Biology; Pmagic gateway entry plasmidAvailable SinceJune 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
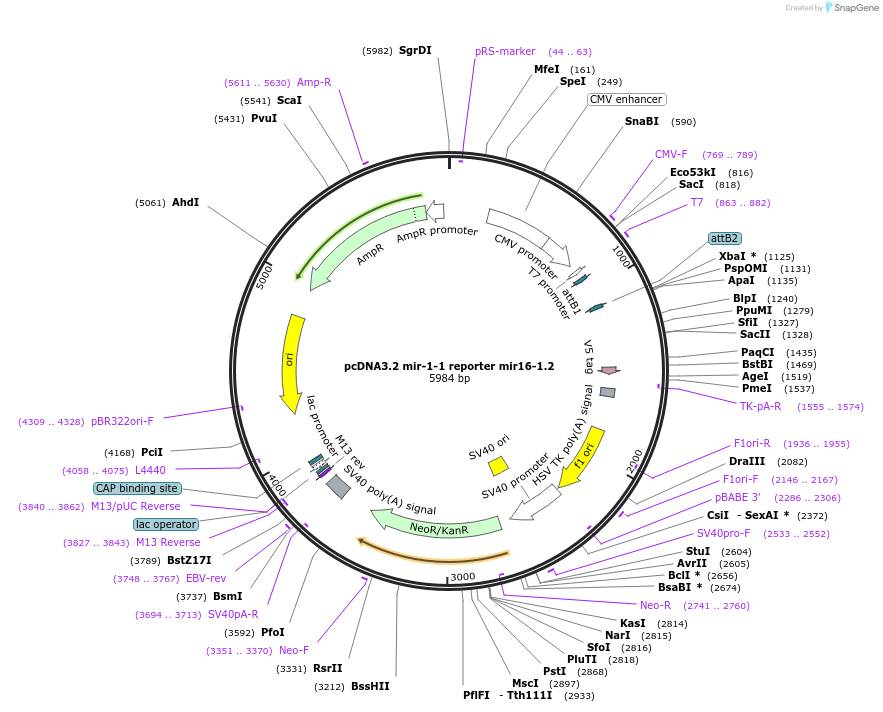
pcDNA3.2 mir-1-1 reporter mir16-1.2
Plasmid#46681PurposeExpresses a mutant version of the wild-type miR-16-1 in mammalian cellsDepositorInsertmir16-1.2 (MIR16-1 Synthetic, Human)
UseRNAiTagsV5 and hsa-mir-1-1ExpressionMammalianMutationsee sequence and publicationPromoterCMVAvailable SinceJune 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
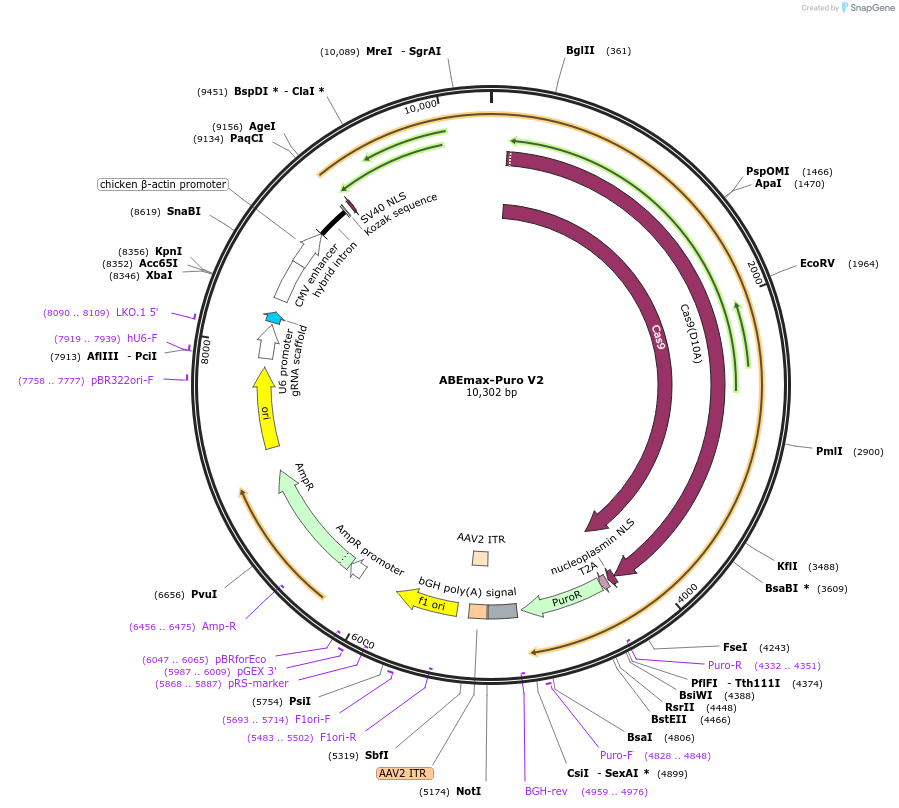
ABEmax-Puro V2
Plasmid#226955PurposeABEmax plasmid enabling A:T to G:C base editing using a single plasmid.DepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceNov. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
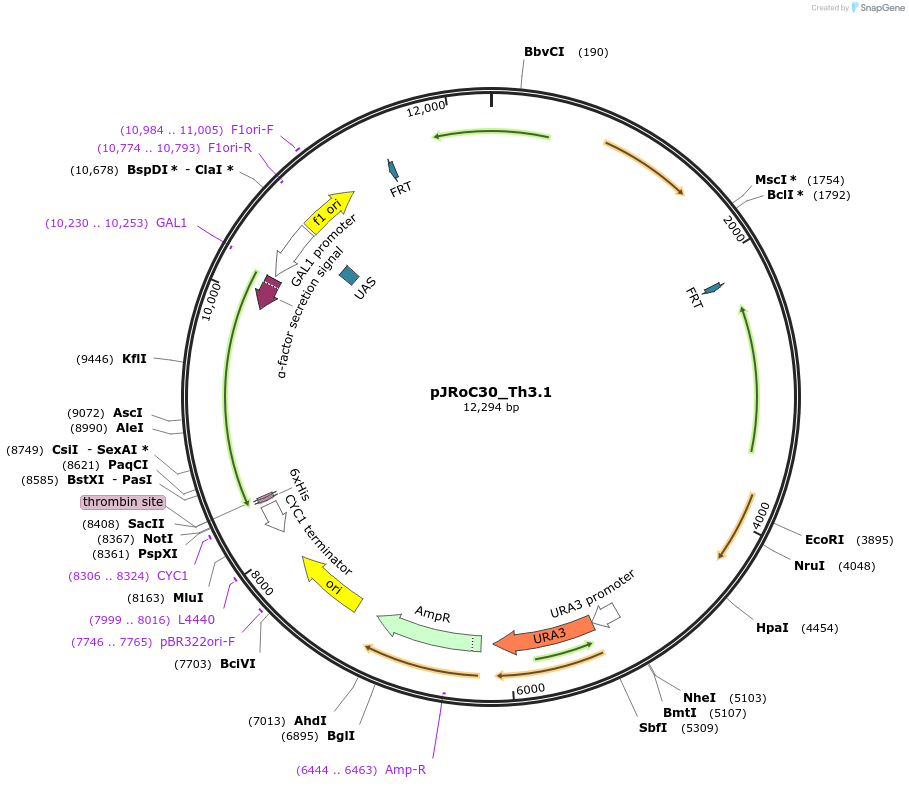
pJRoC30_Th3.1
Plasmid#188065PurposeFuncLib-designed high-redox potential laccase from Trametes hirsuta (Th3.1)DepositorInsertFuncLib-designed high-redox potential laccase from Trametes hirsuta
TagsS. cerevisiae evolved α-factor prepro-leader sign…ExpressionYeastMutation20 PROSS + 4 FuncLib mutations, described in publ…Available SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
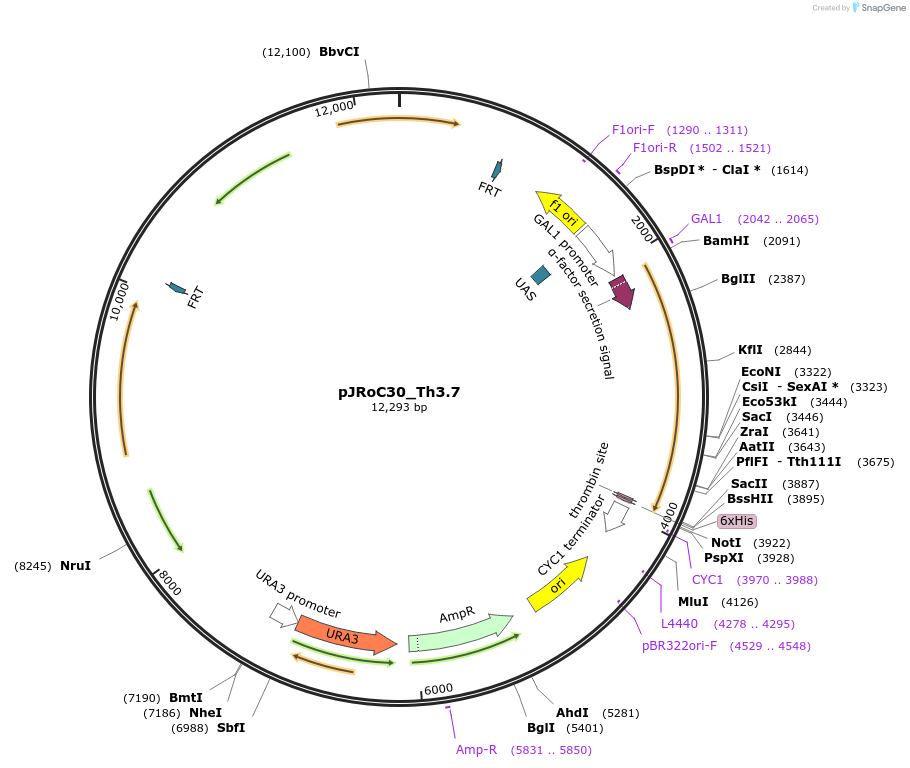
pJRoC30_Th3.7
Plasmid#188066PurposeFuncLib-designed high-redox potential laccase from Trametes hirsuta (Th3.7)DepositorInsertFuncLib-designed high-redox potential laccase from Trametes hirsuta
TagsS. cerevisiae evolved α-factor prepro-leader sign…ExpressionYeastMutation20 PROSS + 3 FuncLib mutations, described in publ…Available SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
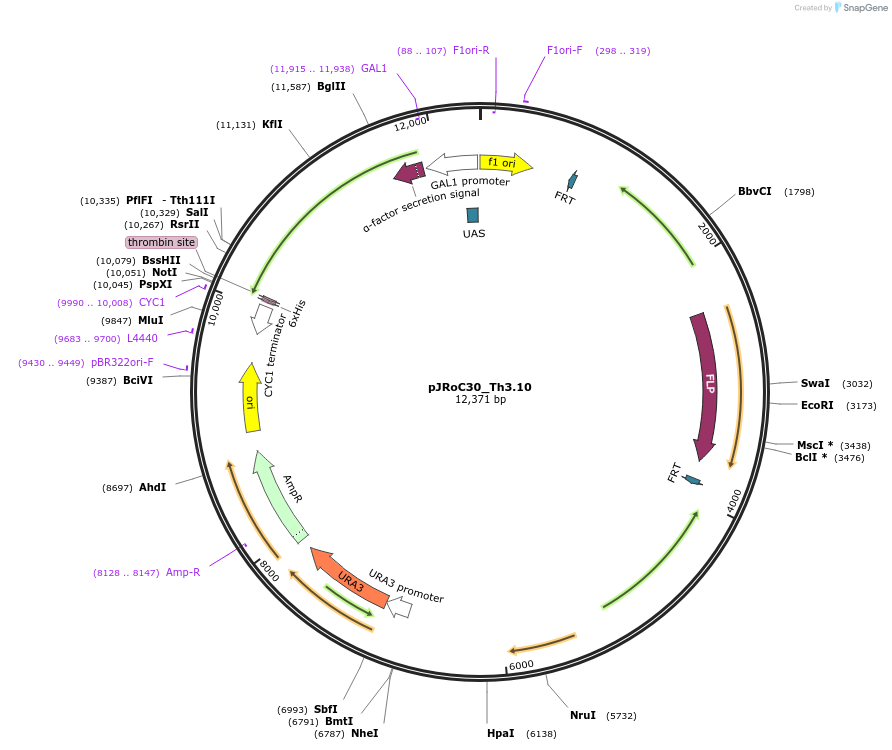
pJRoC30_Th3.10
Plasmid#188067PurposeFuncLib-designed high-redox potential laccase from Trametes hirsuta (Th3.10)DepositorInsertFuncLib-designed high-redox potential laccase from Trametes hirsuta
TagsS. cerevisiae evolved α-factor prepro-leader sign…ExpressionYeastMutation20 PROSS + 3 FuncLib mutations, described in publ…Available SinceOct. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
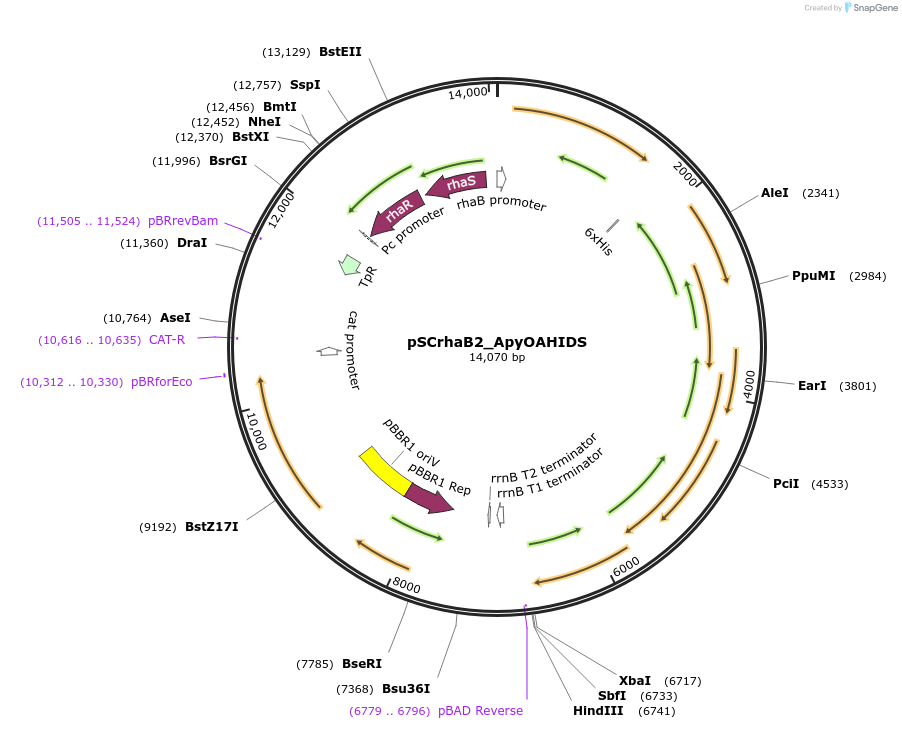
pSCrhaB2_ApyOAHIDS
Plasmid#226246PurposeProduction of ApyA peptide modified by the B12-rSAM enzyme ApyD (optional, refer to publication), the cytochrome P450 enzyme ApyO, and the MNIO enzyme ApyHI, and the methyltransferase ApySDepositorInsertsHis6_ApyA
ApyD
ApyO
ApyH
ApyI
ApyS
TagsHexa-HistidineExpressionBacterialAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
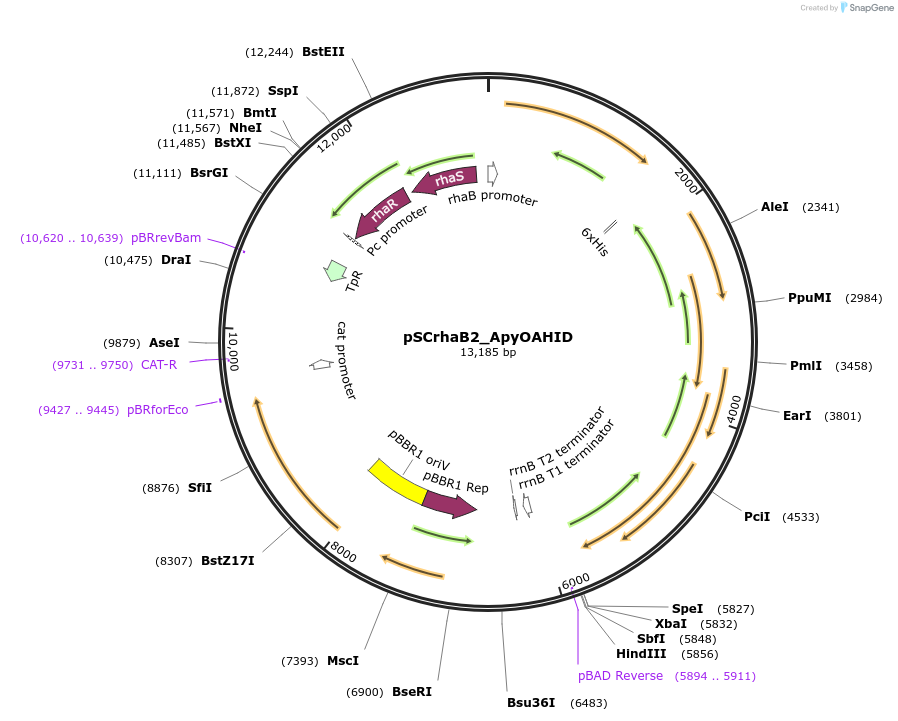
pSCrhaB2_ApyOAHID
Plasmid#226245PurposeProduction of ApyA peptide modified by the B12-rSAM enzyme ApyD (optional, refer to publication), the cytochrome P450 enzyme ApyO, and the MNIO enzyme ApyHIDepositorInsertsHis6_ApyA
ApyD
ApyO
ApyH
ApyI
TagsHexa-HistidineExpressionBacterialAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only