We narrowed to 36,095 results for: Sis;
-
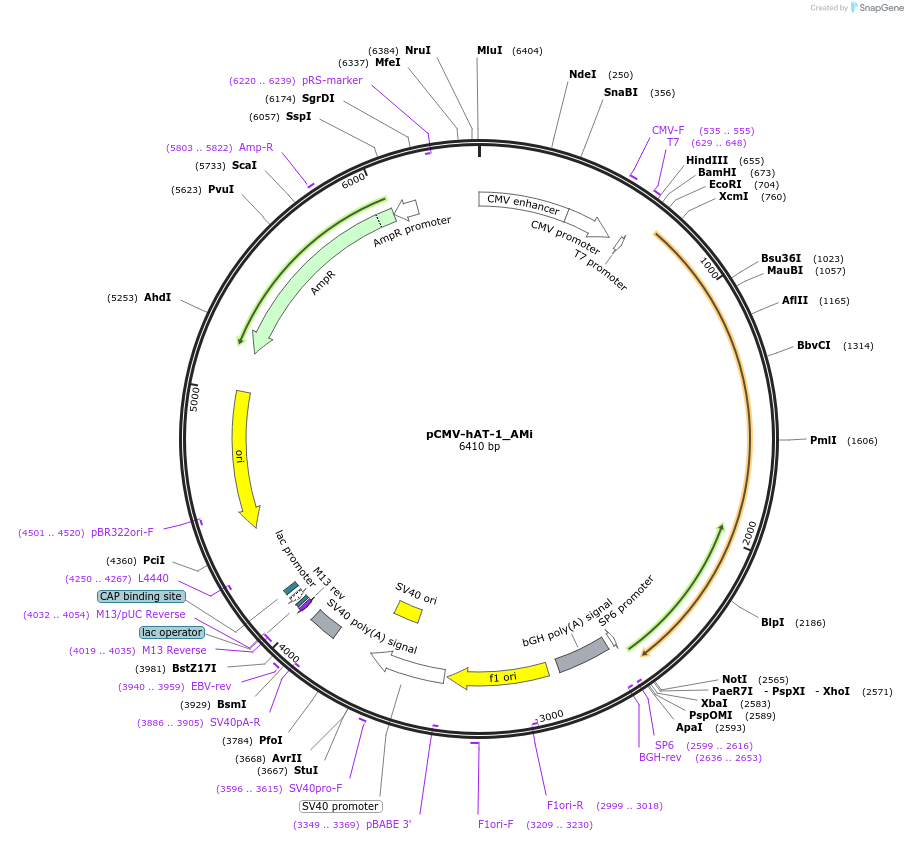
Plasmid#217170PurposeMammalian expression vector for expression of hAT-1_Ami transposase (CMV promoter)DepositorInserthAT-1_AMi transposase
ExpressionMammalianAvailable SinceOct. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCMV-Mariner-2_AMi
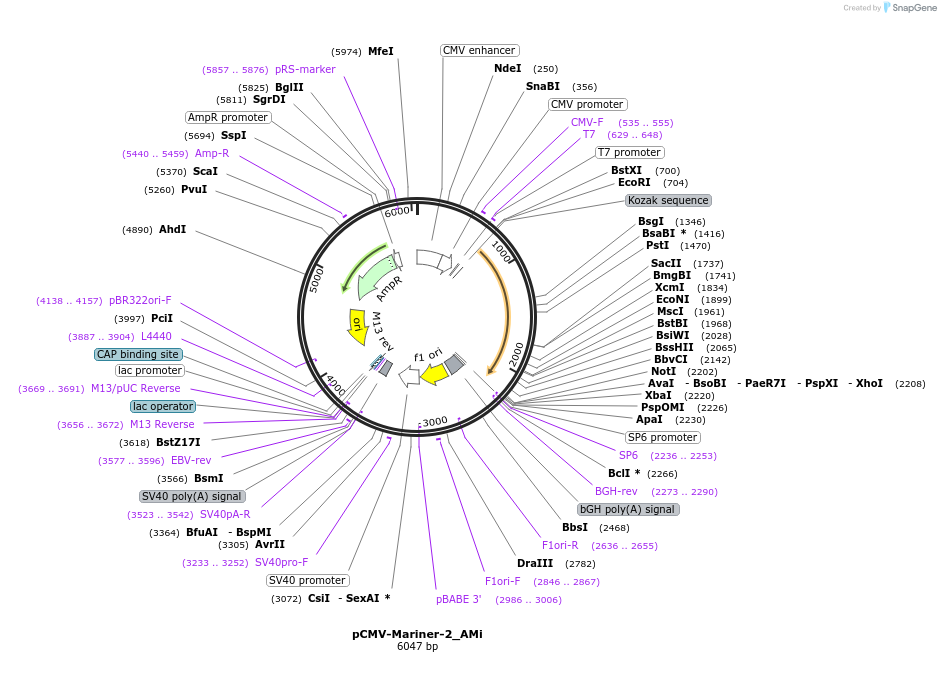
Plasmid#217171PurposeMammalian expression vector for expression of Mariner-2_Ami transposase (CMV promoter)DepositorInsertMariner-2_AMi transposase
ExpressionMammalianAvailable SinceOct. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCMV-hAT-13_AMi
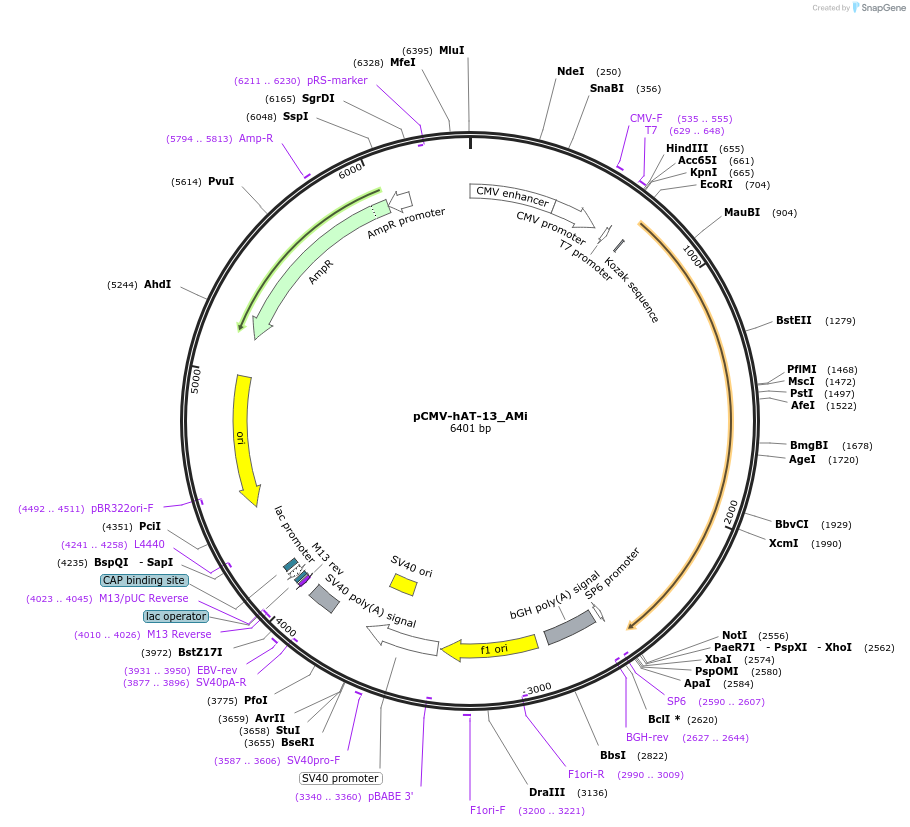
Plasmid#217172PurposeMammalian expression vector for expression of hAT-13_Ami transposase (CMV promoter)DepositorInserthAT-13_AMi transposase
ExpressionMammalianAvailable SinceOct. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1-AtSLAC1 WT
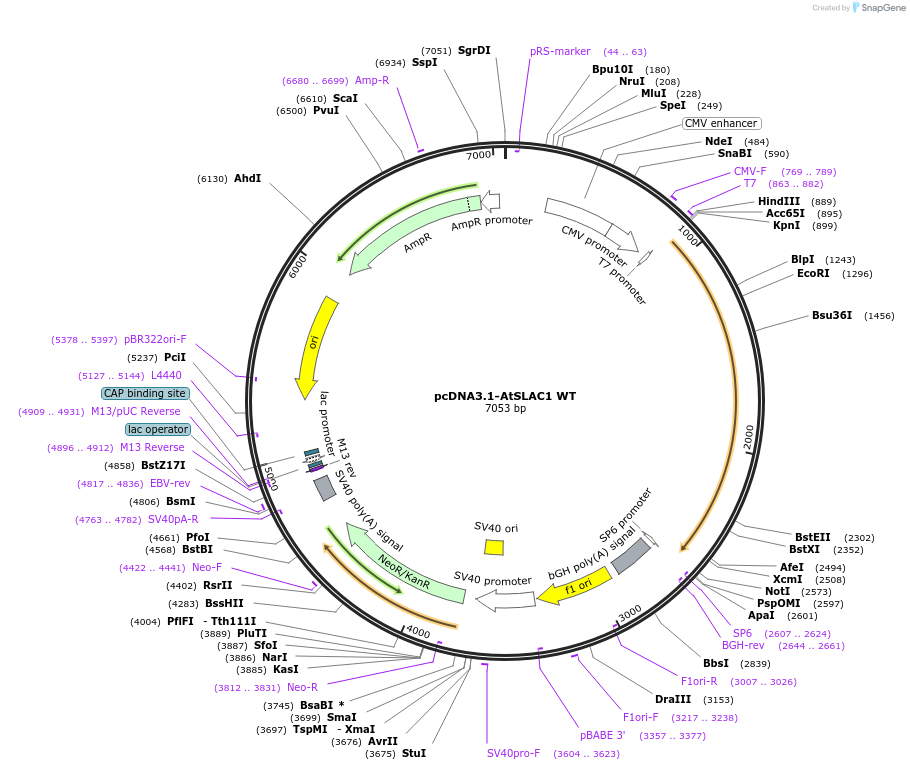
Plasmid#212946PurposeVector used for expression of AtSLAC1 WT in mammalian cellDepositorAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
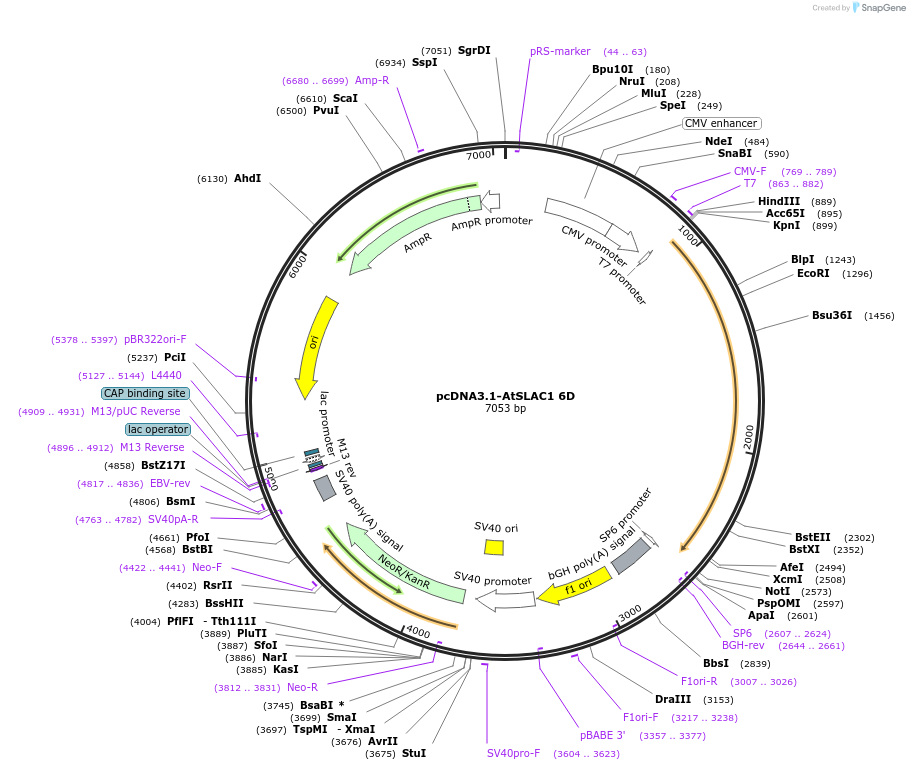
pcDNA3.1-AtSLAC1 6D
Plasmid#212947PurposeVector used for expression of AtSLAC1 6D in mammalian cellDepositorInsertSlow anion channel-associated 1 (OZS1 Mustard Weed)
ExpressionMammalianMutationT62D/S65D/S107D/S124D/S146D/S152DPromoterCMVAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
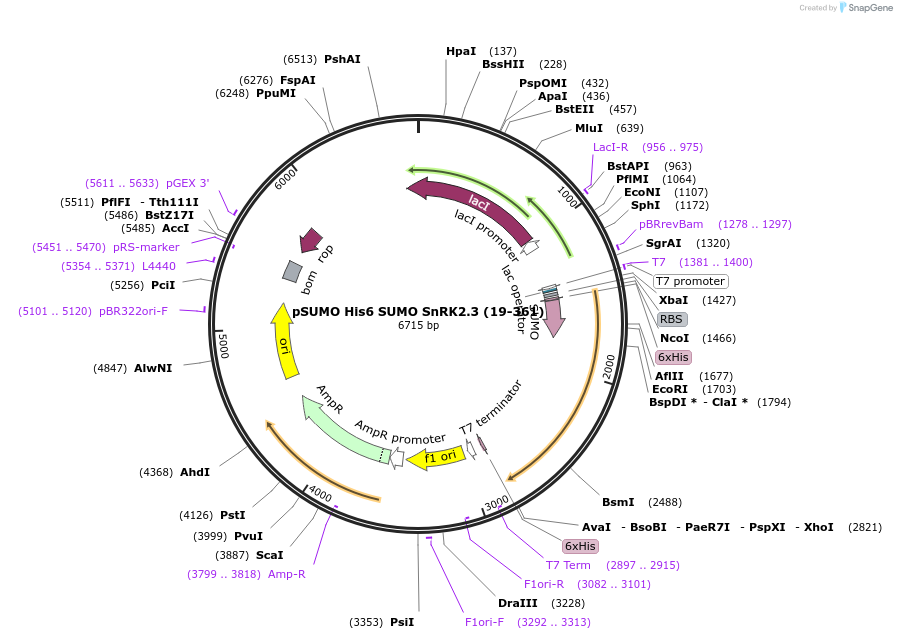
pSUMO His6 SUMO SnRK2.3 (19-361)
Plasmid#177846PurposeBacterial Expression of SnRK2.3DepositorAvailable SinceDec. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
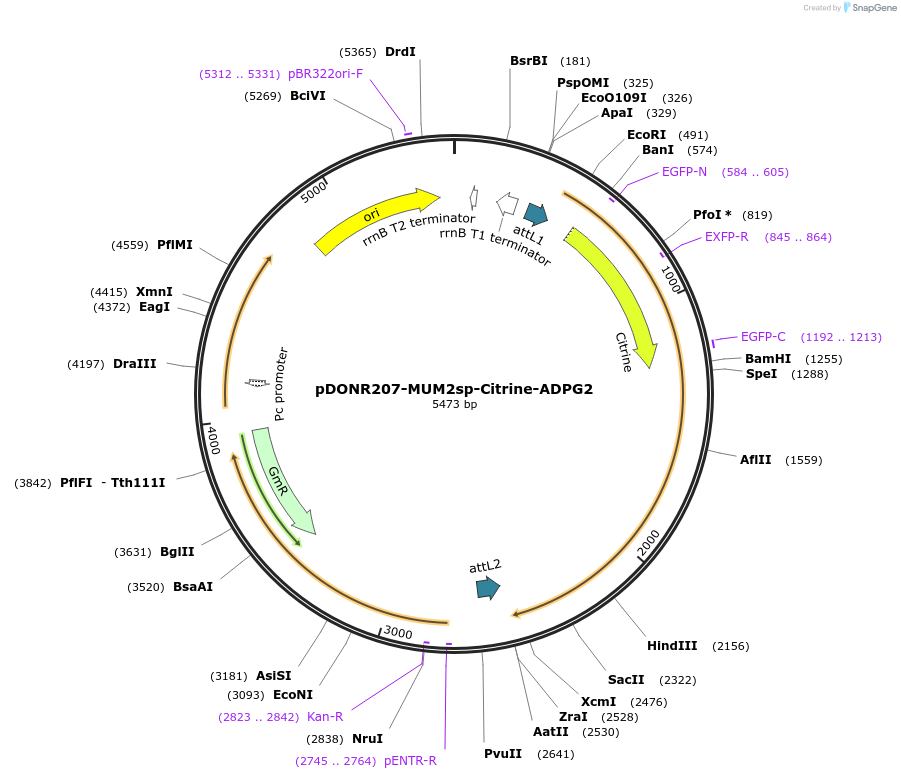
pDONR207-MUM2sp-Citrine-ADPG2
Plasmid#170725PurposeGateway (Invitrogen) entry clone (pDONR207) containing the MUM2 (At5g63800) signal peptide (84bp) fused in-frame to the Citrine and the coding sequence of the HG degrading enzyme ADPG2 (At2g41850.1)DepositorInsertARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2 (PGAZAT Mustard Weed)
UseGateway donor vector / entry cloneTagsCitrine tag (714bp)Available SinceNov. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
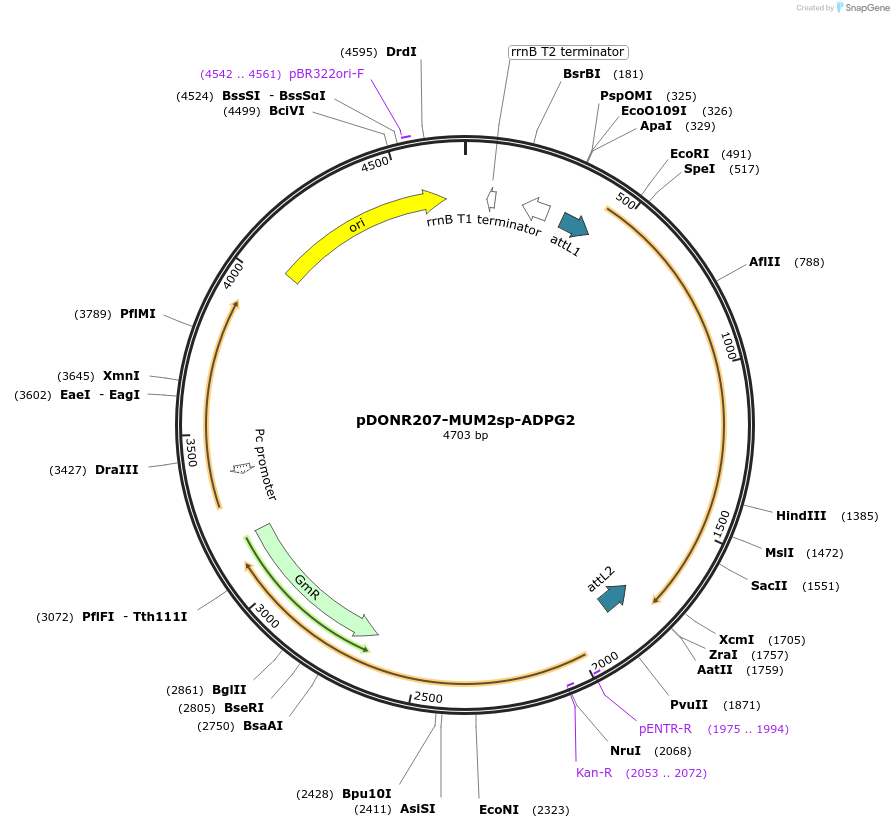
pDONR207-MUM2sp-ADPG2
Plasmid#170723PurposeGateway (Invitrogen) entry clone (pDONR207) containing the MUM2 (At5g63800) signal peptide (84bp) fused in-frame to the coding sequence of the HG degrading enzyme ADPG2 (At2g41850.1)DepositorInsertARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2 (PGAZAT Mustard Weed)
UseGateway donor vector / entry cloneAvailable SinceNov. 11, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
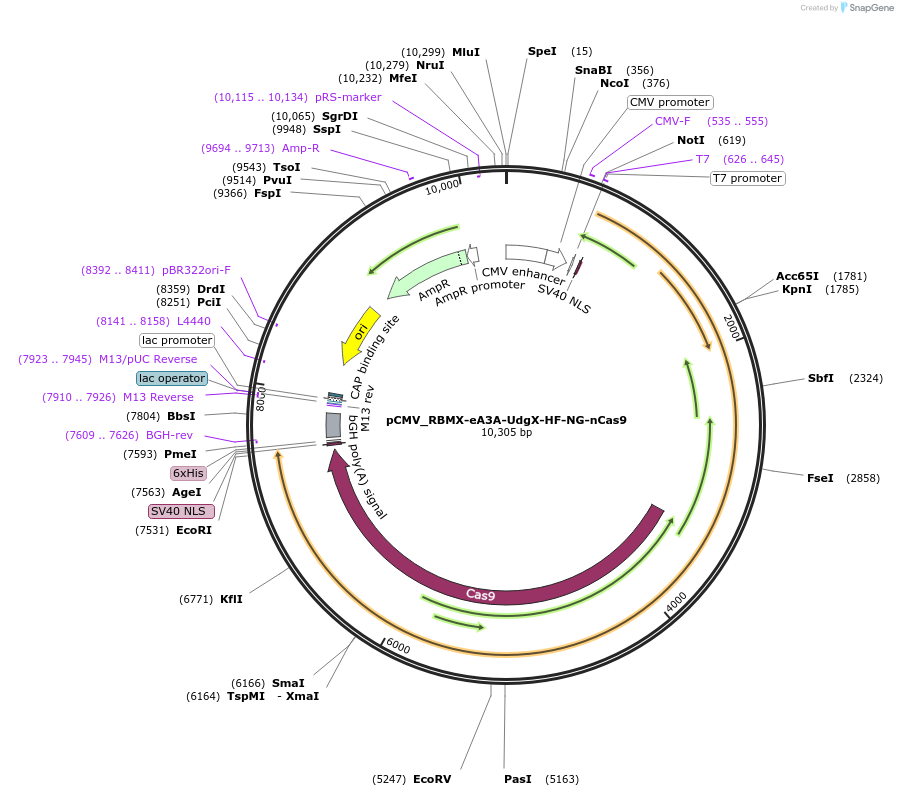
pCMV_RBMX-eA3A-UdgX-HF-NG-nCas9
Plasmid#163550PurposeMammalian CG-to-GC base editingDepositorInsertRBMX-eA3A-UdgX-HF-NG-nCas9
UseCRISPRExpressionMammalianMutationnCas9 (D10A)N497A, R661A, Q695A, Q926A//L111R, D1…Available SinceDec. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
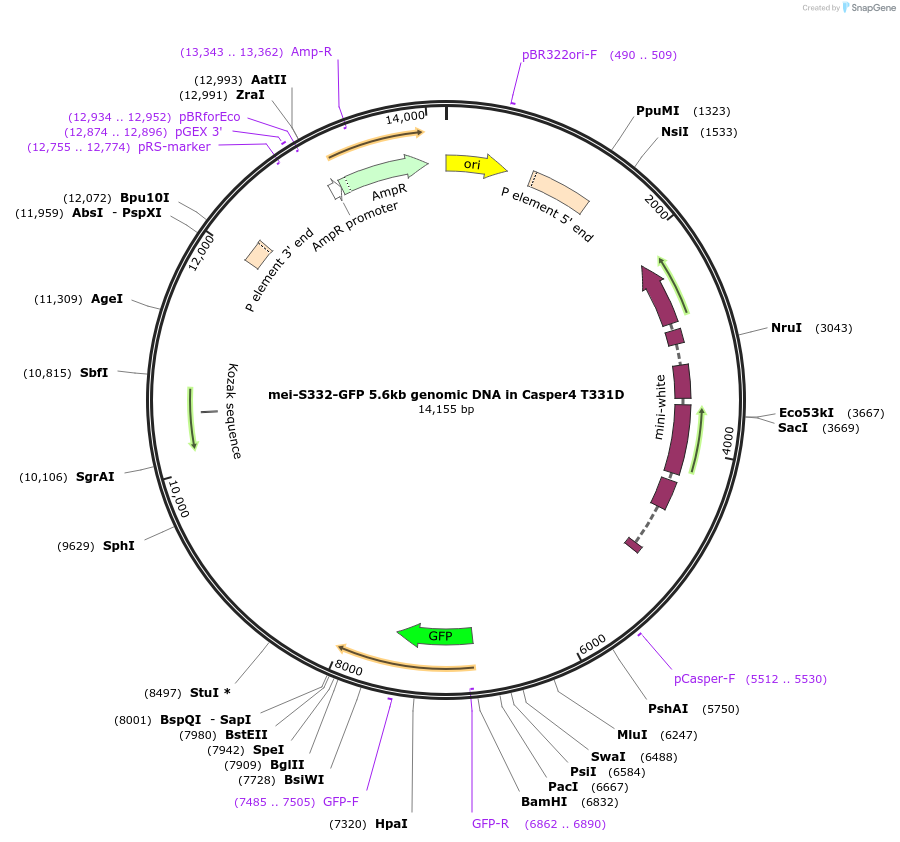
mei-S332-GFP 5.6kb genomic DNA in Casper4 T331D
Plasmid#112986PurposeP element vector for transposon insertion of mei-S332-GFP 5.6kb genomic DNA (T331D) into Drosophila genomeDepositorAvailable SinceAug. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
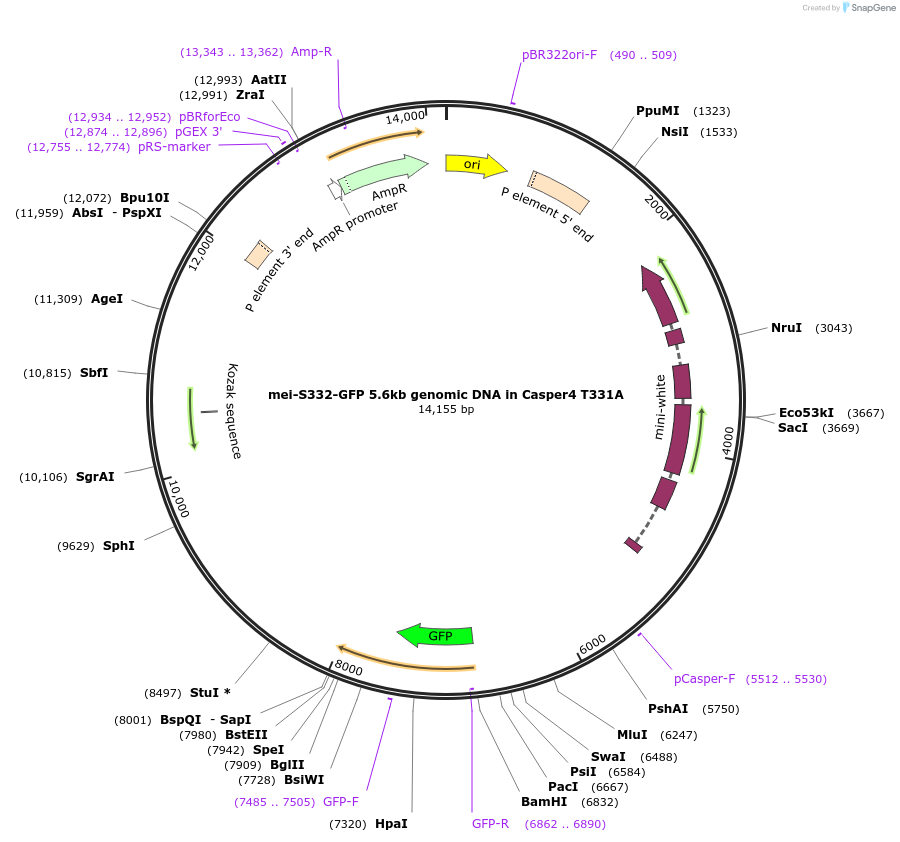
mei-S332-GFP 5.6kb genomic DNA in Casper4 T331A
Plasmid#112988PurposeP element vector for transposon insertion of mei-S332-GFP 5.6kb genomic DNA (T331A) into Drosophila genomeDepositorAvailable SinceAug. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
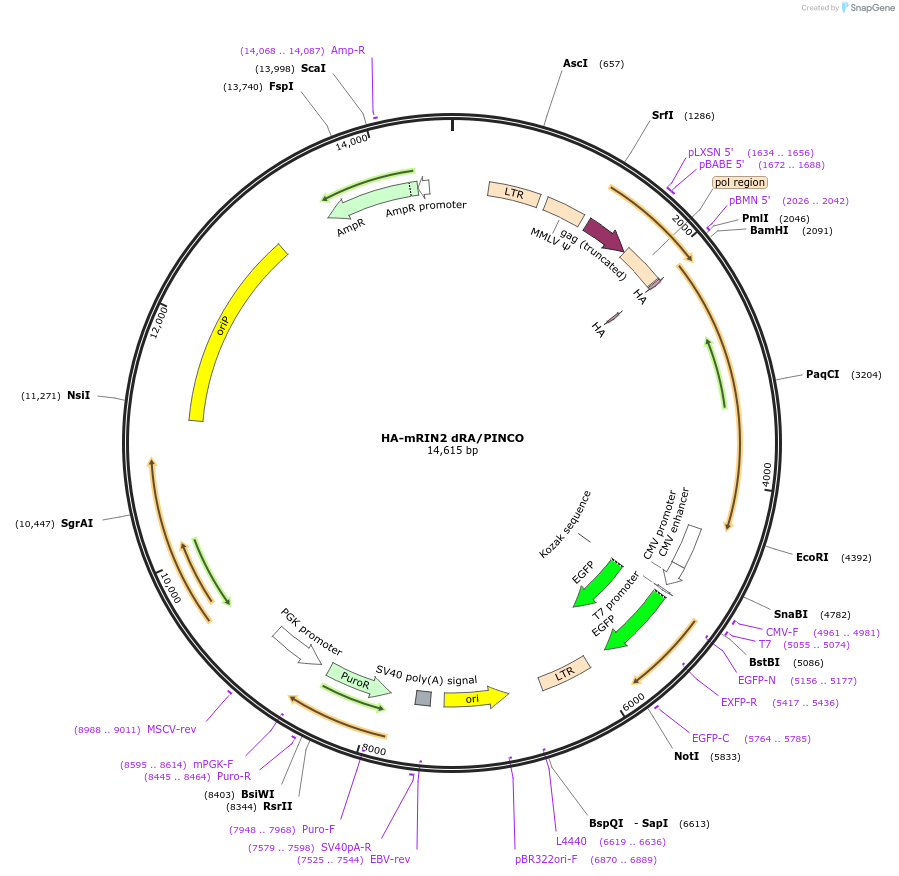
HA-mRIN2 dRA/PINCO
Plasmid#108938PurposeRetroviral expression of mRIN2 dRADepositorInsertRas and Rab interactor 2 (Rin2 Mouse)
UseRetroviralTagsHAExpressionMammalianMutationDeletion of RA domain (aa 751-836)PromoterLTRAvailable SinceJune 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
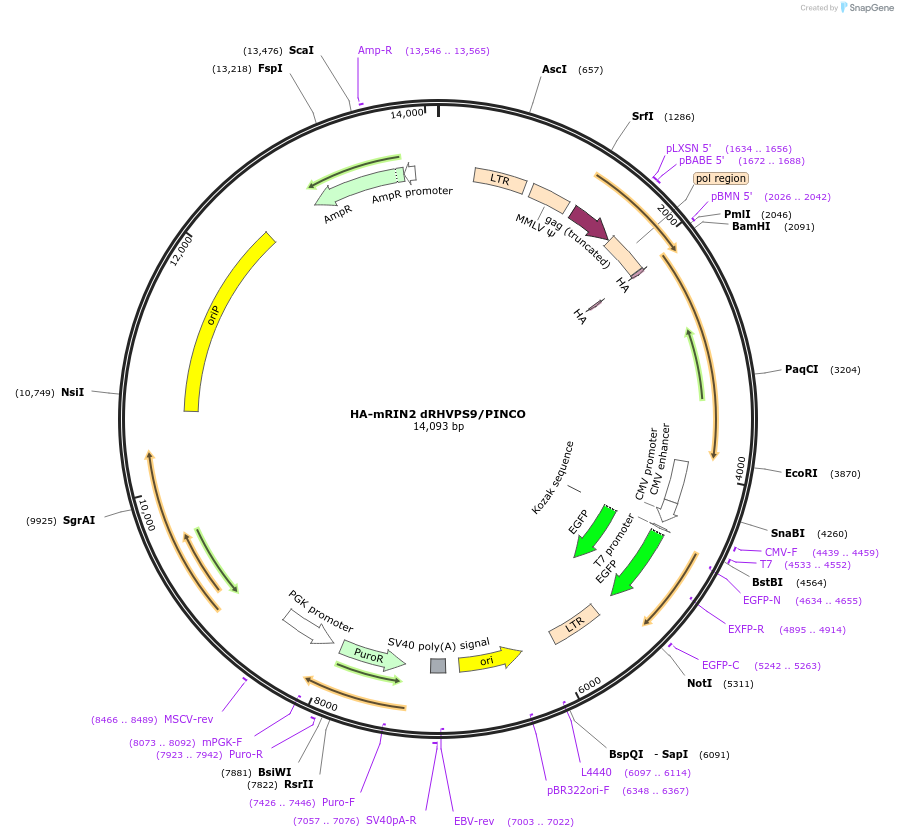
HA-mRIN2 dRHVPS9/PINCO
Plasmid#108939PurposeRetroviral expression of mRIN2 dRHVPS9DepositorInsertRas and Rab interactor 2 (Rin2 Mouse)
UseRetroviralTagsHAExpressionMammalianMutationDeletion of RHVPS9 domain (aa 455-738)PromoterLTRAvailable SinceJune 25, 2018AvailabilityAcademic Institutions and Nonprofits only -
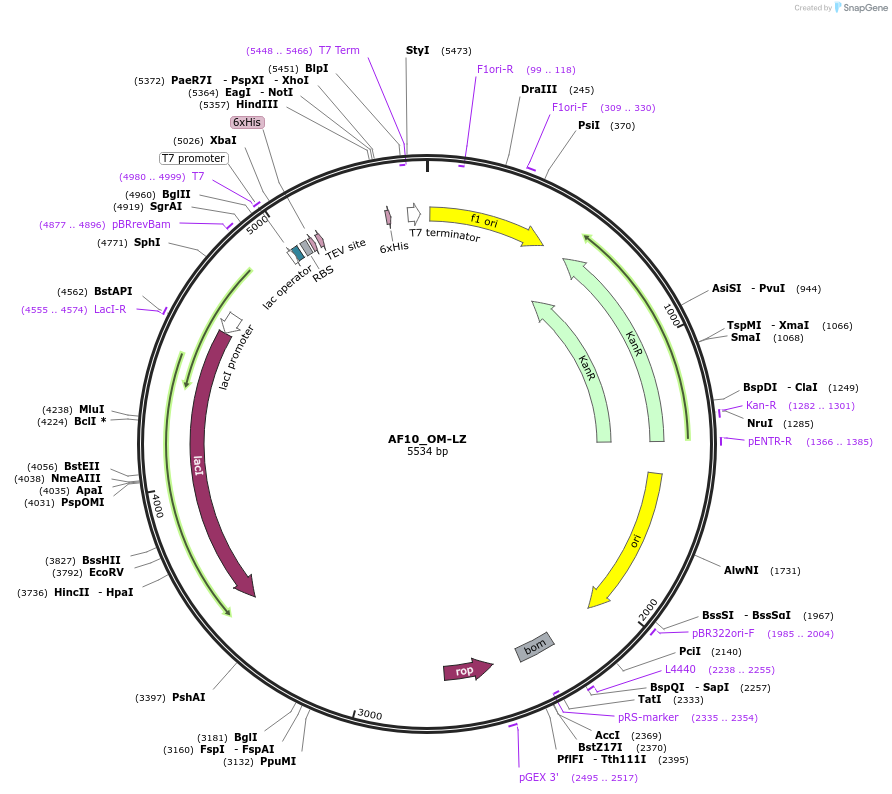
AF10_OM-LZ
Plasmid#111753PurposeBacterial expression for structure determinationDepositorInsertMLLT10 (MLLT10 Human)
Tags6x His - TEVExpressionBacterialMutationContains the OM-LZ region of AF10 (residues 720-7…PromoterT7Available SinceJune 14, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
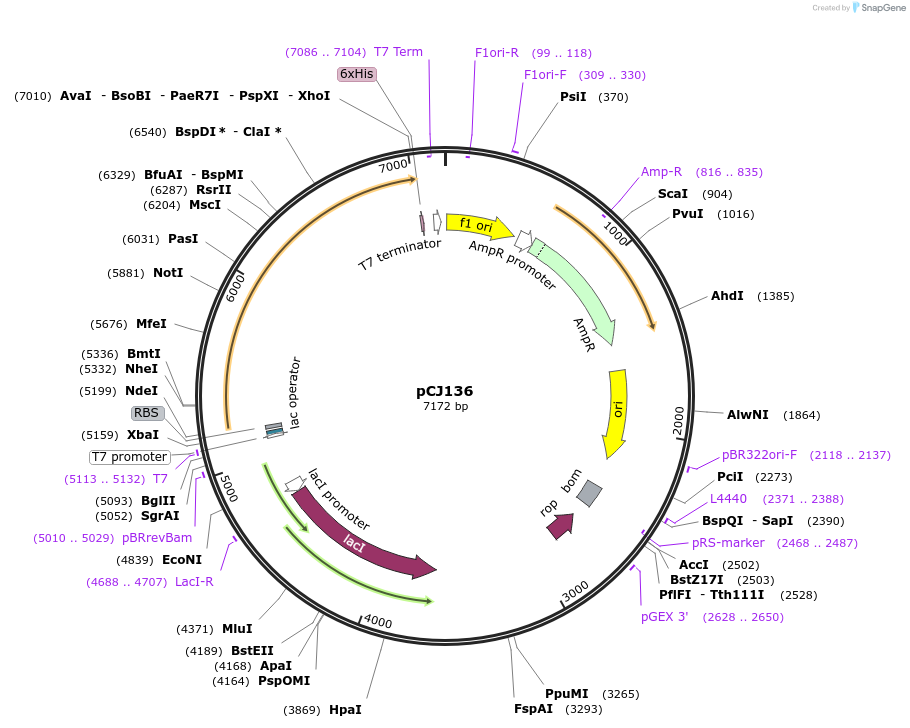
pCJ136
Plasmid#162665PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
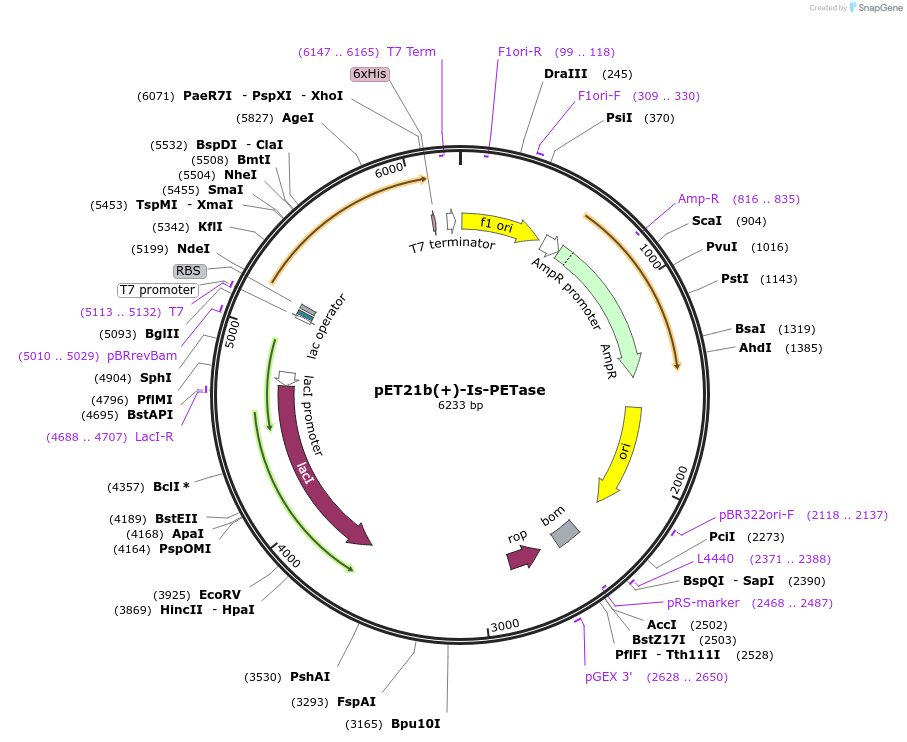
pET21b(+)-Is-PETase
Plasmid#112202PurposepET-21b(+) based plasmid for expression of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1), codon optimized for expression in E. coli K12DepositorInsertPETase gene from Ideonella sakaiensis 201-F6, codon optimized for expression in E. coli K12
Tags6XHISExpressionBacterialPromoterN/AAvailable SinceJuly 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
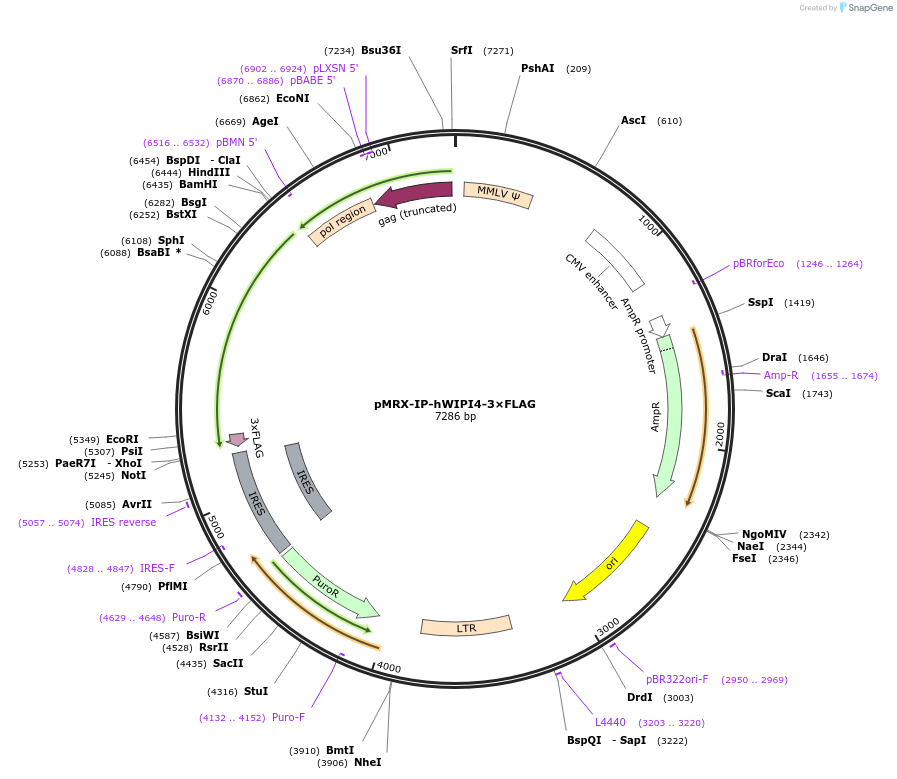
pMRX-IP-hWIPI4-3×FLAG
Plasmid#215512PurposeExpresses FLAG tagged human WIPI4.DepositorInsertWD-repeat protein interacting with phosphoinositides 4 (WDR45 Human)
UseRetroviralTags3×FLAGExpressionMammalianAvailable SinceMay 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
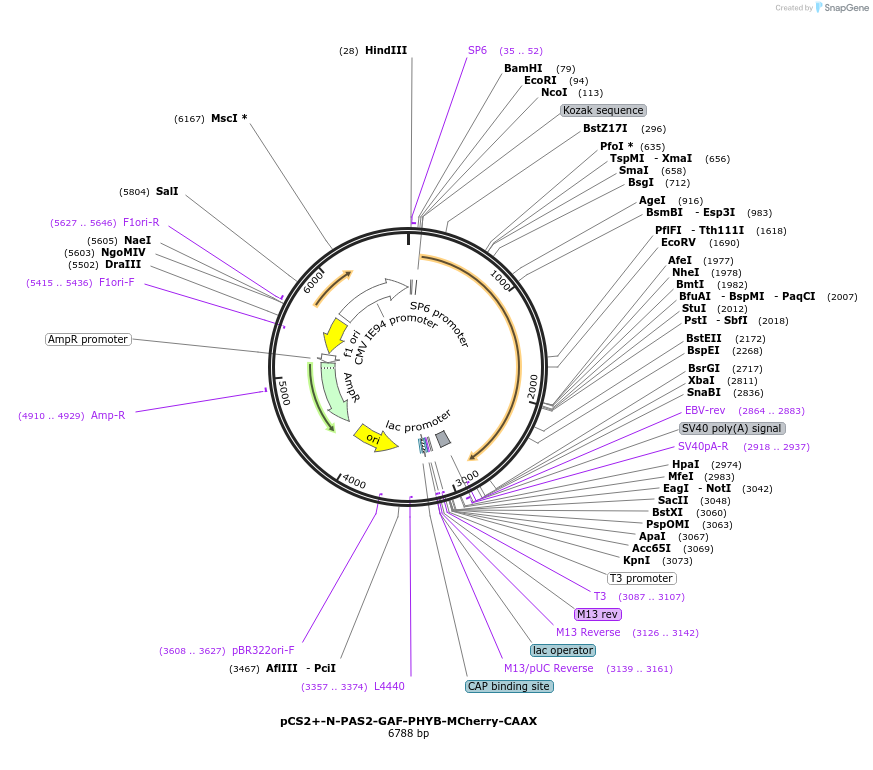
pCS2+-N-PAS2-GAF-PHYB-MCherry-CAAX
Plasmid#154910Purpose1-621 amino acids of Arabidopsis PHYB (Gene ID: 816394), tagged with mCherry fluorescence protein and the CAAX membrane moiety. Mammalian codon optimised.DepositorInsertPhytochrome B (PHYB Mustard Weed, Synthetic)
UseXenopus/avian/zebrafishTagsCAAX membrane moiety and mCherryExpressionMammalianMutation1-621 amino acids of Arabidopsis PHYBPromoterCMVAvailable SinceNov. 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
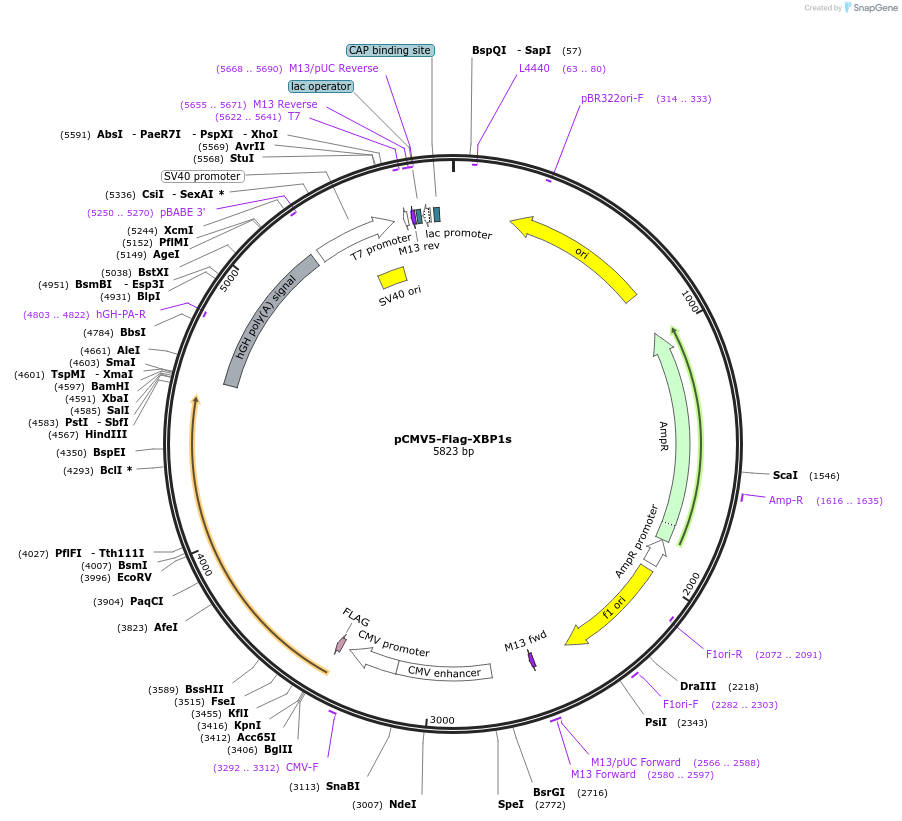
pCMV5-Flag-XBP1s
Plasmid#63680PurposeMammalian expression of Flag tagged XBP1sDepositorAvailable SinceMarch 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
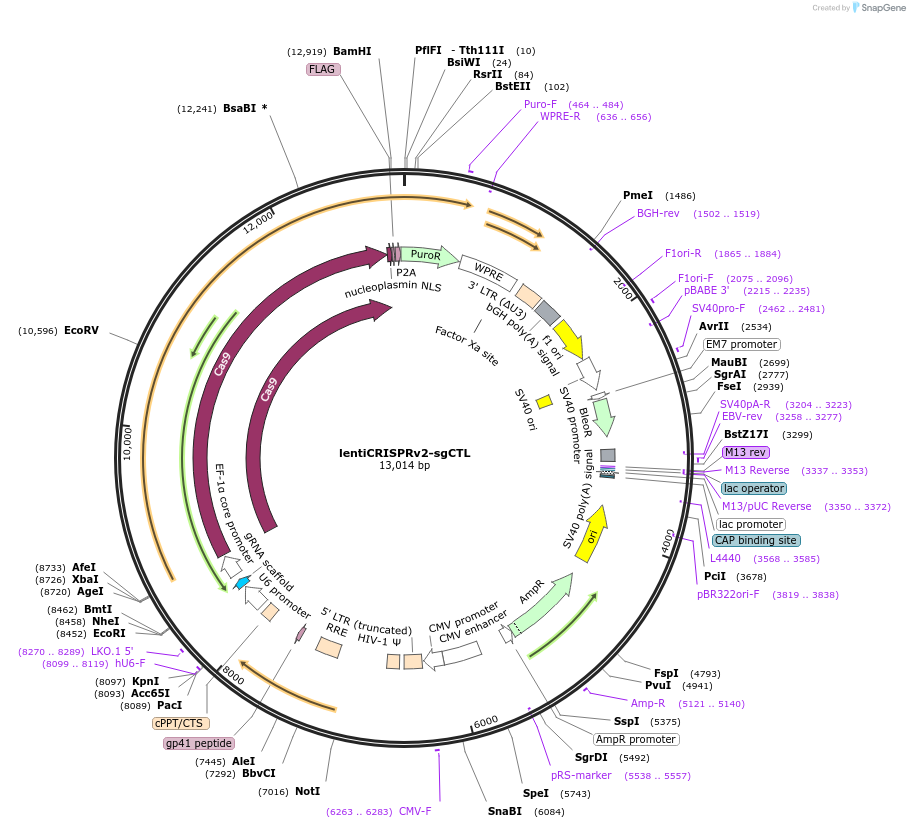
lentiCRISPRv2-sgCTL
Plasmid#234771PurposeNon Targeting ControlDepositorInsertNon Targeting Control sgRNA
UseLentiviralAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only