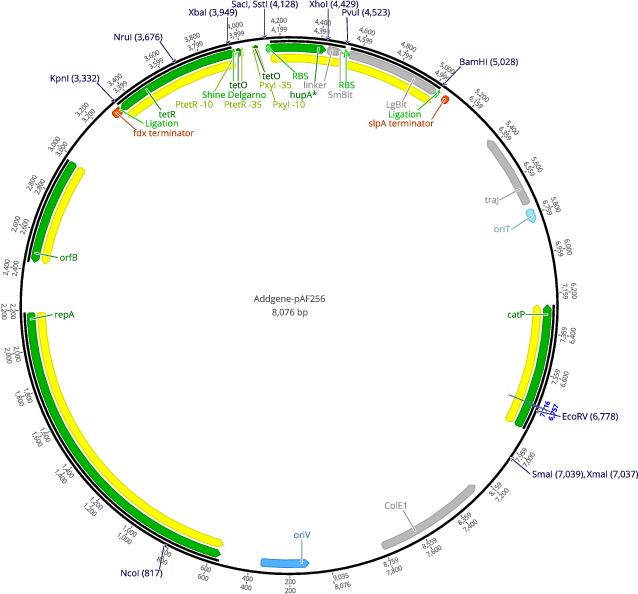
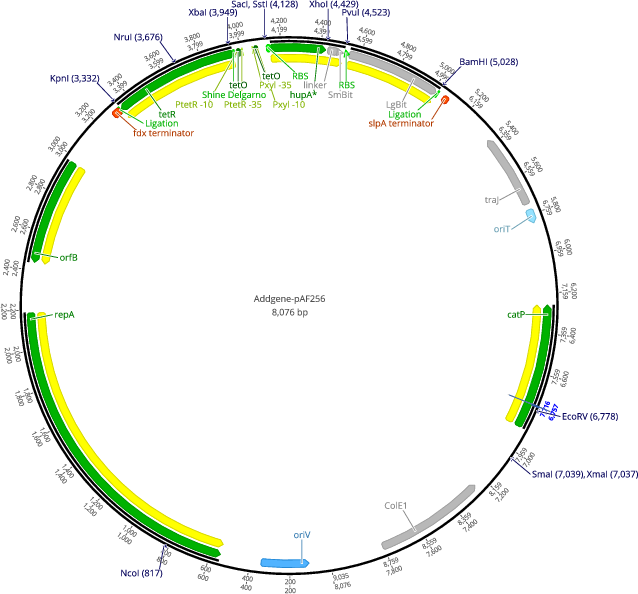
pAF256
(Plasmid
#105496)
-
PurposepRPF185 derived plasmid encoding an anhydrotetracyclin inducible HupA-SmBiT and LgBiT
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 105496 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepRPF185
-
Backbone manufacturerR.P. Fagan, University of Sheffield
- Total vector size (bp) 8076
-
Vector typeATc-dependent expression in C. difficile
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol, 25 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameHupA-SmBiT/LgBiT
-
SpeciesC. difficile / synthetic
-
Insert Size (bp)900
- Promoter Ptet
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site SacI (not destroyed)
- 3′ cloning site BamHI (not destroyed)
- 5′ sequencing primer CTGGACTTCATGAAAAACTAAAAAAAATATTG
- 3′ sequencing primer CACCGACGAGCAAGGCAAGACCG (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please visit https://www.biorxiv.org/content/early/2018/09/27/426809 for bioRxiv preprint.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pAF256 was a gift from Wiep Klaas Smits (Addgene plasmid # 105496 ; http://n2t.net/addgene:105496 ; RRID:Addgene_105496) -
For your References section:
The Bacterial Chromatin Protein HupA Can Remodel DNA and Associates with the Nucleoid in Clostridium difficile. Oliveira Paiva AM, Friggen AH, Qin L, Douwes R, Dame RT, Smits WK. J Mol Biol. 2019 Jan 8. pii: S0022-2836(18)31160-4. doi: 10.1016/j.jmb.2019.01.001. 10.1016/j.jmb.2019.01.001 PubMed 30633871