We narrowed to 25,974 results for: CHI
-
Plasmid#215499PurposeA pLys3 backbone integrating plasmid expressing mECitrine under the inducible promoter 41nmt1DepositorInsertmECitrine
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
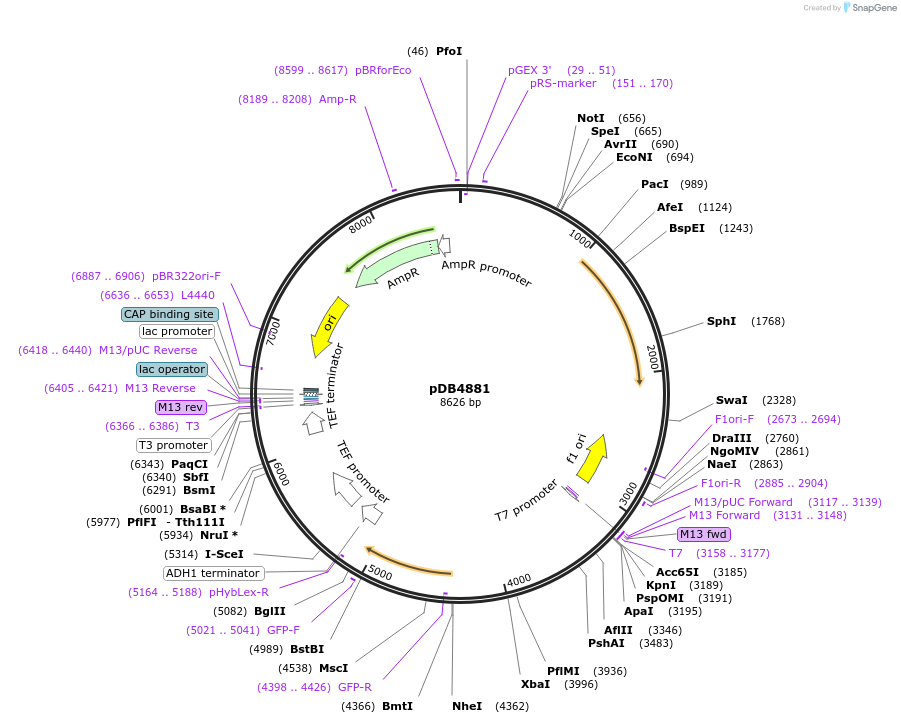
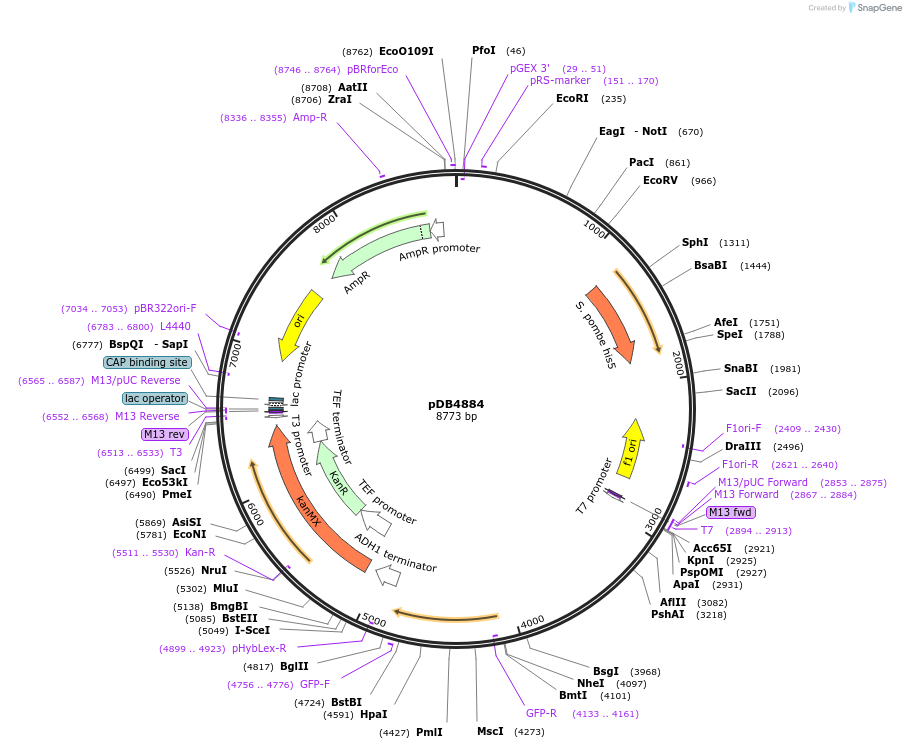
pDB4884
Plasmid#215500PurposeA pHis5 backbone integrating plasmid expressing mECitrine under the inducible promoter 41nmt1DepositorInsertmECitrine
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
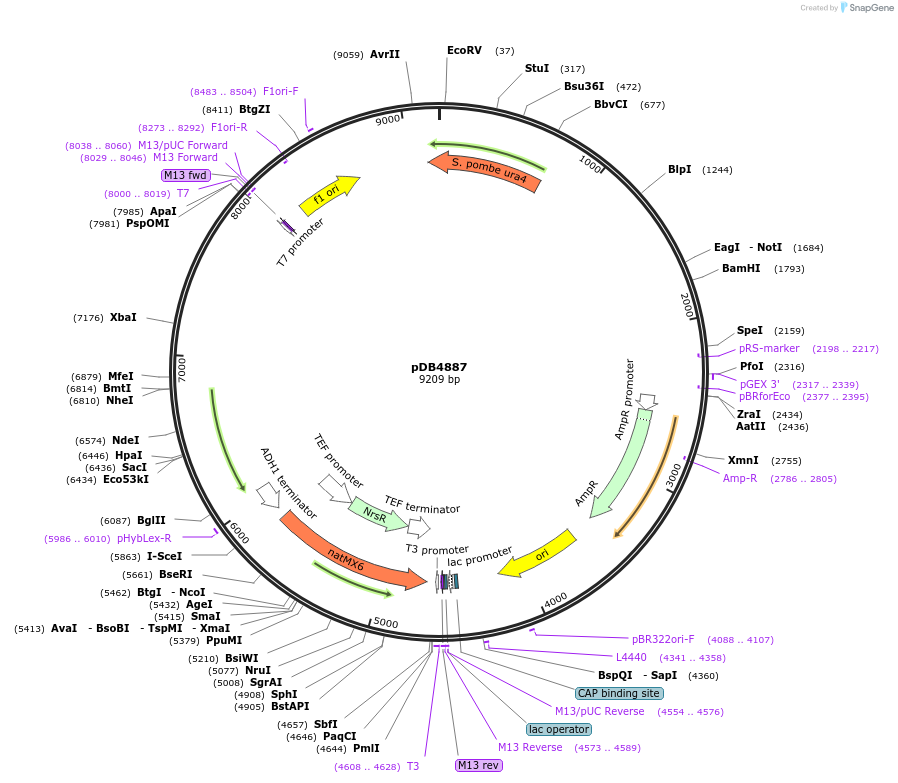
pDB4887
Plasmid#215502PurposeA pUra4 backbone integrating plasmid expressing mTurquoise2 under the inducible promoter 41nmt1DepositorInsertmTurquoise2
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
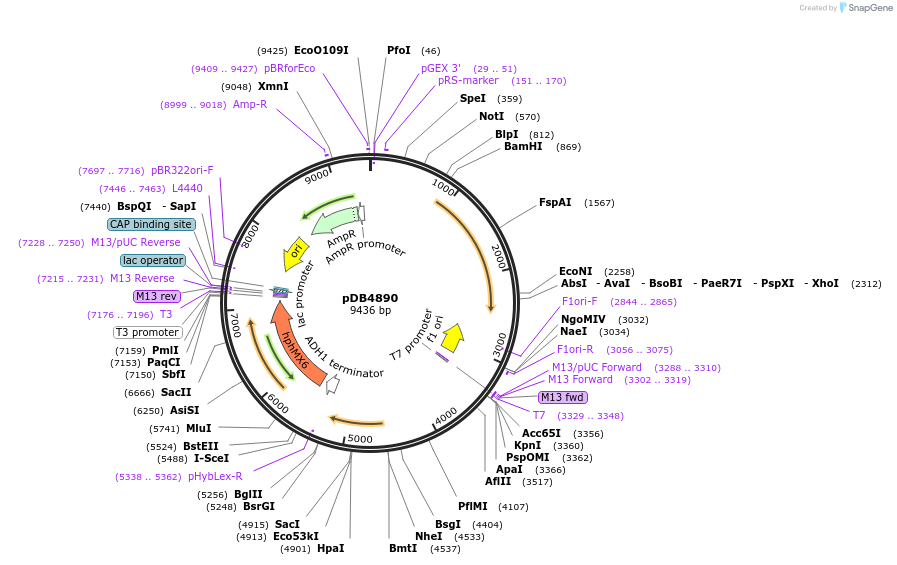
pDB4890
Plasmid#215503PurposeA pAde6 backbone integrating plasmid expressing mTurquoise2 under the inducible promoter 41nmt1DepositorInsertmTurquoise2
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
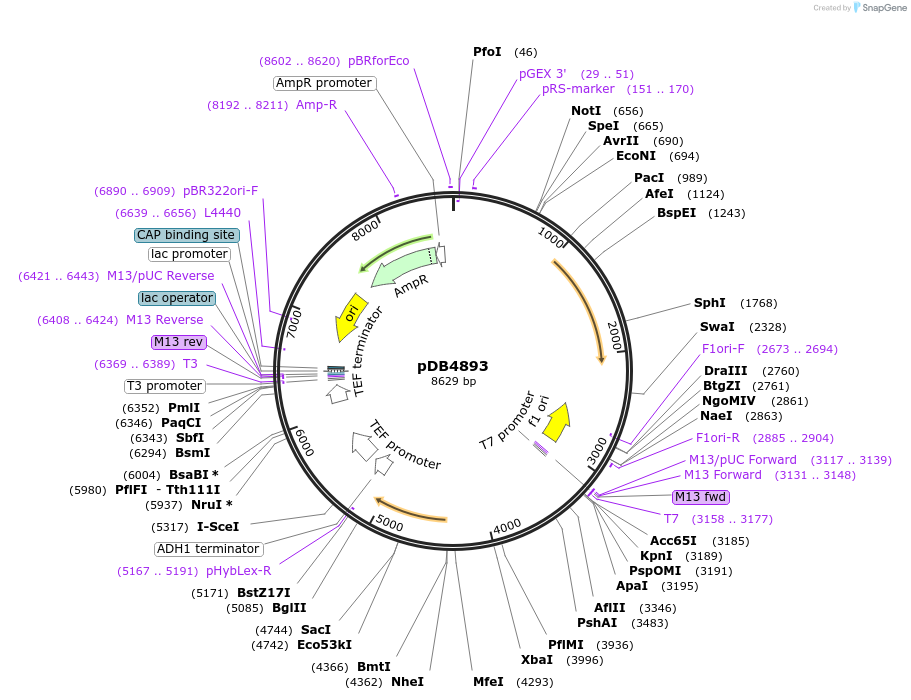
pDB4893
Plasmid#215504PurposeA pLys3 backbone integrating plasmid expressing mTurquoise2 under the inducible promoter 41nmt1DepositorInsertmTurquoise2
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
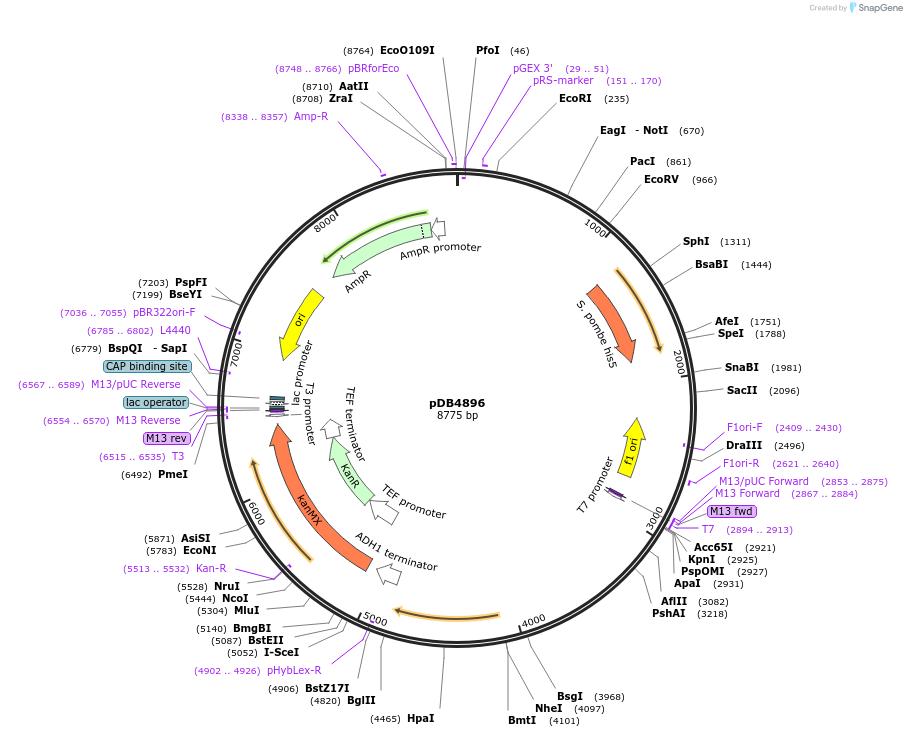
pDB4896
Plasmid#215505PurposeA pHis5 backbone integrating plasmid expressing mTurquoise2 under the inducible promoter 41nmt1DepositorInsertmTurquoise2
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
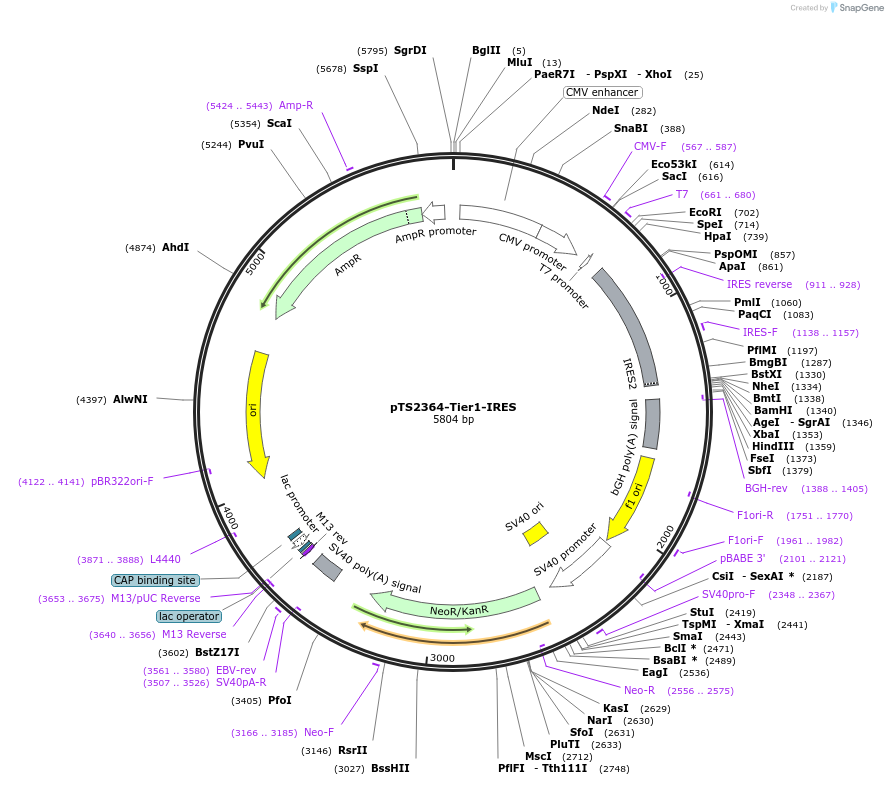
pTS2364-Tier1-IRES
Plasmid#169562PurposeTier-1 vector carrying an internal ribosome entry site (IRES-pA)DepositorTypeEmpty backboneExpressionMammalianAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
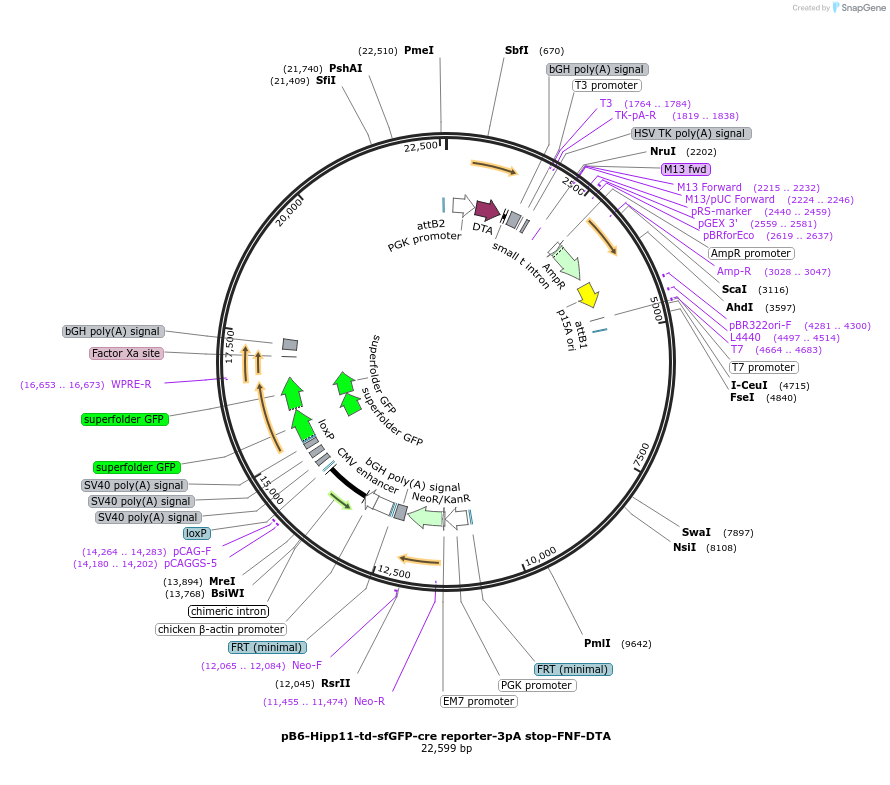
pB6-Hipp11-td-sfGFP-cre reporter-3pA stop-FNF-DTA
Plasmid#183026PurposeB6J Hipp11-FNF-pCAG-loxP-3xSV40pA-loxP-td-sfGFP-WPRE-bpA allele targeting vector, low copy numberDepositorInserttd-sfGFP
UseCre/Lox and Mouse TargetingExpressionBacterial and MammalianAvailable SinceApril 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
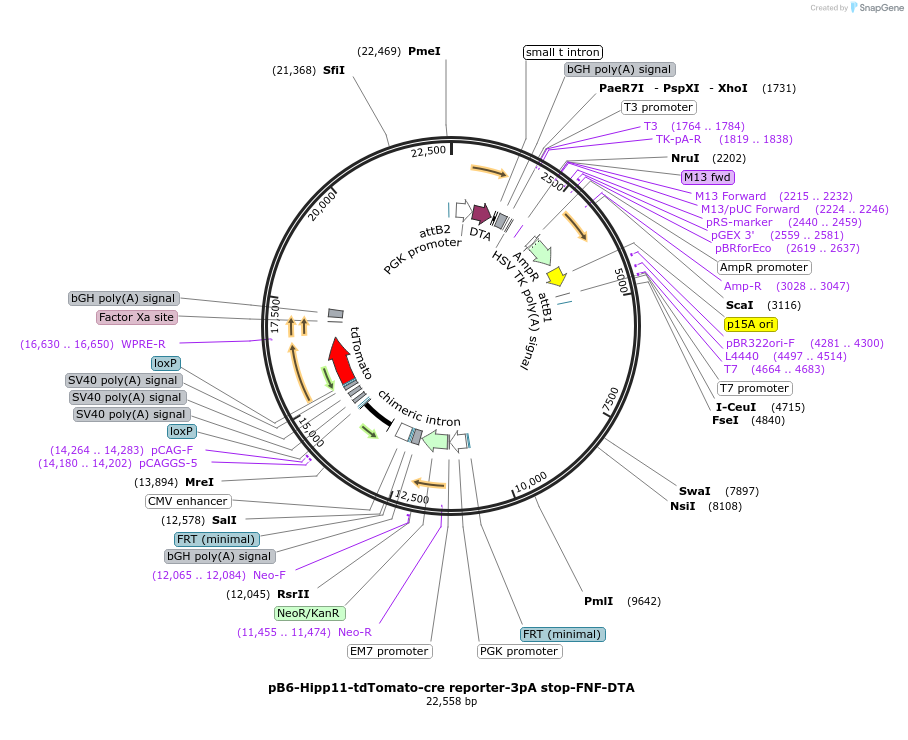
pB6-Hipp11-tdTomato-cre reporter-3pA stop-FNF-DTA
Plasmid#183025PurposeB6J Hipp11-FNF-pCAG-loxP-3xSV40pA-loxP-tdTomato-WPRE-bpA allele targeting vector, low copy numberDepositorInserttdTomato
UseCre/Lox and Mouse TargetingExpressionBacterial and MammalianAvailable SinceApril 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
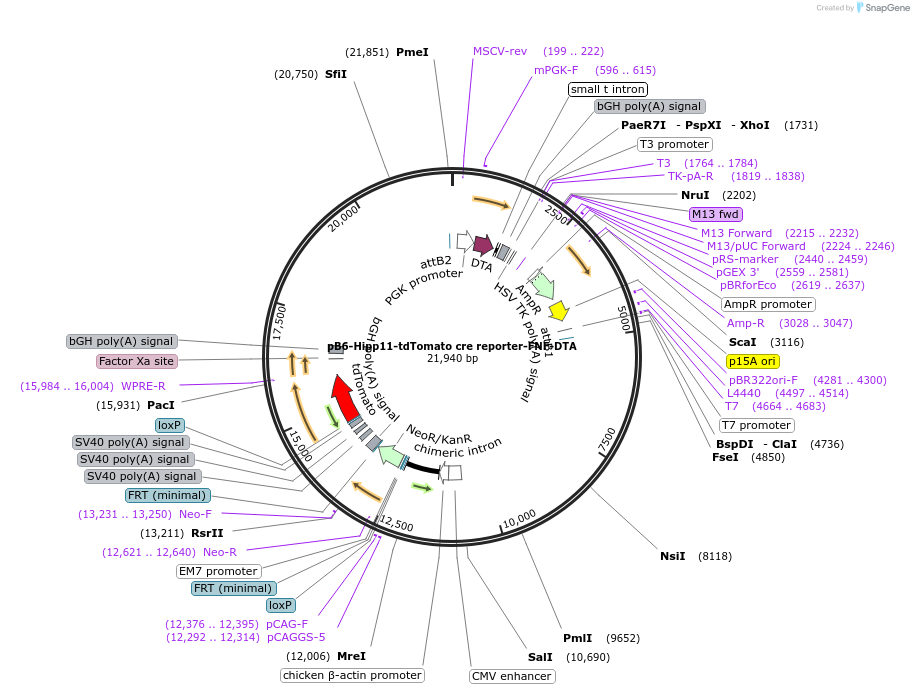
pB6-Hipp11-tdTomato cre reporter-FNF-DTA
Plasmid#183024PurposeB6J Hipp11-pCAG-loxP-FNF-bGHpA-3xSV40pA-loxP-tdTomato-WPRE-bpA allele targeting vector, low copy numberDepositorInserttdTomato
UseCre/Lox and Mouse TargetingExpressionBacterial and MammalianAvailable SinceApril 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
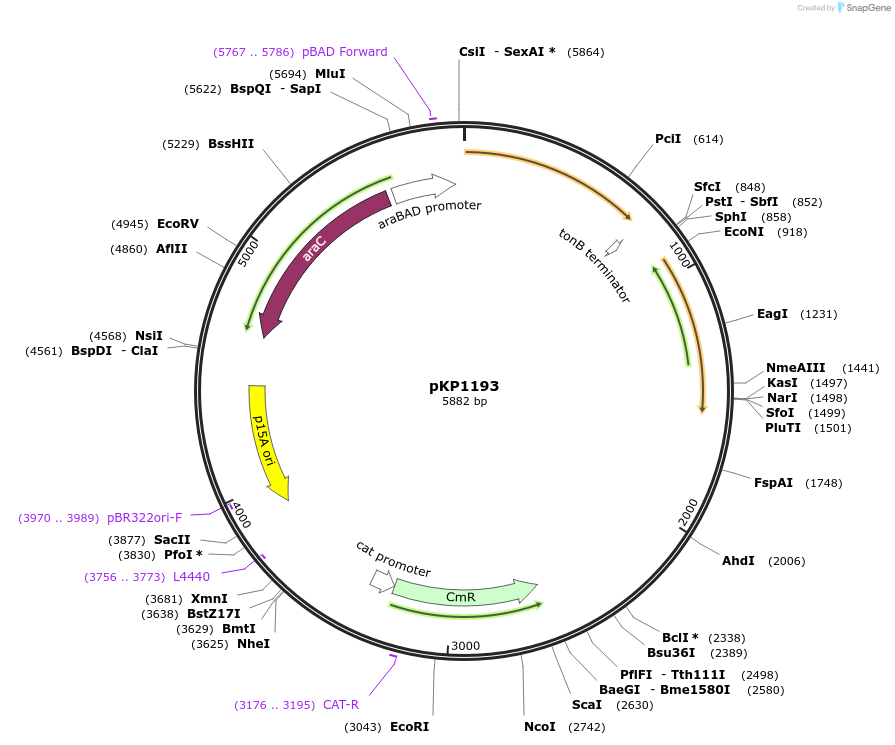
pKP1193
Plasmid#167614PurposeExpress TonB H20NDepositorInserttonB H20N
ExpressionBacterialMutationtonB with H20N mutationAvailable SinceJuly 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
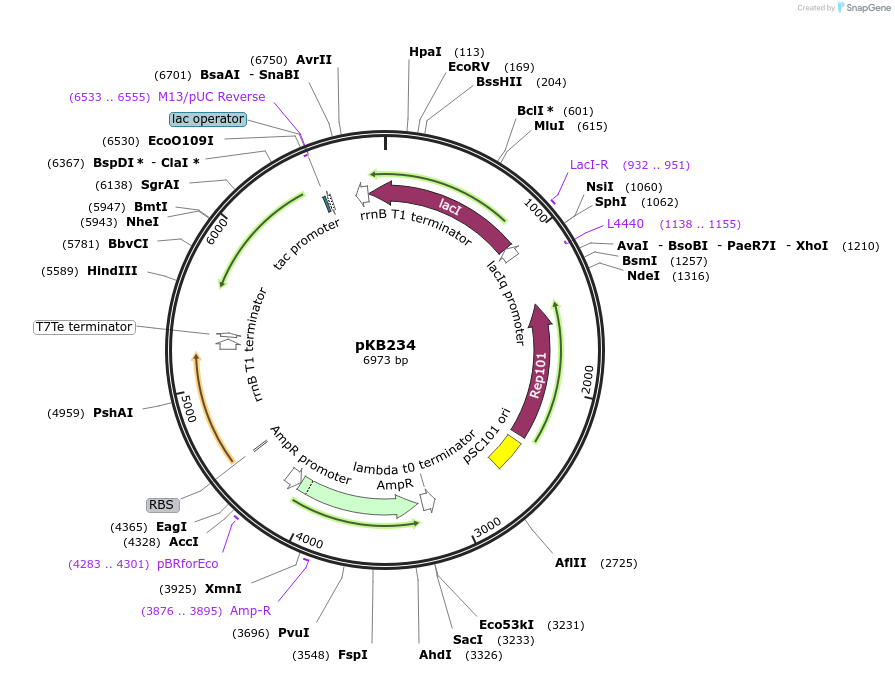
pKB234
Plasmid#170021PurposeEncodes LacI-controlled expression of surface-displayed hBD3 and PvirK-mNeonGreen reporterDepositorArticleInsertsmNeonGreen
hBD3
UseSynthetic BiologyTagsLpp, OmpA, and TetherExpressionBacterialPromoterPtac and PvirKAvailabilityAcademic Institutions and Nonprofits only -
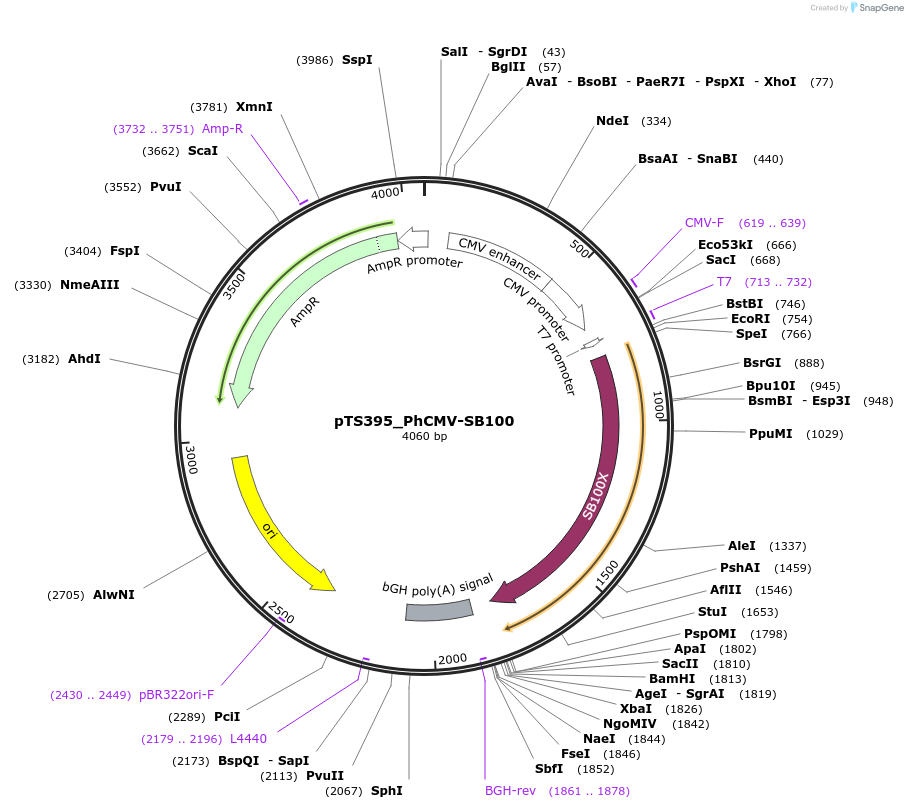
pTS395_PhCMV-SB100
Plasmid#169633PurposeExpression vector encoding PhCMV driven SB100 sleeping beauty transposase (PhCMV-SB100-pA)DepositorInsertPCMV-driven SB100
ExpressionMammalianPromoterPhCMVAvailable SinceJune 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
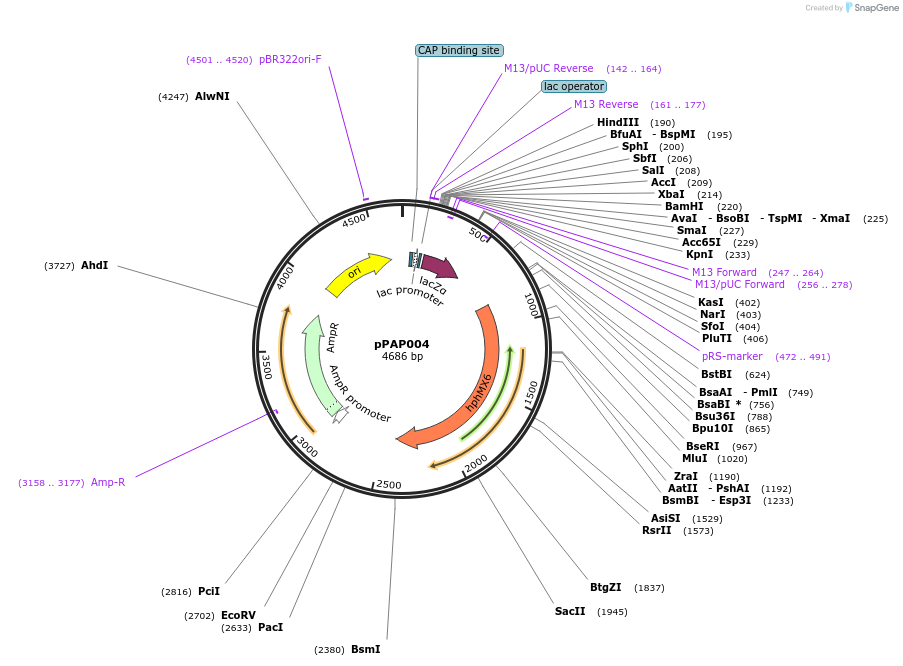
pPAP004
Plasmid#160911PurposeE.coli/ P.pastoris shuttle plasmid for episomal expression in P.pastorisDepositorInsertlacZ selection cassette
ExpressionYeastAvailable SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
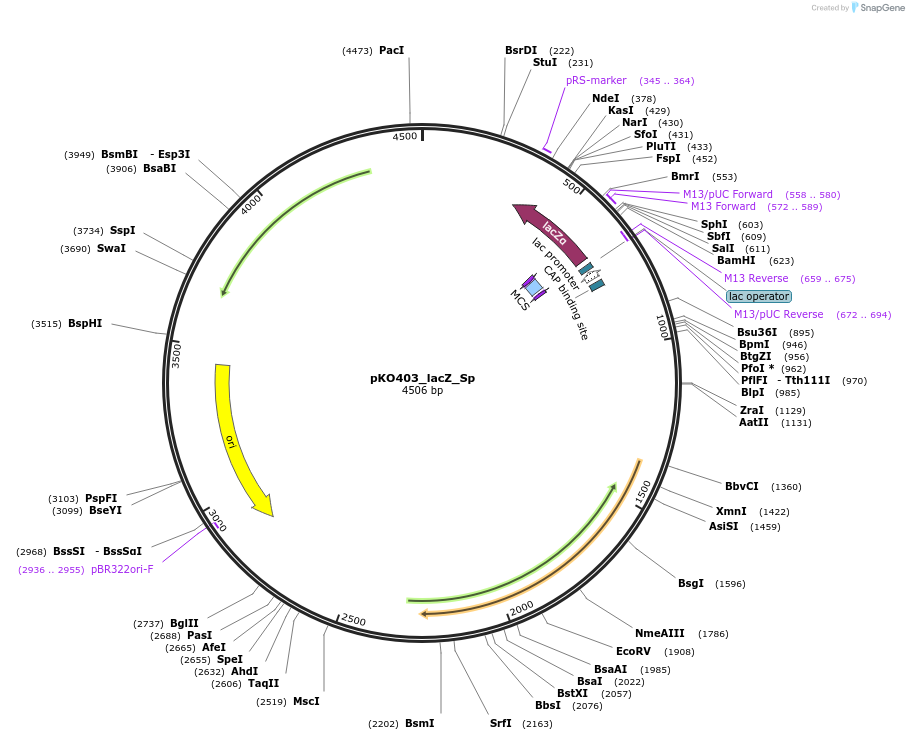
pKO403_lacZ_Sp
Plasmid#174726PurposeBifidobacterium-E.coli shuttle vector for seamless gene cloningDepositorTypeEmpty backboneExpressionBacterialAvailable SinceMarch 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
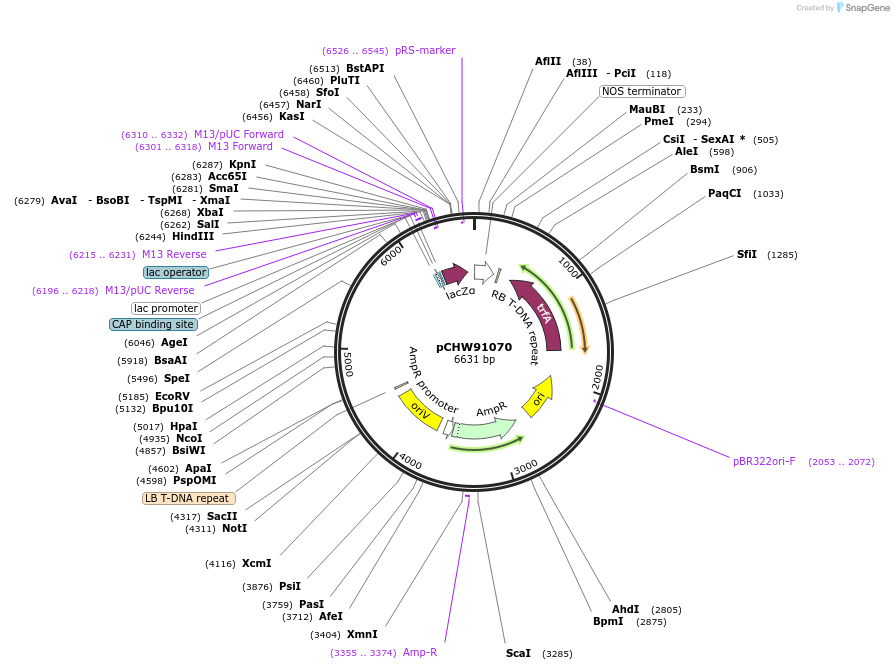
pCHW91070
Plasmid#222463PurposeLevel 1 acceptor, Position 3 containing CBS4-CMV1::mCherry2-HyP5SM-LacZ::nosT.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionPlantMutationnoPromoterCBS4-CMV1 (Cucumber Mosaic Virus)Available SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
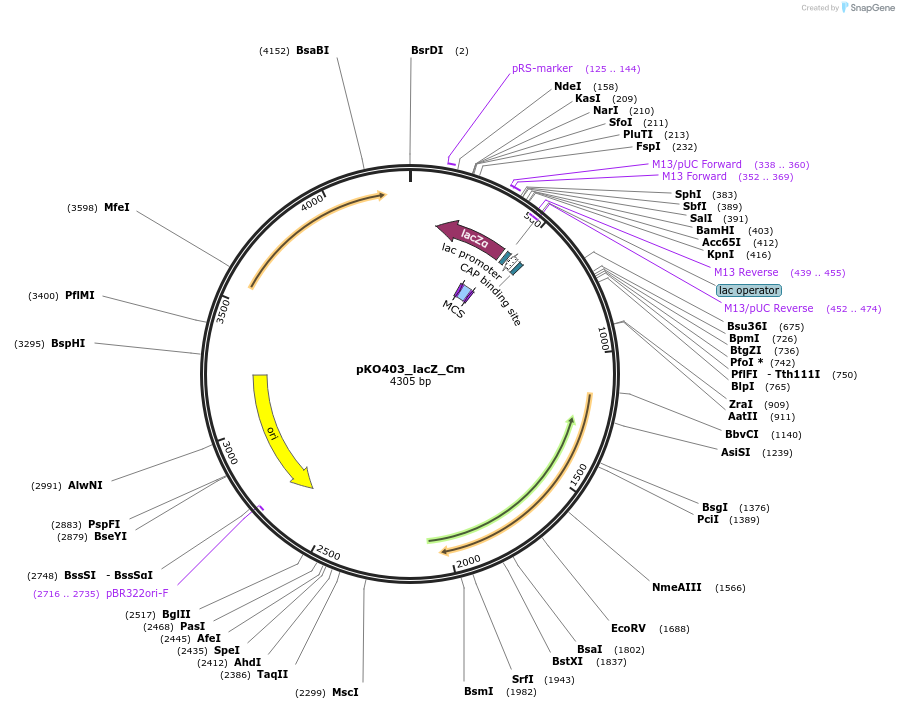
pKO403_lacZ_Cm
Plasmid#174725PurposeBifidobacterium-E.coli shuttle vector for seamless gene cloningDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJune 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
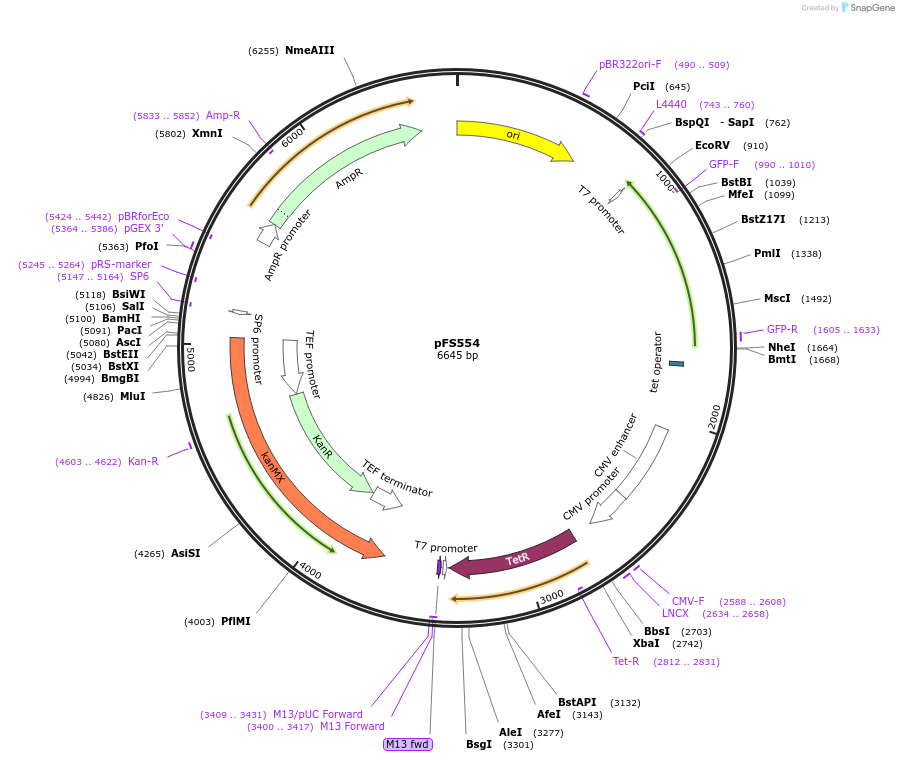
pFS554
Plasmid#242854PurposeThis plasmid will be used to replace the promoter of a gene with the PenotetSW3 promoter at its endogenous locusDepositorInsertsTetR
mECitrine
ExpressionYeastPromoterCMV promoter and enotetSW3 promoterAvailable SinceSept. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
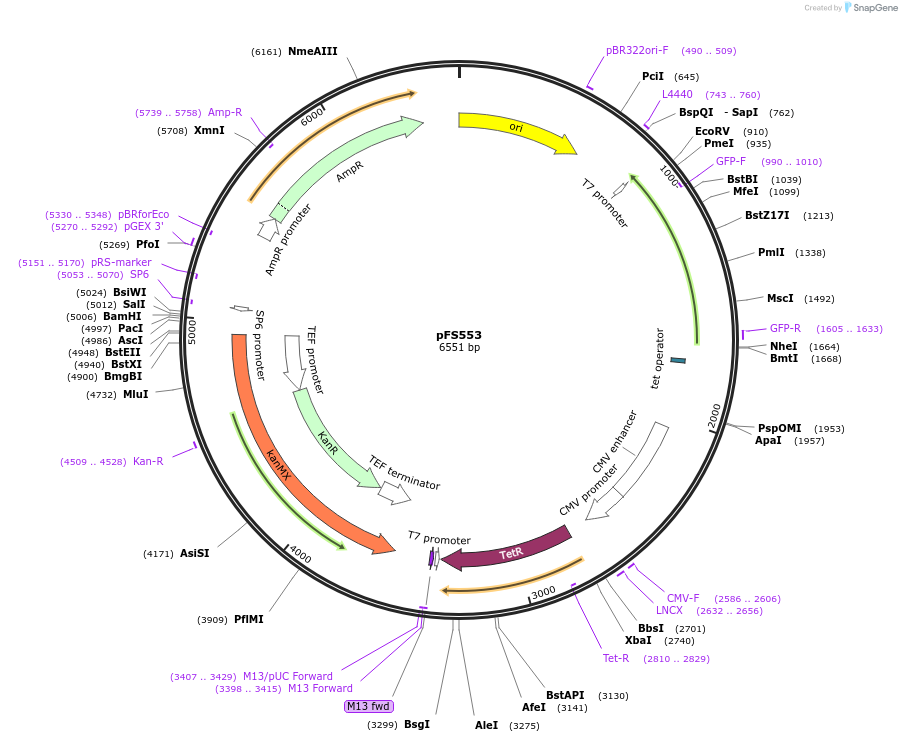
pFS553
Plasmid#242853PurposeThis plasmid will be used to replace the promoter of a gene with the PenotetSW2 promoter at its endogenous locusDepositorInsertsTetR
mECitrine
ExpressionYeastPromoterCMV promoter and enotetSW2 promoterAvailable SinceAug. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
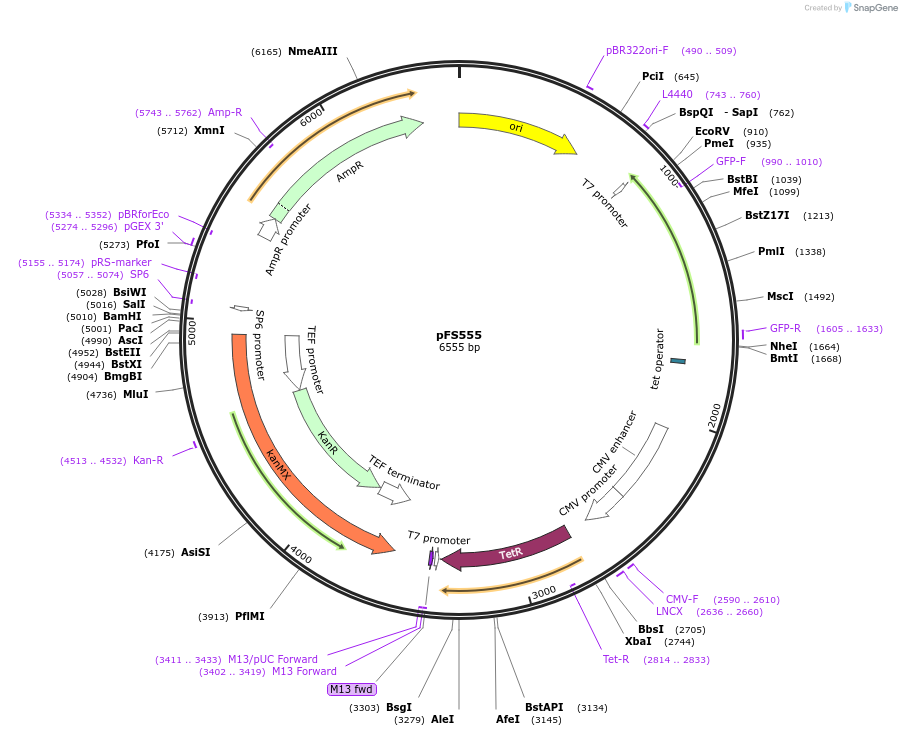
pFS555
Plasmid#242855PurposeThis plasmid will be used to replace the promoter of a gene with the PenotetSW4 promoter at its endogenous locusDepositorInsertsTetR
mECitrine
ExpressionYeastPromoterCMV promoter and enotetSW4 promoterAvailable SinceAug. 25, 2025AvailabilityAcademic Institutions and Nonprofits only