CHLOROMODAS Kit
(Kit #
1000000269
)
Depositing Lab: Tobias Erb
This plasmid collection provides a modular cloning (MoClo) toolkit for engineering the Chlamydomonas reinhardtii chloroplast. The set includes standardized vectors and genetic parts that enable efficient assembly and testing of synthetic constructs for chloroplast expression. Together, these resources support streamlined design and implementation of chloroplast engineering projects within the research community.
This kit will be sent as bacterial glycerol stocks in 96-well plate format.
Original Publication
A modular high-throughput approach for advancing synthetic biology in the chloroplast of Chlamydomonas. Inckemann RM, Chotel T, Burgis M, Brinkmann CK, Andreas L, Baumann J, Sharma P, Klose M, Barrett J, Ries F, Paczia N, Glatter T, Mackinder LCM, Willmund F, Erb TJ. Nat Plants. 2025; 11(11):2332-2349. doi: 10.1038/s41477-025-02126-2. PubMed (Link opens in a new window) Article (Link opens in a new window) bioRxiv Preprint (Link opens in a new window)
Description
The Chloroplast Modular Assembly System (CHLOROMODAS) provides a versatile toolkit for engineering the C. reinhardtii chloroplast genome. It uses a hierarchical Golden Gate-based cloning strategy to assemble standardized genetic parts — promoters, UTRs, coding sequences, terminators, and specialized connectors — into single transcription units (Level 1) or multigene constructs (Level 2). The system incorporates chloroplast-specific homology regions for precise site-directed integration, enabling flexible and efficient genome editing.
Unique to this system are connector and operon parts that allow users to control the order, orientation, and polycistronic arrangement of transcription units. Reverse connectors enable inversion of transcription units to prevent transcriptional read-through, while operon connectors and intercistronic expression elements (IEEs) support the construction of polycistronic gene clusters for coordinated expression of multiple genes from a single transcript.
The kit also includes placeholder parts for streamlined library construction and exchange of components, along with detailed assembly maps, selectable markers, and optimized protocols for cloning and chloroplast transformation. Together, these resources enable rapid design, assembly, and functional testing of synthetic chloroplast constructs.
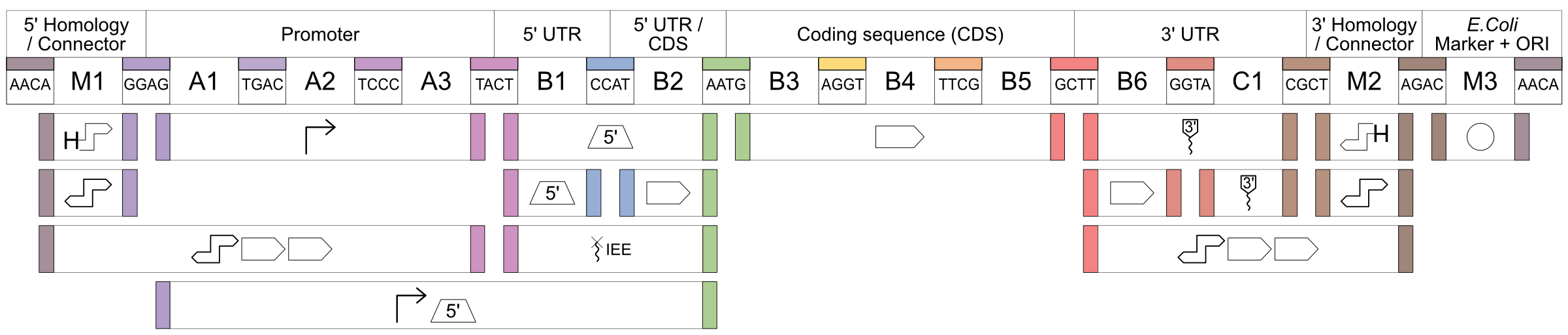
Figure 1: Architecture of the novel chloroplast MoClo system and overview of the created genetic parts. The architecture follows the Phytobrick standard and conserves the cloning overhangs of this system, as well as the nomenclature for the part types (A1–C1). The positions M1–3 allow for higher modularity as reported by Stukenberg et al. (2021) (Link opens in a new window). Number of parts are indicated for each position.
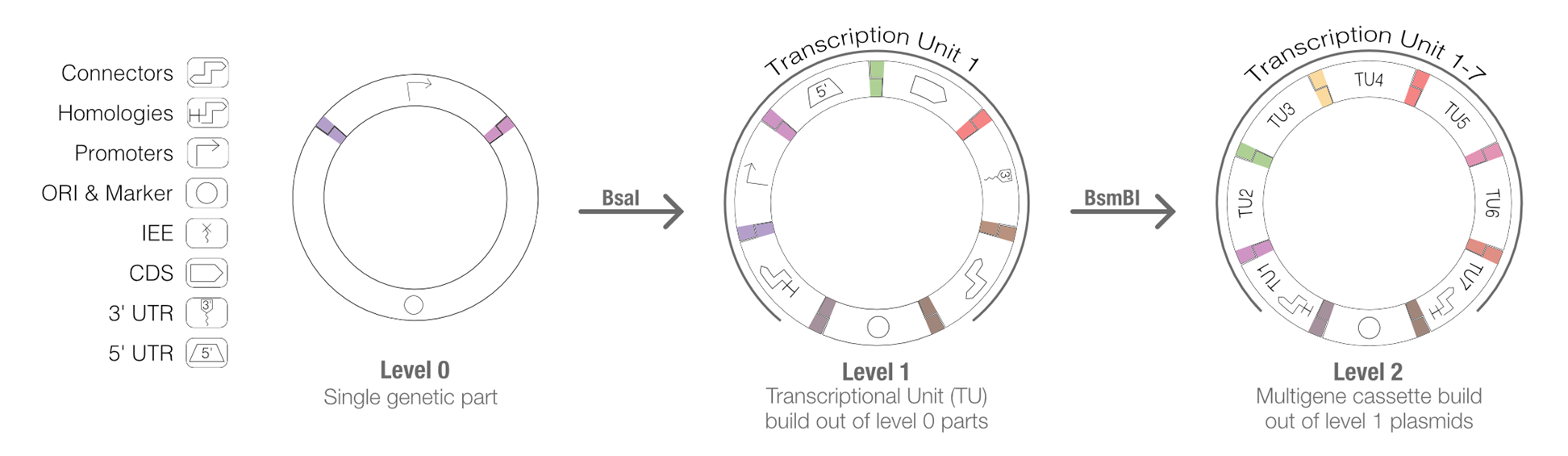
Figure 2: New parts are first domesticated and assembled into a universal acceptor vector (Level 0). These parts can then be combined in a one-pot reaction to create a Level 1 plasmid containing a single transcriptional unit. Optionally, these single transcriptional units can be further assembled into a multigene cassette (Level 2).
Kit Documentation
Additional information about the collection and the GenBank files are available on the Erb Lab GitHub page (Link opens in a new window).
How to Cite this Kit
These plasmids were created by your colleagues. Please acknowledge the Depositing Lab, cite the article in which they were created, and include Addgene in the Materials and Methods of your future publications.
For your Materials and Methods section:
"The CHLOROMODAS Kit was a gift from Tobias Erb (Addgene kit #1000000269)."
For your Reference section:
A modular high-throughput approach for advancing synthetic biology in the chloroplast of Chlamydomonas. Inckemann RM, Chotel T, Burgis M, Brinkmann CK, Andreas L, Baumann J, Sharma P, Klose M, Barrett J, Ries F, Paczia N, Glatter T, Mackinder LCM, Willmund F, Erb TJ. Nat Plants. 2025; 11(11):2332-2349. doi: 10.1038/s41477-025-02126-2. PubMed (Link opens in a new window) Article (Link opens in a new window)
CHLOROMODAS Kit - #1000000269
- Resistance Color Key
Each circle corresponds to a specific antibiotic resistance in the kit plate map wells.
- Inventory
Searchable and sortable table of all plasmids in kit. The Well column links to its kit plate map on the right, as well as lists the plasmid well location in its plate. The Plasmid column links to a plasmid's individual web page.
- Kit Plate Map
96-well plate map for plasmid layout. Hovering over a well reveals the plasmid name, while clicking on a well opens the plasmid page.
Resistance Color Key
| Chloramphenicol | |
| Ampicillin | |
| Kanamycin |


