We narrowed to 9,164 results for: Bre;
-
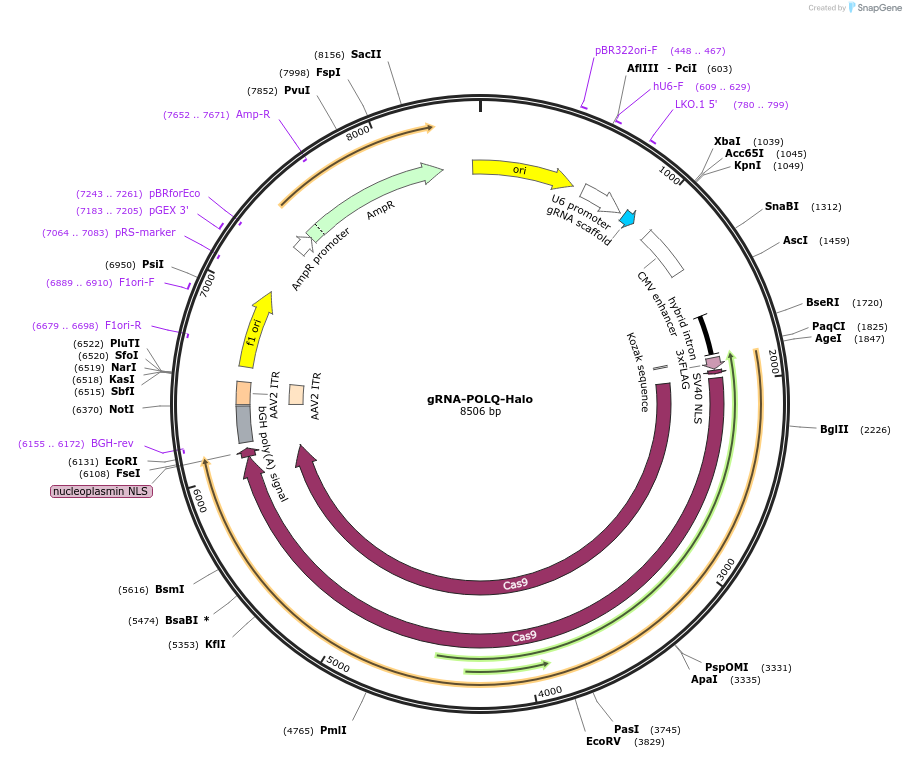
Plasmid#211636PurposesgRNA against POLQDepositorInsertsgRNA POLQ
UseCRISPRExpressionMammalianPromoterU6Available SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
gRNA-POLQ-Halo
Plasmid#211636PurposesgRNA against POLQDepositorInsertsgRNA POLQ
UseCRISPRExpressionMammalianPromoterU6Available SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSKB2-TRex
Plasmid#31349DepositorInsertThermus aquaticus Redox Sensing Repressor
TagsHisExpressionBacterialAvailable SinceAug. 18, 2011AvailabilityAcademic Institutions and Nonprofits only -
pSKB2-TRex
Plasmid#31349DepositorInsertThermus aquaticus Redox Sensing Repressor
TagsHisExpressionBacterialAvailable SinceAug. 18, 2011AvailabilityAcademic Institutions and Nonprofits only -
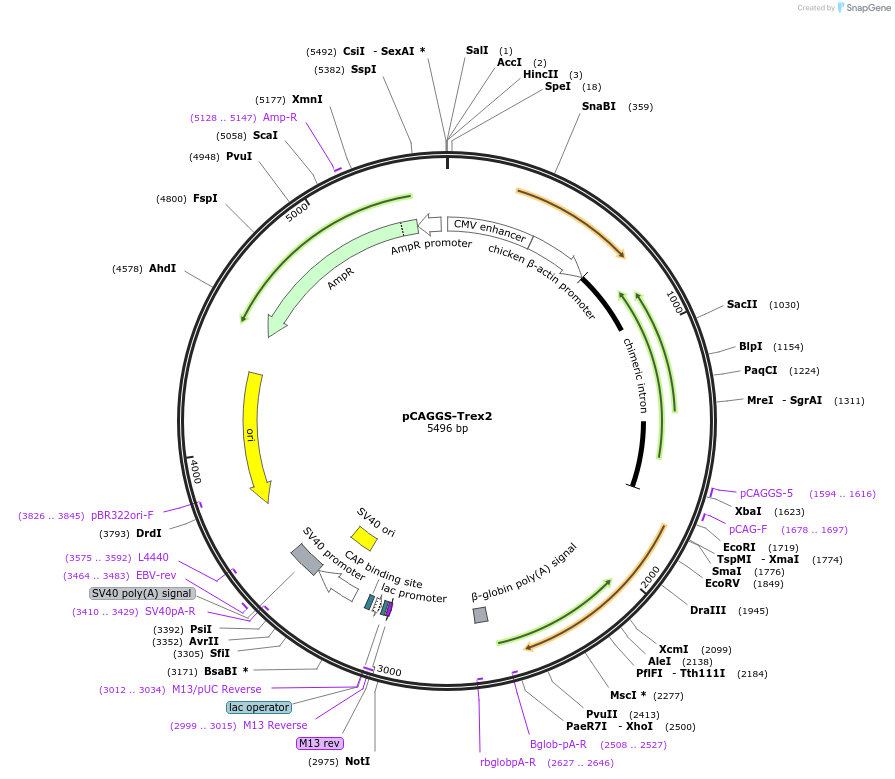
pCAGGS-Trex2
Plasmid#79246PurposeExpression vector for Trex2, which is a non-processive 3' exonuclease.DepositorAvailable SinceAug. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCAGGS-Trex2
Plasmid#79246PurposeExpression vector for Trex2, which is a non-processive 3' exonuclease.DepositorAvailable SinceAug. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
pBS mScx (CT#404)
Plasmid#13958DepositorInsertScleraxis (Scx Mouse)
ExpressionBacterialAvailable SinceFeb. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
pBS mScx (CT#404)
Plasmid#13958DepositorInsertScleraxis (Scx Mouse)
ExpressionBacterialAvailable SinceFeb. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
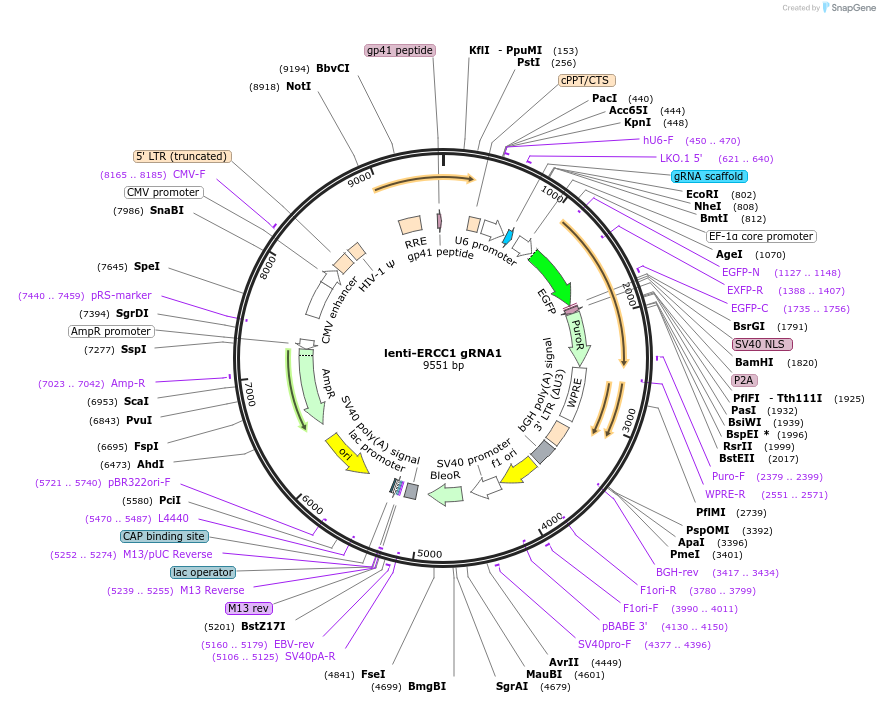
lenti-ERCC1 gRNA1
Plasmid#211630PurposesgRNA-1 against ERCC1DepositorInsertsgRNA ERCC1
UseCRISPRExpressionMammalianPromoterU6Available SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
lenti-ERCC1 gRNA1
Plasmid#211630PurposesgRNA-1 against ERCC1DepositorInsertsgRNA ERCC1
UseCRISPRExpressionMammalianPromoterU6Available SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
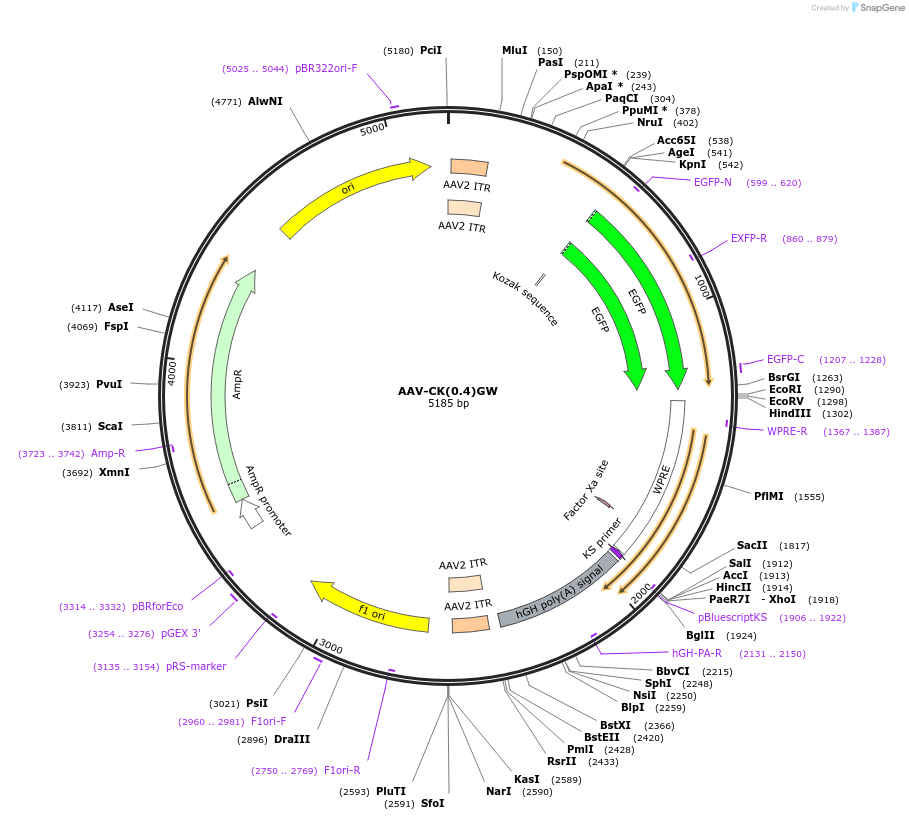
AAV-CK(0.4)GW
Plasmid#27226DepositorInsertEGFP
UseAAVTagsEGFPAvailable SinceMarch 11, 2011AvailabilityAcademic Institutions and Nonprofits only -
AAV-CK(0.4)GW
Plasmid#27226DepositorInsertEGFP
UseAAVTagsEGFPAvailable SinceMarch 11, 2011AvailabilityAcademic Institutions and Nonprofits only -
aCE120
Plasmid#204491PurposeFigure 1. reporter circuitDepositorInsertsEBFP2
mKate2
EYFP
UseSynthetic BiologyTagsnoneExpressionBacterial and MammalianPromoterhEF1aAvailable SinceDec. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
aCE120
Plasmid#204491PurposeFigure 1. reporter circuitDepositorInsertsEBFP2
mKate2
EYFP
UseSynthetic BiologyTagsnoneExpressionBacterial and MammalianPromoterhEF1aAvailable SinceDec. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
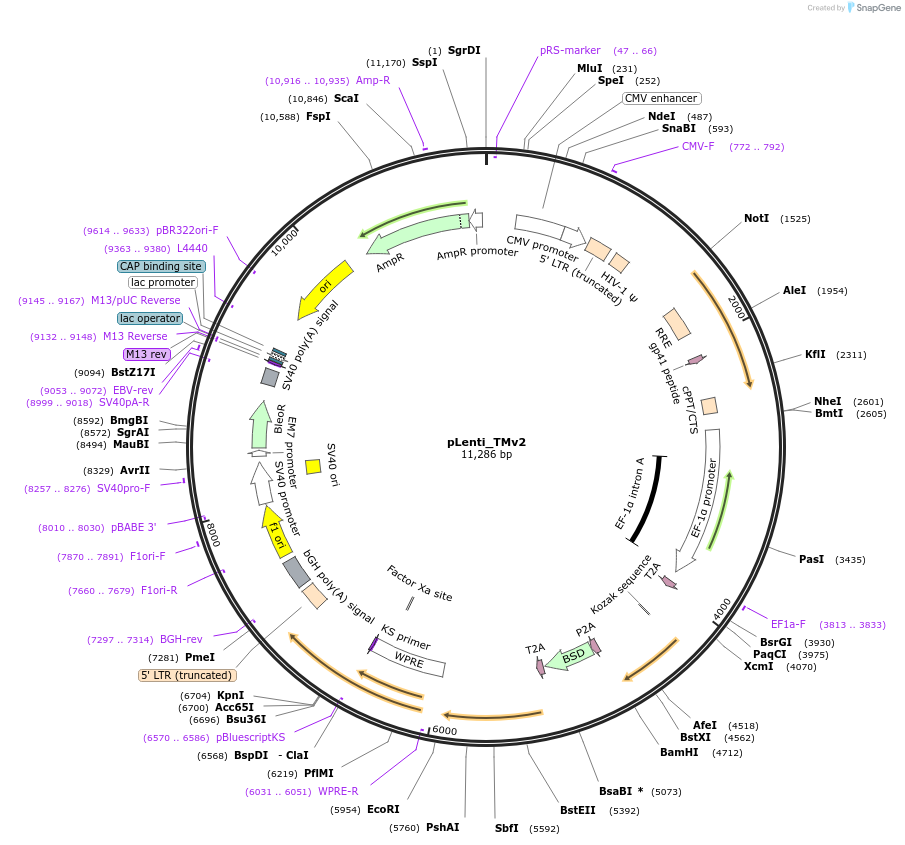
pLenti_TMv2
Plasmid#131761Purposeframeshift reporter for barcode retrievalDepositorTypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
pLenti_TMv2
Plasmid#131761Purposeframeshift reporter for barcode retrievalDepositorTypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
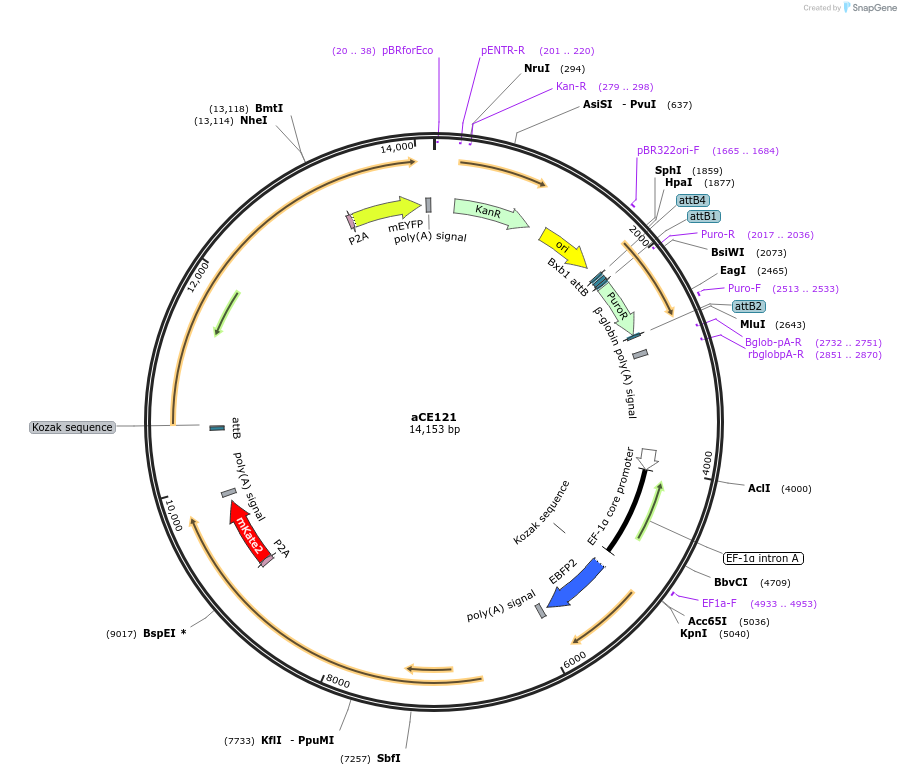
aCE121
Plasmid#204492PurposeCadherin circuitDepositorInsertsEBFP2
Cdh6-mKate2
Cdh1-EYFP
UseSynthetic BiologyTags2AExpressionBacterial and MammalianPromoterhEF1aAvailable SinceDec. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
aCE121
Plasmid#204492PurposeCadherin circuitDepositorInsertsEBFP2
Cdh6-mKate2
Cdh1-EYFP
UseSynthetic BiologyTags2AExpressionBacterial and MammalianPromoterhEF1aAvailable SinceDec. 19, 2023AvailabilityAcademic Institutions and Nonprofits only -
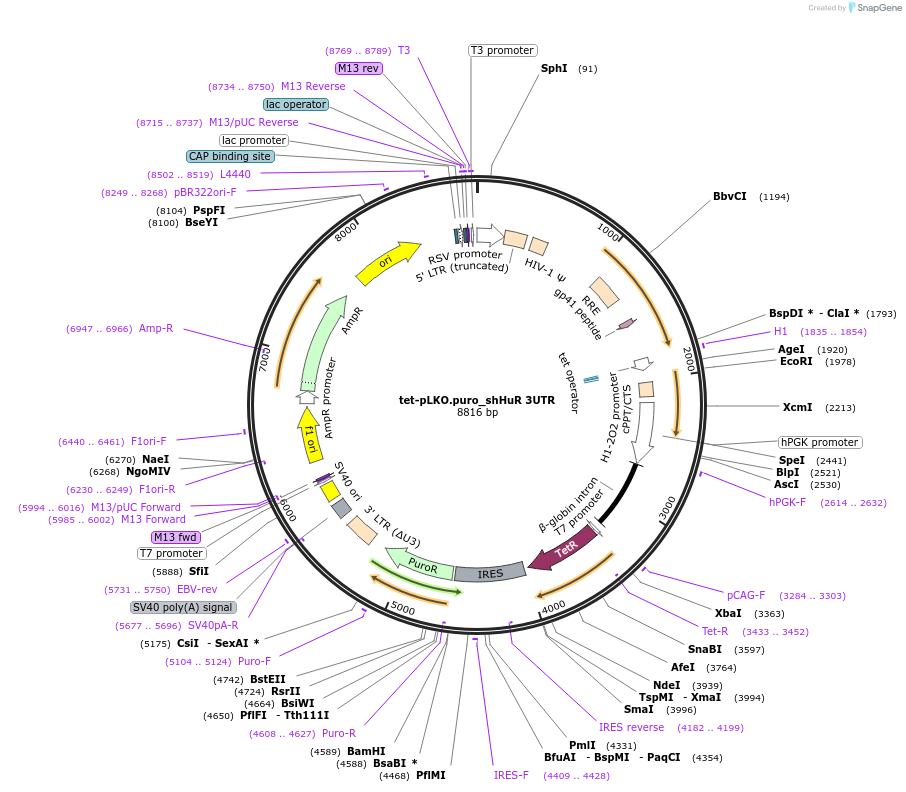
tet-pLKO.puro_shHuR 3UTR
Plasmid#110412PurposeTRCN0000276186 (Target TTGTTAGTGTACAACTCATTT), silence human ELAVL1 (HuR) gene, doxycycline inducible, puromycin selectionDepositorInsertELAVL1 (HuR)
UseLentiviral and RNAiExpressionMammalianPromoterH1/TO (RNA PolIII)Available SinceFeb. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
tet-pLKO.puro_shHuR 3UTR
Plasmid#110412PurposeTRCN0000276186 (Target TTGTTAGTGTACAACTCATTT), silence human ELAVL1 (HuR) gene, doxycycline inducible, puromycin selectionDepositorInsertELAVL1 (HuR)
UseLentiviral and RNAiExpressionMammalianPromoterH1/TO (RNA PolIII)Available SinceFeb. 23, 2021AvailabilityAcademic Institutions and Nonprofits only