We narrowed to 4,541 results for: ALK
-
Plasmid#207139PurposeAssembled Tn10 with ptac promoter driving gfpDepositorInsertTn10-Ptac-GFP
UseSynthetic BiologyExpressionBacterialPromoterptacAvailable SinceJuly 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
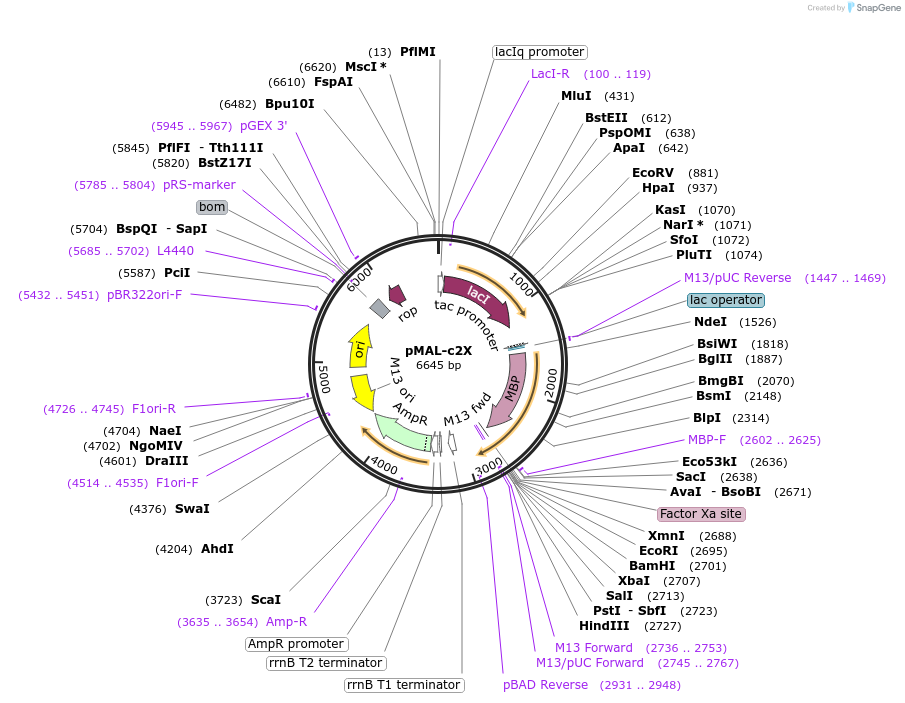
pMAL-c2X
Plasmid#75286PurposeExpresses proteins in the cytoplasm as fusions to maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 18, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
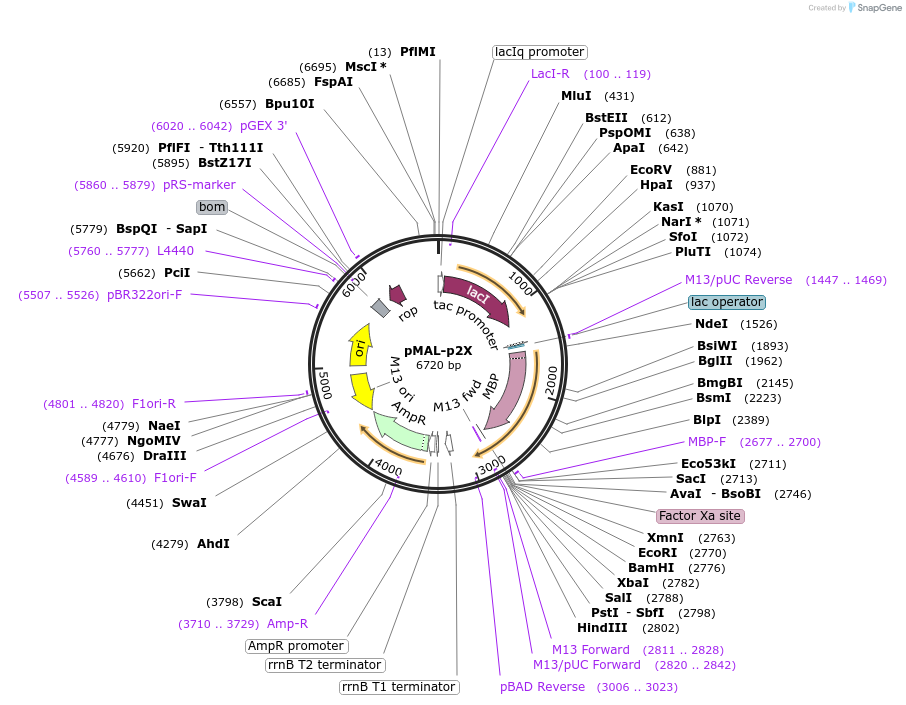
pMAL-p2X
Plasmid#75287PurposeExpresses proteins in the periplasm as fusions to maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 18, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
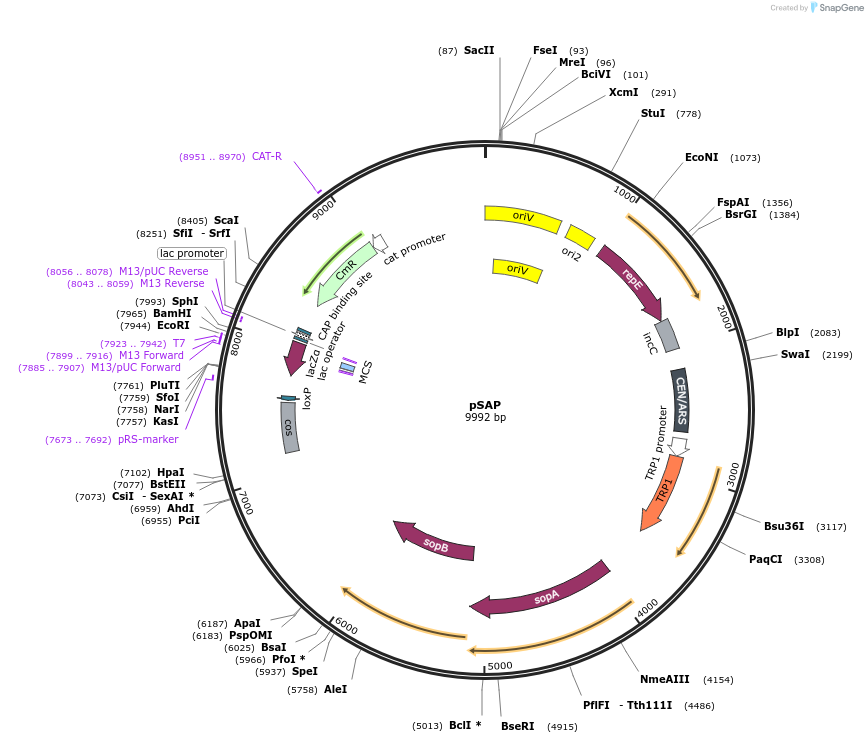
pSAP
Plasmid#206429PurposeA vector backbone domesticated for SapI (pCC1BAC-based), contains elements for selection, maintenance, and propagation in S. cerevisiae (TRP1) and E. coli (cat).DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
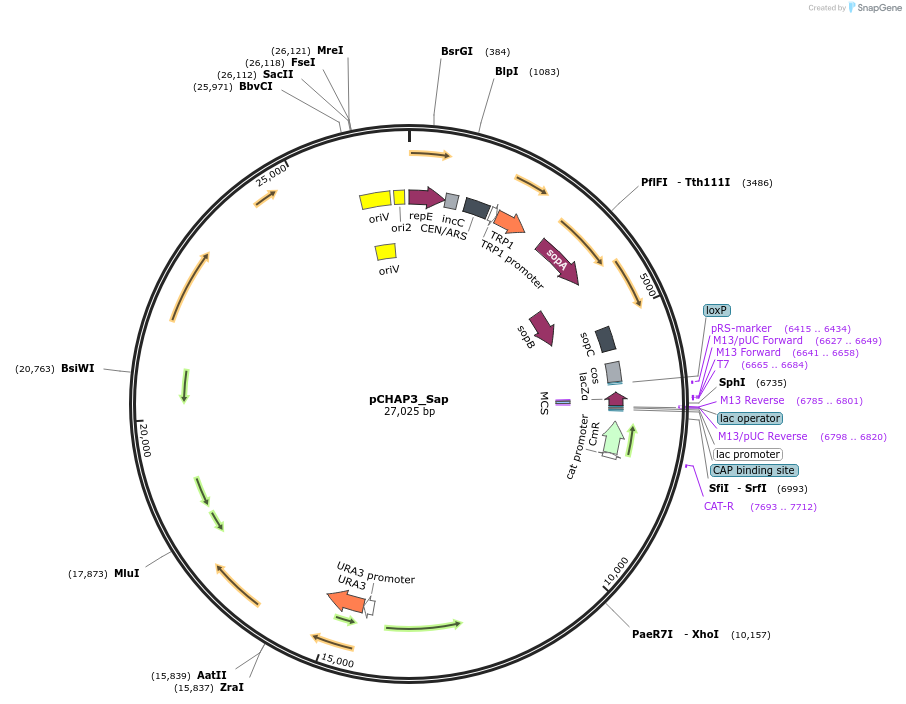
pCHAP3_Sap
Plasmid#206849PurposeA plasmid containing a 17,271 bp fragment of the P. tricornutum chloroplast genome. The yeast marker URA3 was domesticated for SapI and integrated into the chloroplast fragment.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
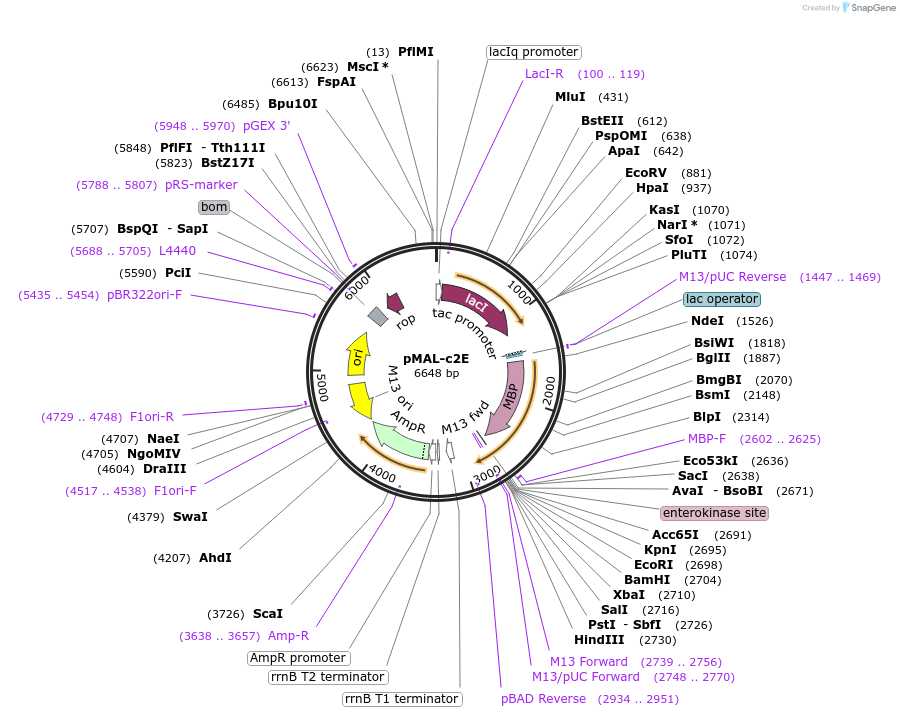
pMAL-c2E
Plasmid#75291PurposeExpresses proteins in the cytoplasm as fusions to maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 20, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
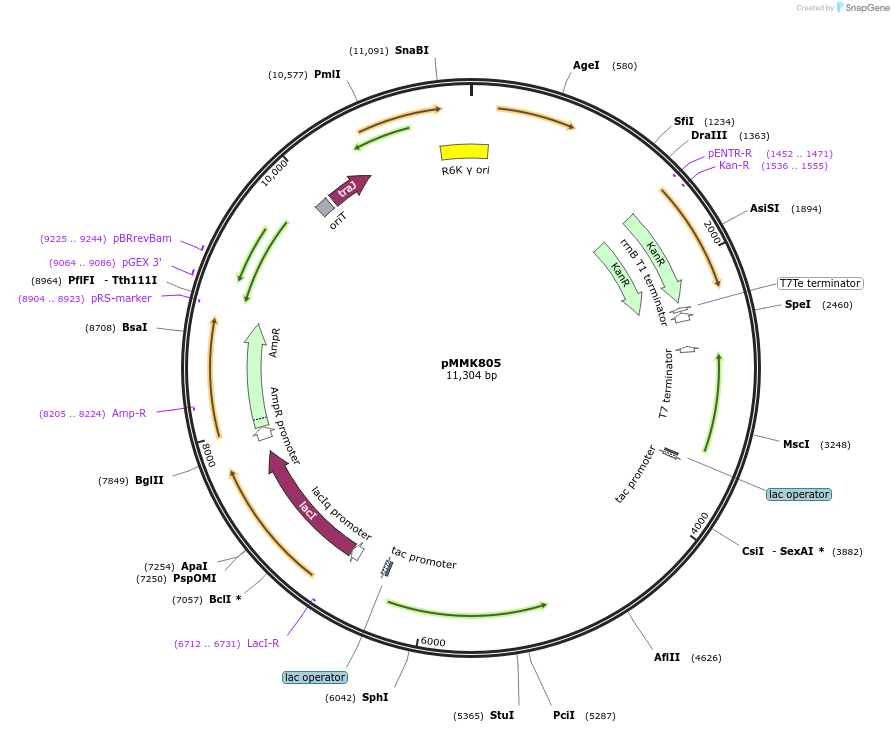
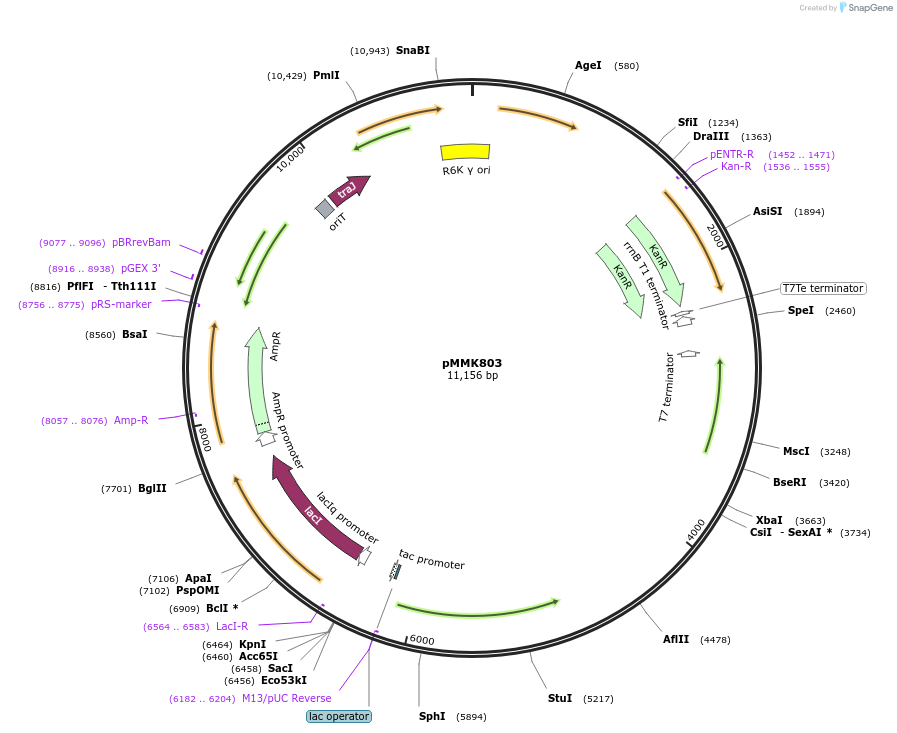
pMMK803
Plasmid#207138PurposeAssembled Tn10 with CP25 promoter driving gfpDepositorInsertTn10-CP25-GFP
UseSynthetic BiologyExpressionBacterialPromoterCP25Available SinceSept. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
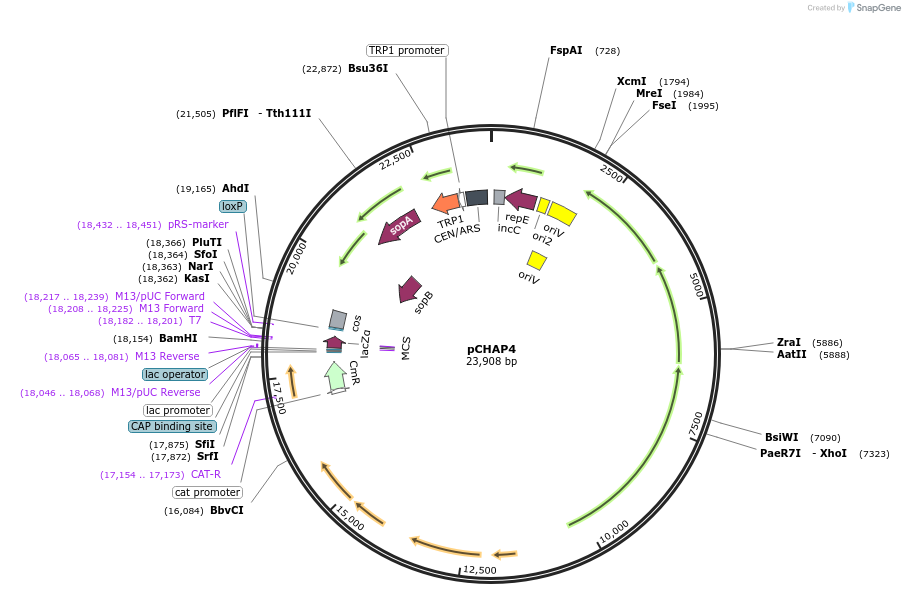
pCHAP4
Plasmid#206850PurposeA plasmid containing a 16,121 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
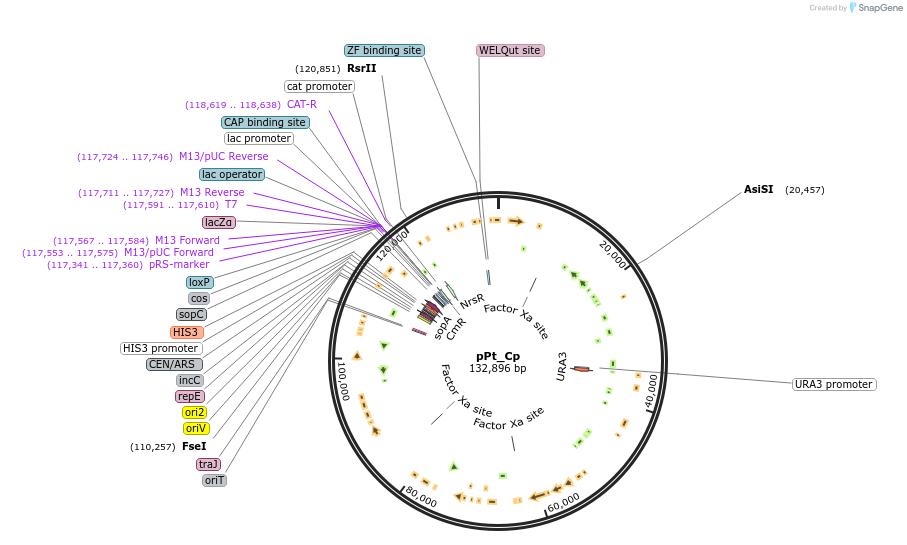
pPt_Cp
Plasmid#206855PurposeA plasmid containing the whole P. tricornutum chloroplast genome, cloned with a pCC1BAC-based backbone. The marker URA3 was integrated into the genome.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
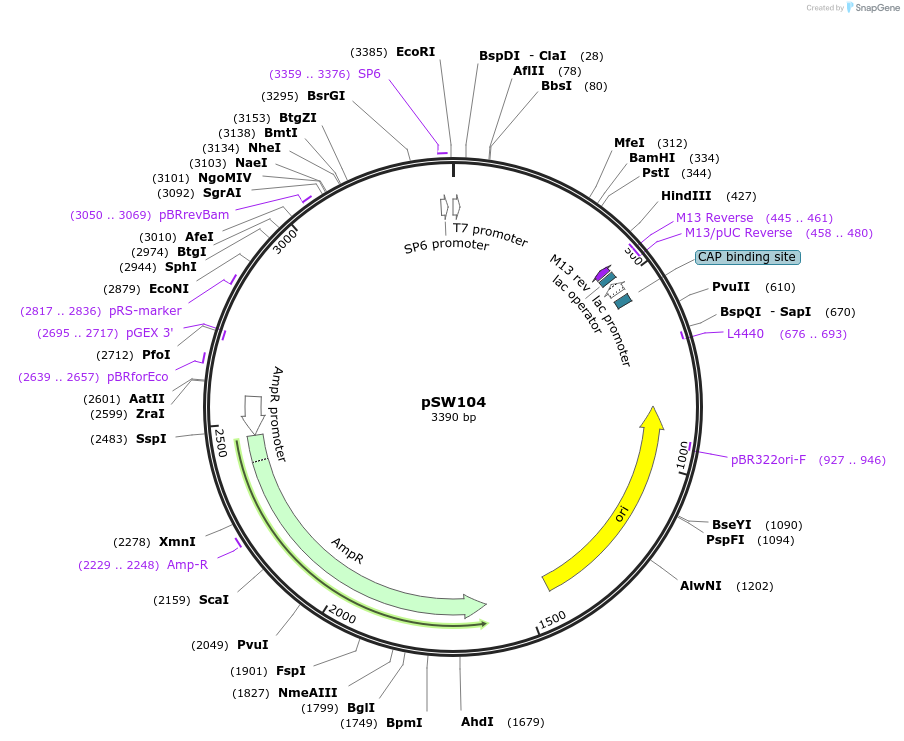
pSW104
Plasmid#122295PurposeFor run-off transcription of yeast RPL41A mRNA with and without an 82 nucleotide poly(A) tail.DepositorInsertRPL41A
UseIn vitro transcriptionAvailable SinceJuly 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMON-HsNMT2
Plasmid#67476PurposeExpression of human N-myristoyltransferase 2 for N-myristoylation of ARF family proteins in bacteriaDepositorInsertHsNMT2 (NMT2 Human)
ExpressionBacterialAvailable SinceOct. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
JMC02A-C10
Plasmid#58754Purposebacterial expression of PDB 4O61DepositorInsertALKBH5 (ALKBH5 Human)
TagsHisExpressionBacterialMutationBacterial expression for structure determination;…PromoterT7Available SinceAug. 4, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
pOX + kqt2
Plasmid#16137DepositorInsertCaenorhabditis elegans potassium channel (kqt-2 Nematode)
UseXenopus oocyte expressionMutationDeletion of aa1-38 compared to GenBank NP_509392.3Available SinceJan. 10, 2008AvailabilityAcademic Institutions and Nonprofits only -
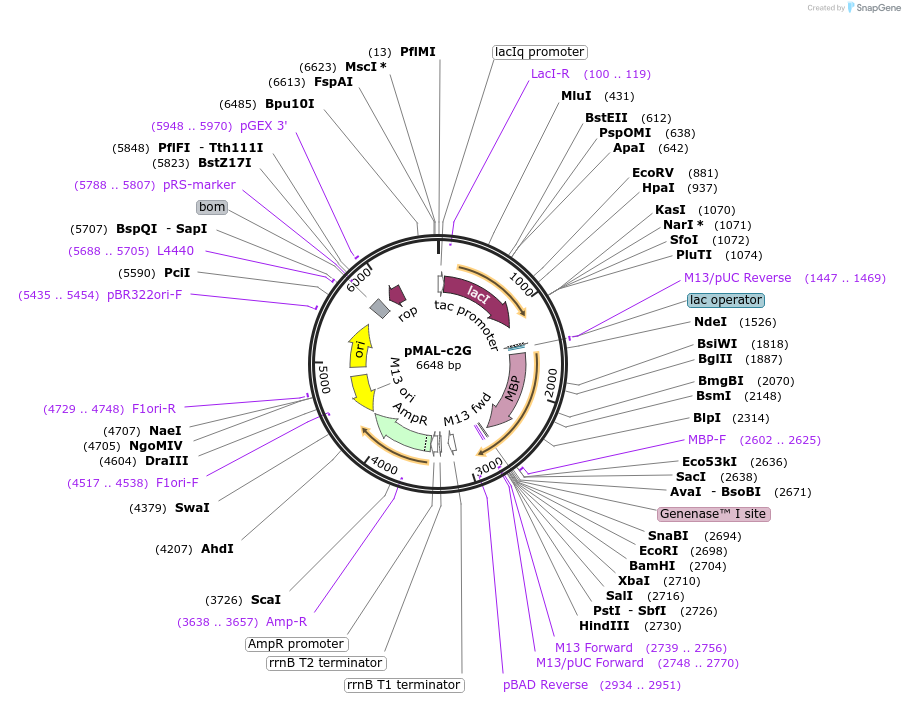
pMAL-c2G
Plasmid#75290PurposeExpresses proteins in the cytoplasm as fusions to maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 20, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
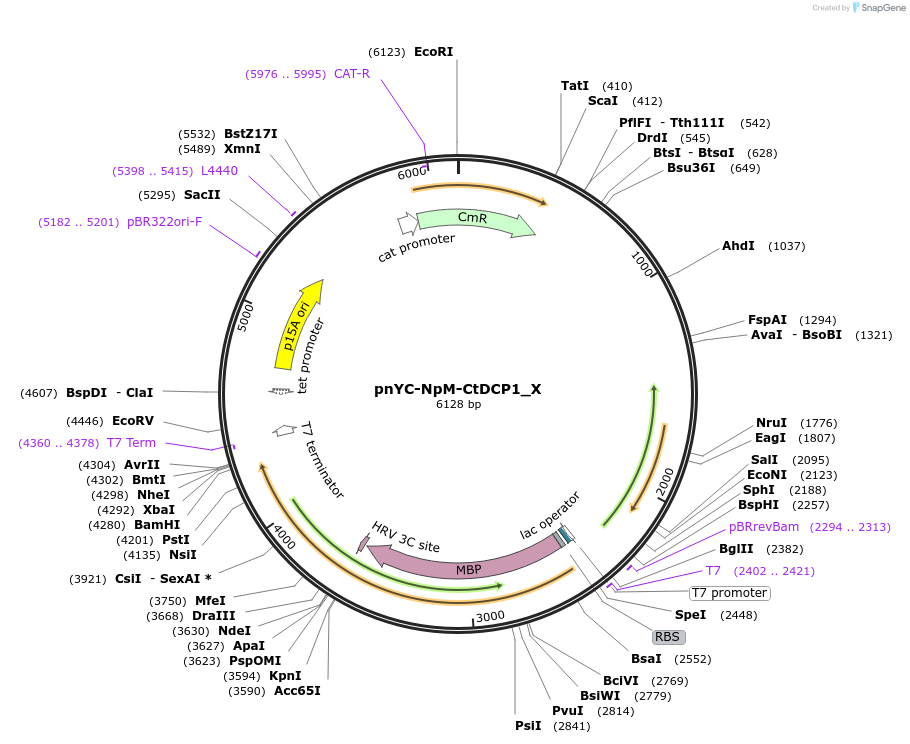
pnYC-NpM-CtDCP1_X
Plasmid#148006PurposeBacterial Expression of CtDCP1DepositorInsertCtDCP1
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pOX + JShal
Plasmid#16147DepositorInsertPolyorchis penicillatus potassium channel alpha subunit (Shal jelly fish)
UseXenopus oocyte expressionMutationN/AAvailable SinceJan. 10, 2008AvailabilityAcademic Institutions and Nonprofits only -
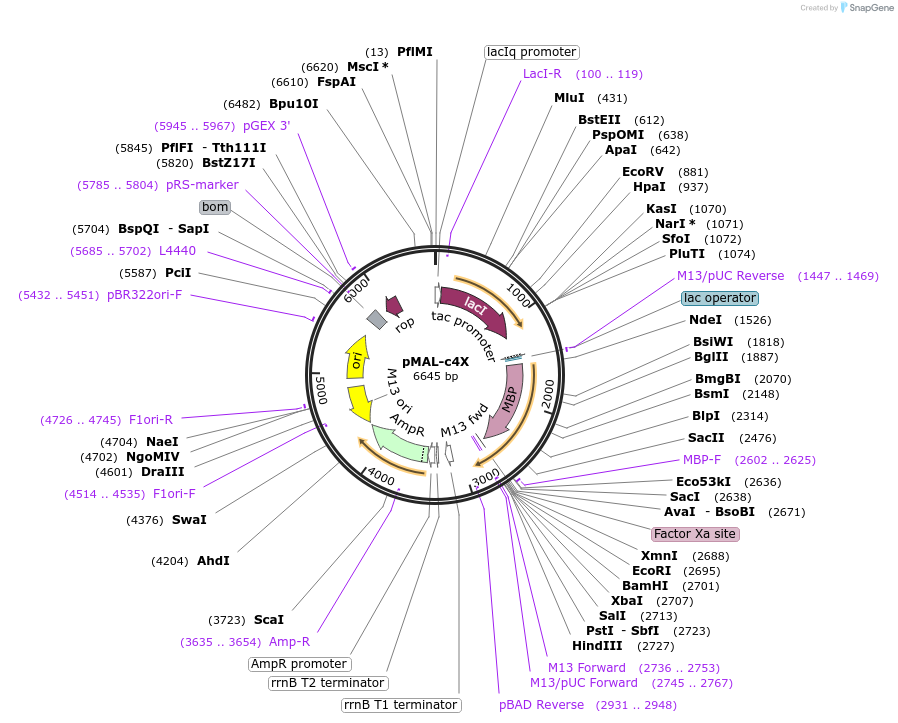
pMAL-c4X
Plasmid#75288PurposeExpresses proteins in the cytoplasm as fusions to a high-binding mutant (A313V) maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 19, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
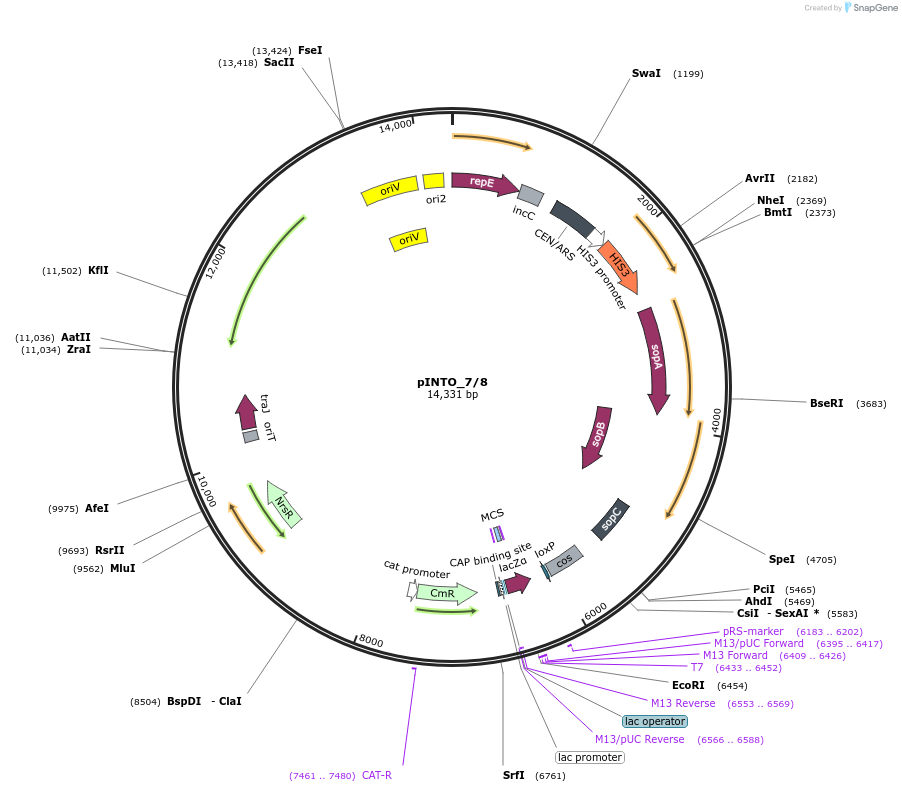
pINTO_7/8
Plasmid#206431PurposeA vector backbone (pCC1BAC-based) containing a single SapI recognition site. When digested, it exposes overhangs that allow integration of the vector into the P. tricornutum chloroplast genome.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pMAL-p4X
Plasmid#75289PurposeExpresses proteins in the periplasm as fusions to a high-binding mutant (A313V) maltose-binding proteinDepositorTypeEmpty backboneTagsmaltose-binding proteinExpressionBacterialPromotertacAvailable SinceJuly 19, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
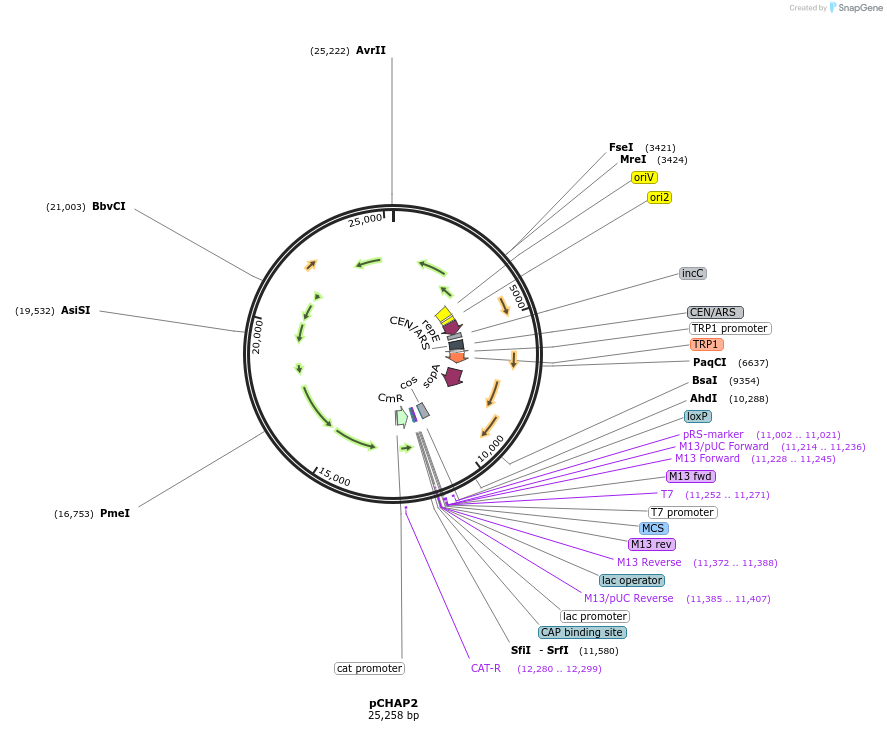
pCHAP2
Plasmid#206847PurposeA plasmid containing a 15,249 bp fragment of the P. tricornutum chloroplast genome. The fragment is flanked by SapI recognition sites, allowing it to be released from the cloning vector pSAP1.DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 23, 2024AvailabilityAcademic Institutions and Nonprofits only