We narrowed to 11,024 results for: cat.1
-
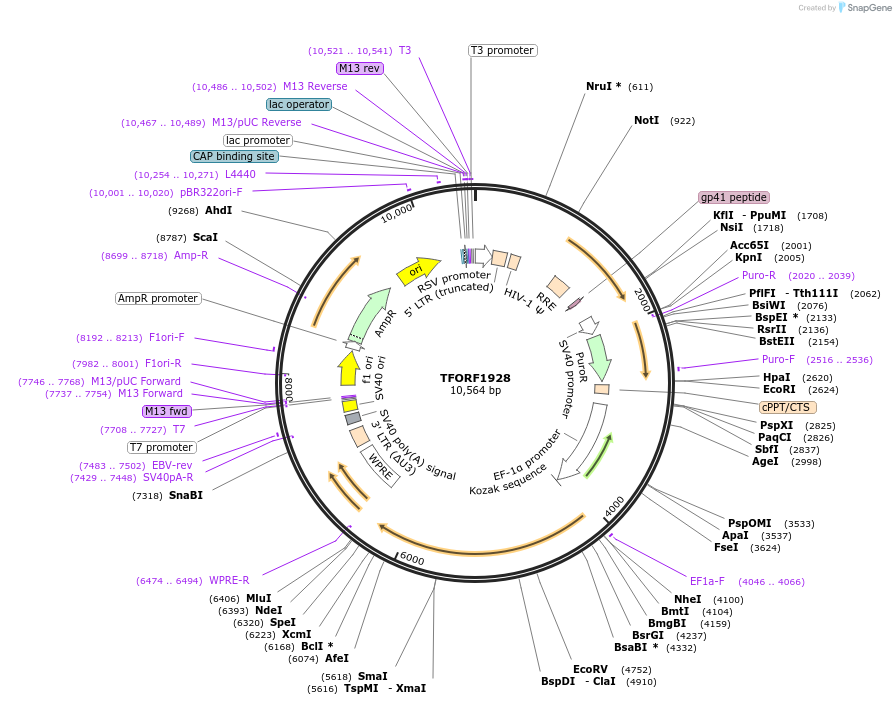
Plasmid#142291PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceMarch 28, 2024AvailabilityAcademic Institutions and Nonprofits only
-
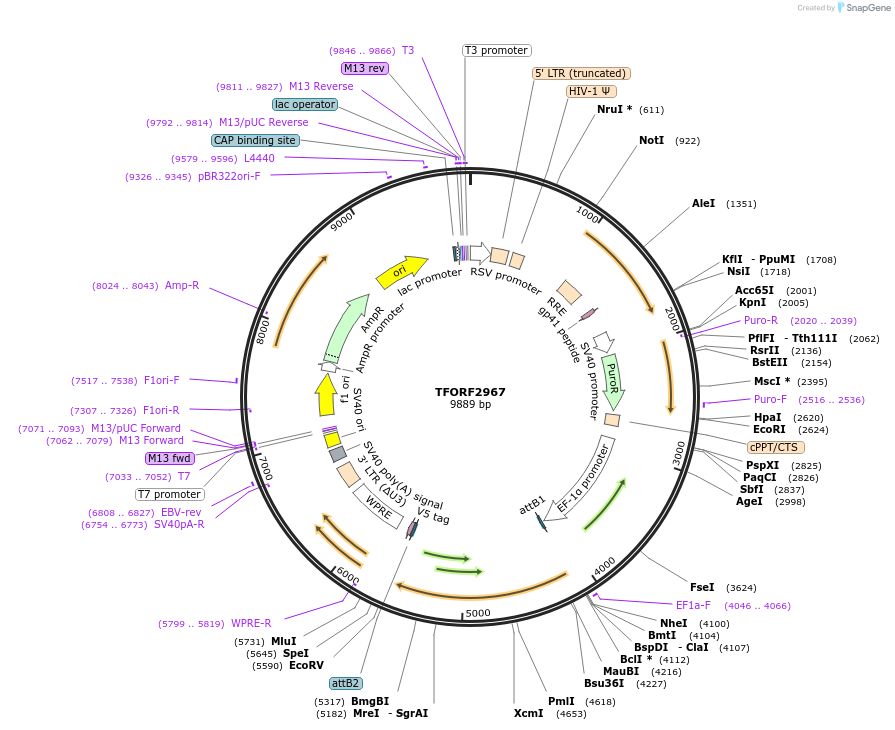
TFORF2967
Plasmid#144443PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceSept. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
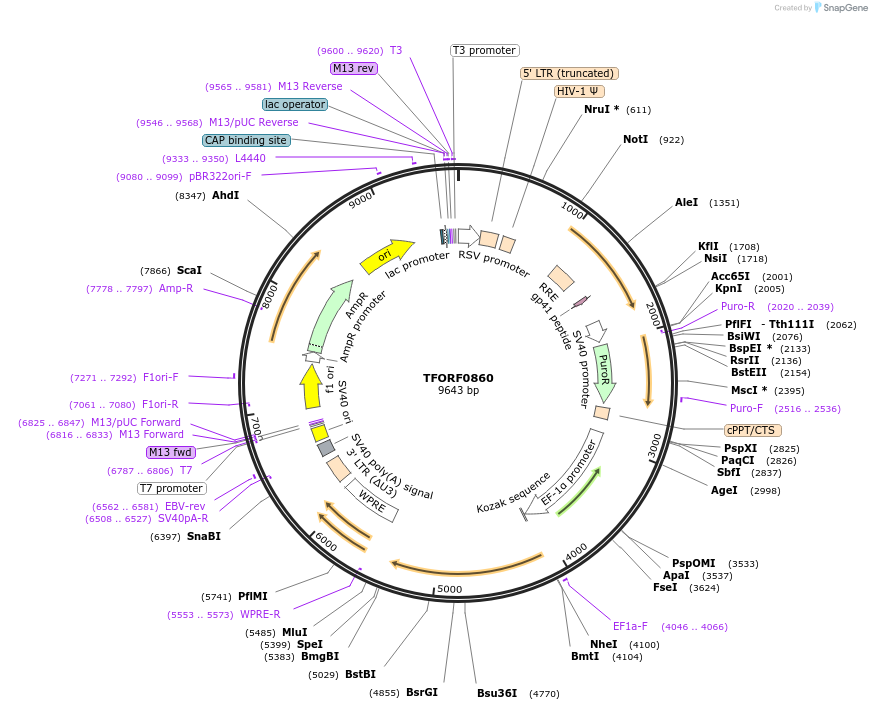
TFORF0860
Plasmid#142699PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
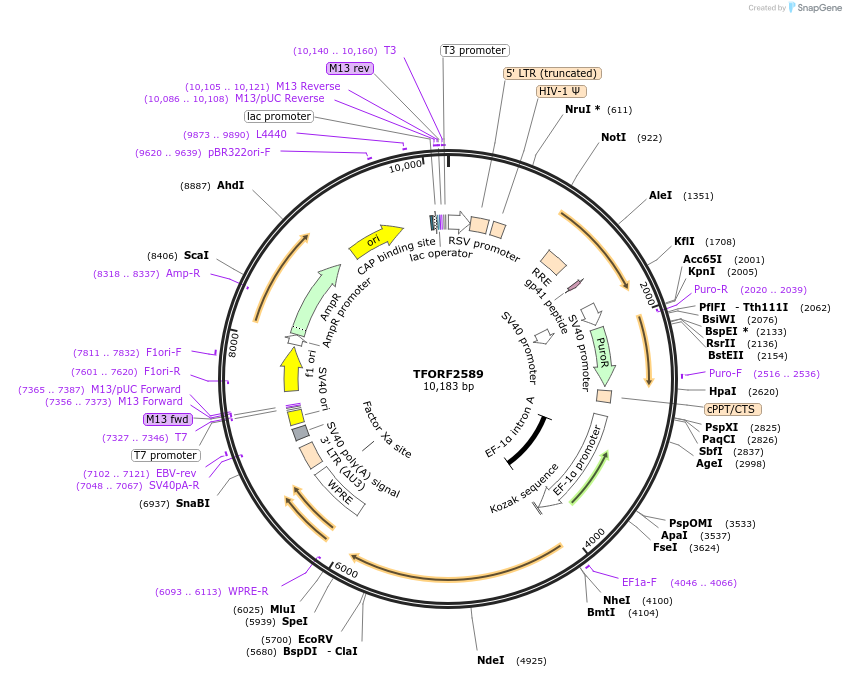
TFORF2589
Plasmid#142333PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF1915
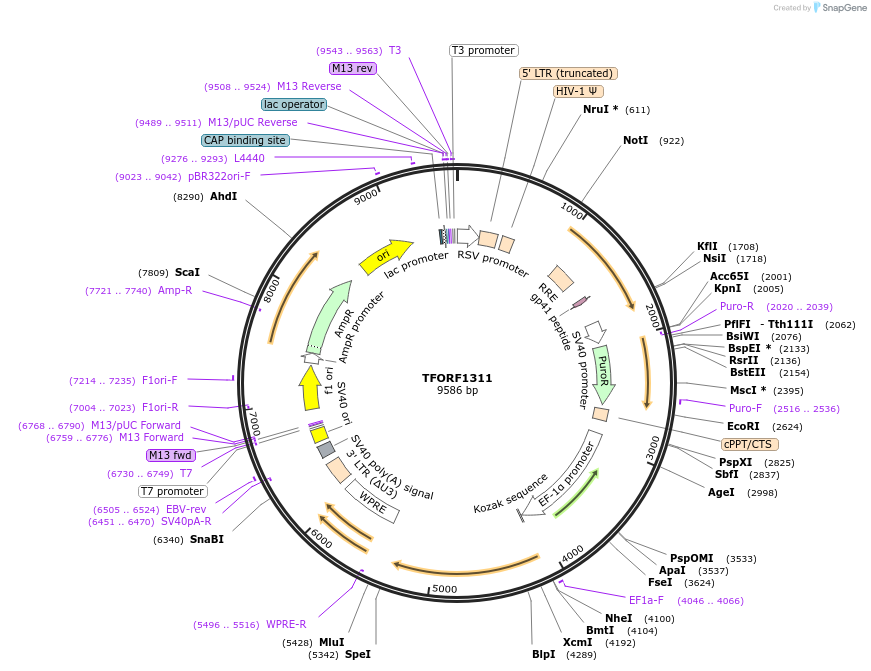
Plasmid#143860PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF1311
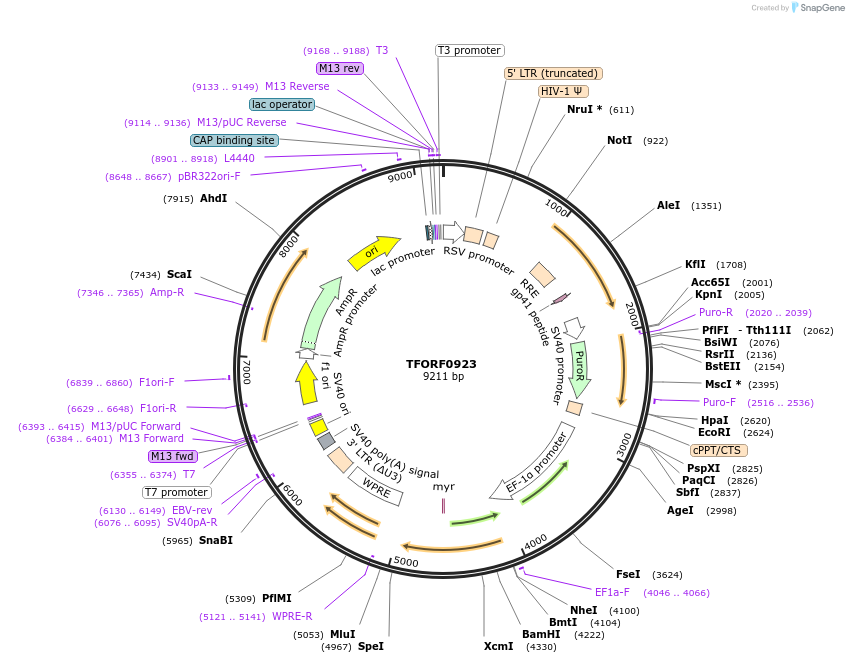
Plasmid#143786PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF0923
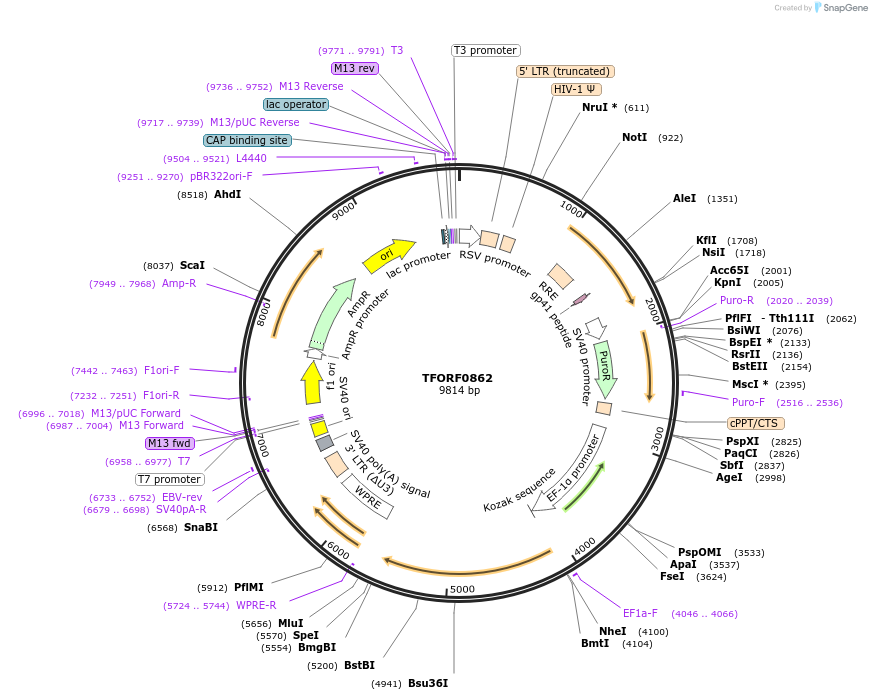
Plasmid#141756PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF0862
Plasmid#141741PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
JA1217
Plasmid#49954PurposeExpresses a TALE-TET1CD (catalytic domain of TET1) fusion protein engineered to bind a site in the human beta-globin (HBB) gene and demethylate CpGs adjacent to the binding siteDepositorAvailable SinceDec. 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
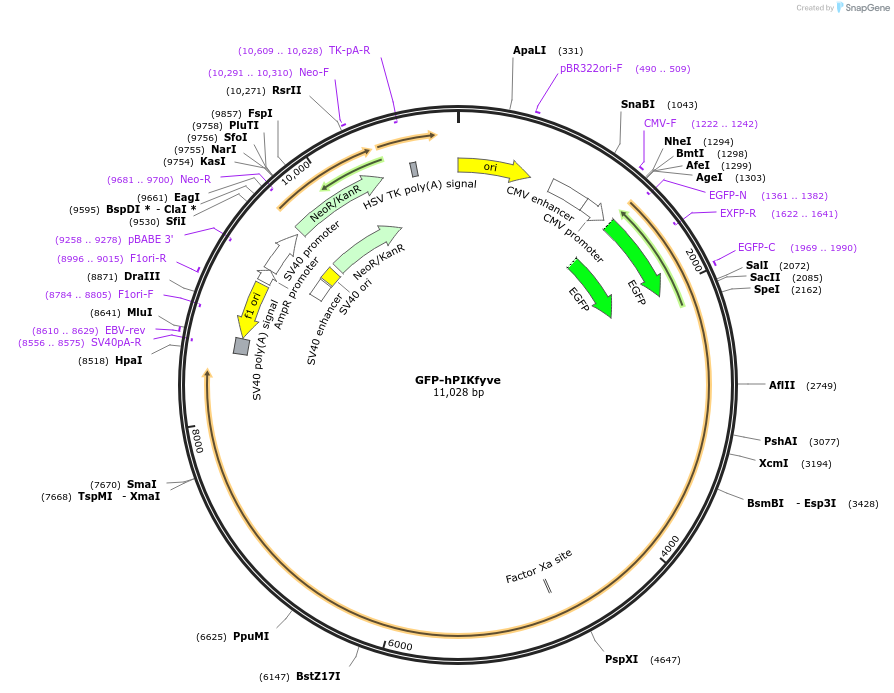
GFP-hPIKfyve
Plasmid#121148PurposeExpresses GFP-tagged human PIKfyve in mammalian cells.DepositorInsert1-phosphatidylinositol 3-phosphate 5-kinase isoform 2 (PIKFYVE Human)
TagsGFPExpressionMammalianPromoterCMVAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
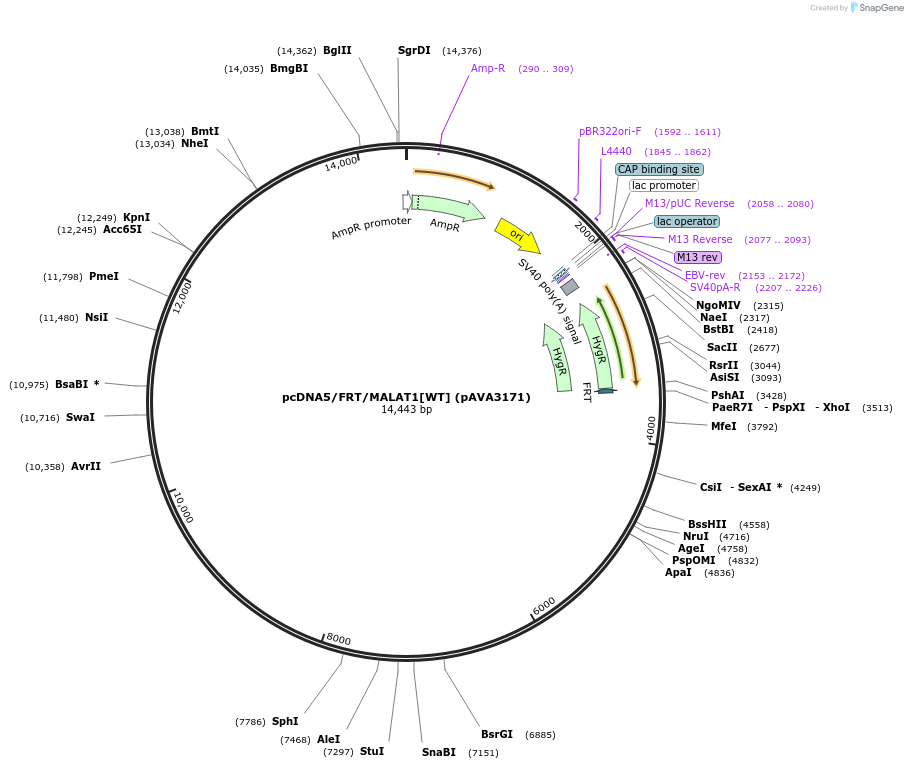
pcDNA5/FRT/MALAT1[WT] (pAVA3171)
Plasmid#239347PurposeExpresses full-length, driven by its own promoter MALAT1 with wild-type ENE and an 11-nt insertion (cgctcgacgta).DepositorInsert10,843 bp of MALAT1 of genomic locus amplified from genomic DNA of Hela cells (MALAT1 Human)
ExpressionMammalianMutationAn 11-nt insertion (cgctcgacgta)Available SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
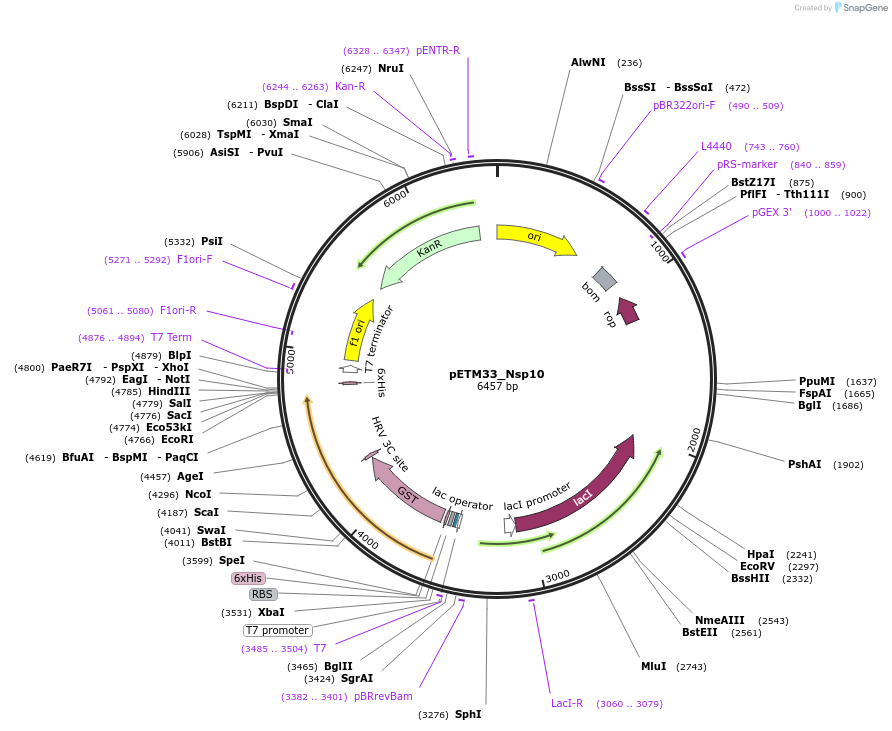
pETM33_Nsp10
Plasmid#156479PurposeBacterial expression of Sars-CoV2 Nsp10 protein with His-tag and GST-tagDepositorInsertNsp10 (ORF1ab Severe acute respiratory syndrome coronavirus 2, Synthetic)
TagsHis-GSTExpressionBacterialMutationGene insert is codon optimized for Ecoli.PromoterT7/LacOAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
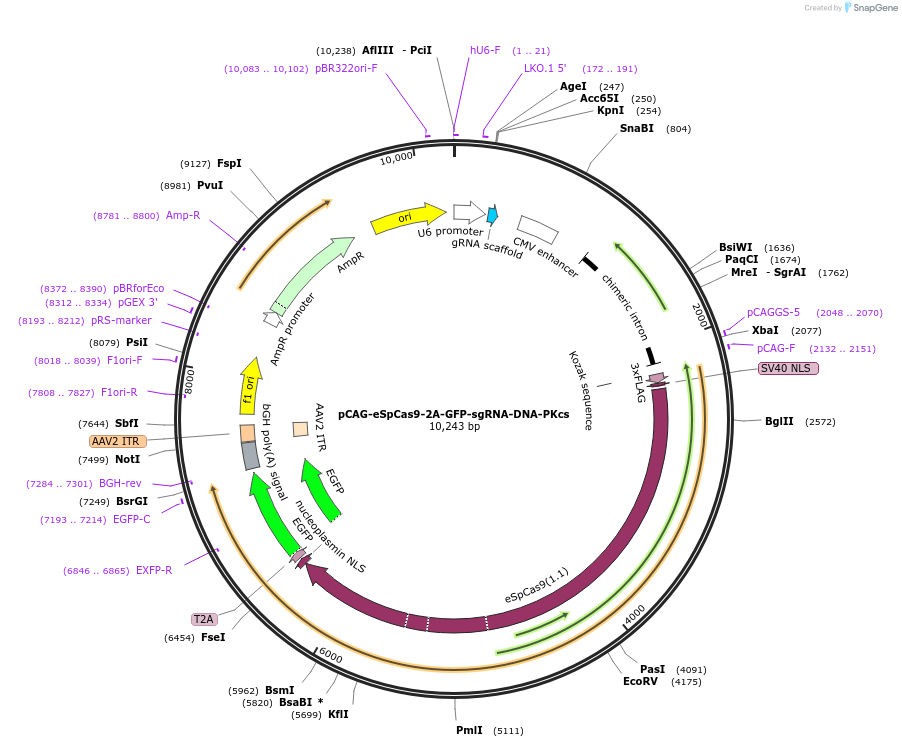
pCAG-eSpCas9-2A-GFP-sgRNA-DNA-PKcs
Plasmid#220493PurposeTo generate DNA-PKcs KO in human cells. Co-expresses eSpCas9(1.1), GFP and a guide RNA against DNA-PKcs exon 1.DepositorAvailable SinceJuly 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
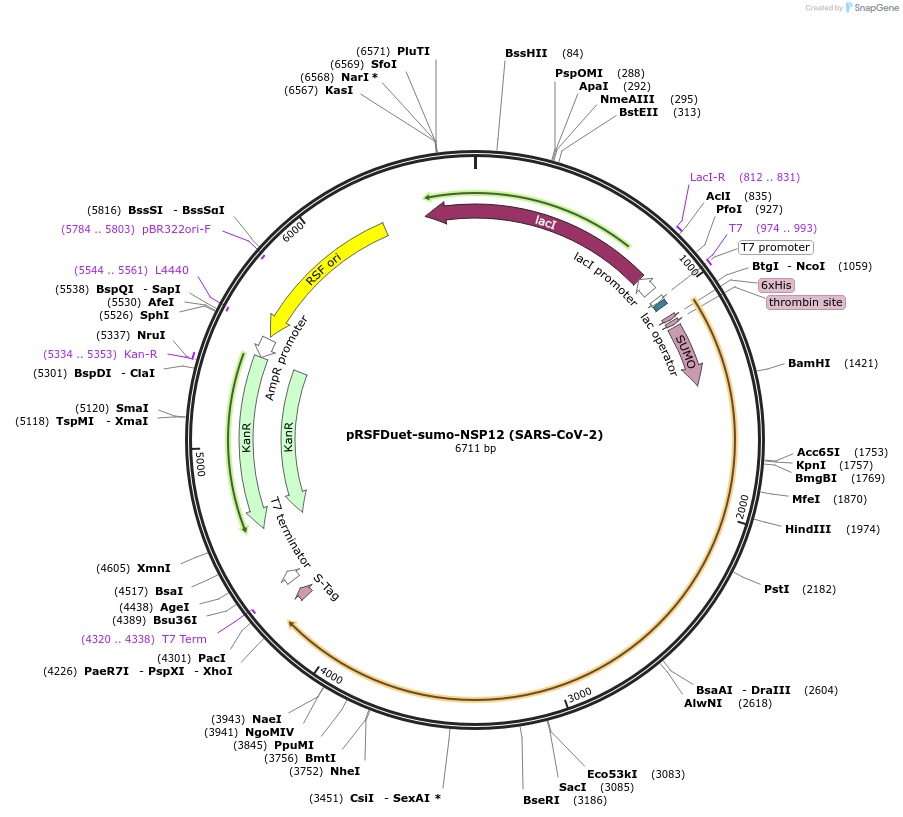
pRSFDuet-sumo-NSP12 (SARS-CoV-2)
Plasmid#159107PurposeThe SARS-CoV-2 NSP12 coding sequence was cloned into a modified pRSFDuet-1 vector (Novagen) bearing an N-terminal His6-SUMO-tag which is cleavable by the ubiquitin-like protease (ULP1).DepositorInsertNSP12
TagsHis-SUMO tagExpressionBacterialMutationNonePromoterT7Available SinceSept. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
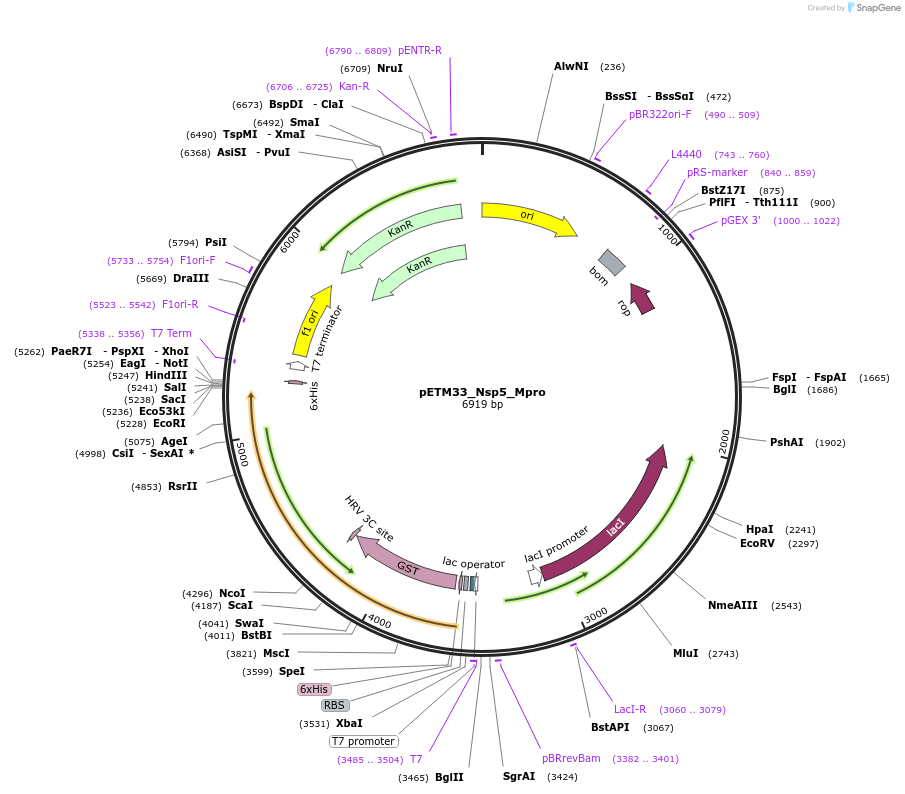
pETM33_Nsp5_Mpro
Plasmid#156475PurposeBacterial expression of Sars-CoV2 Nsp5_Mpro protein with His-tag and GST-tagDepositorInsertNsp5_Mpro (ORF1ab Severe acute respiratory syndrome coronavirus 2, Synthetic)
TagsHis-GSTExpressionBacterialMutationGene insert is codon optimized for Ecoli.PromoterT7/LacOAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
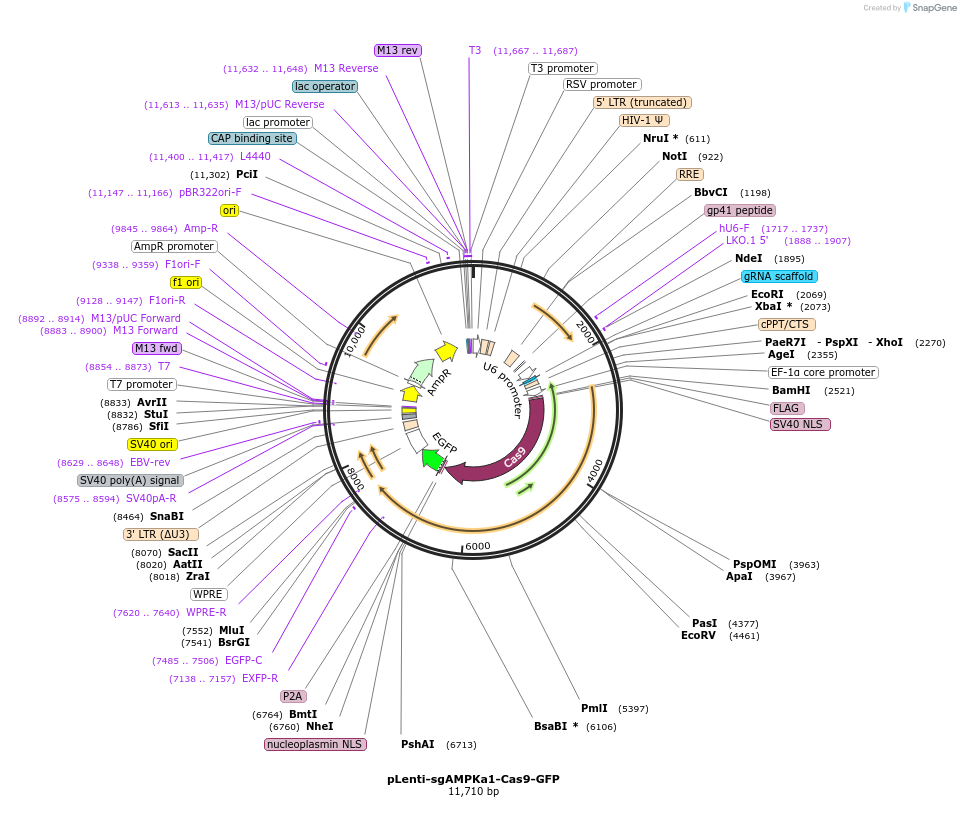
pLenti-sgAMPKa1-Cas9-GFP
Plasmid#208049PurposeLentiviral vector expressing Cas9 and an sgRNA targeting AMPKa1DepositorAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
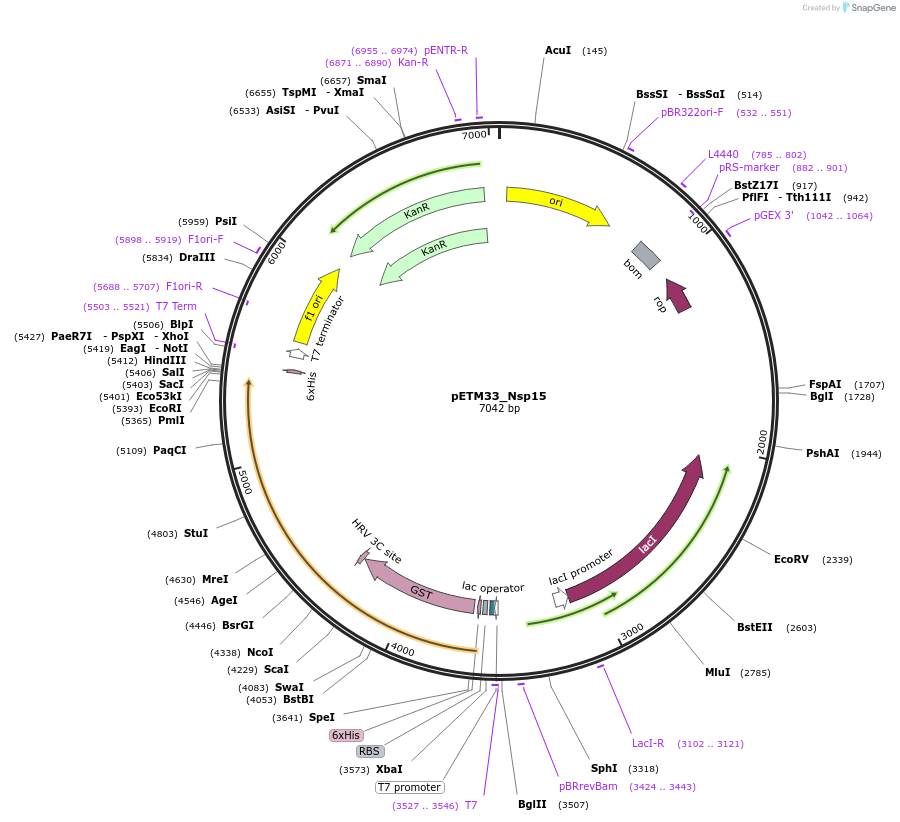
pETM33_Nsp15
Plasmid#156481PurposeBacterial expression of Sars-CoV2 Nsp15 protein with His-tag and GST-tagDepositorInsertNsp15 (ORF1ab Severe acute respiratory syndrome coronavirus 2, Synthetic)
TagsHis-GSTExpressionBacterialMutationGene insert is codon optimized for Ecoli.PromoterT7/LacOAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
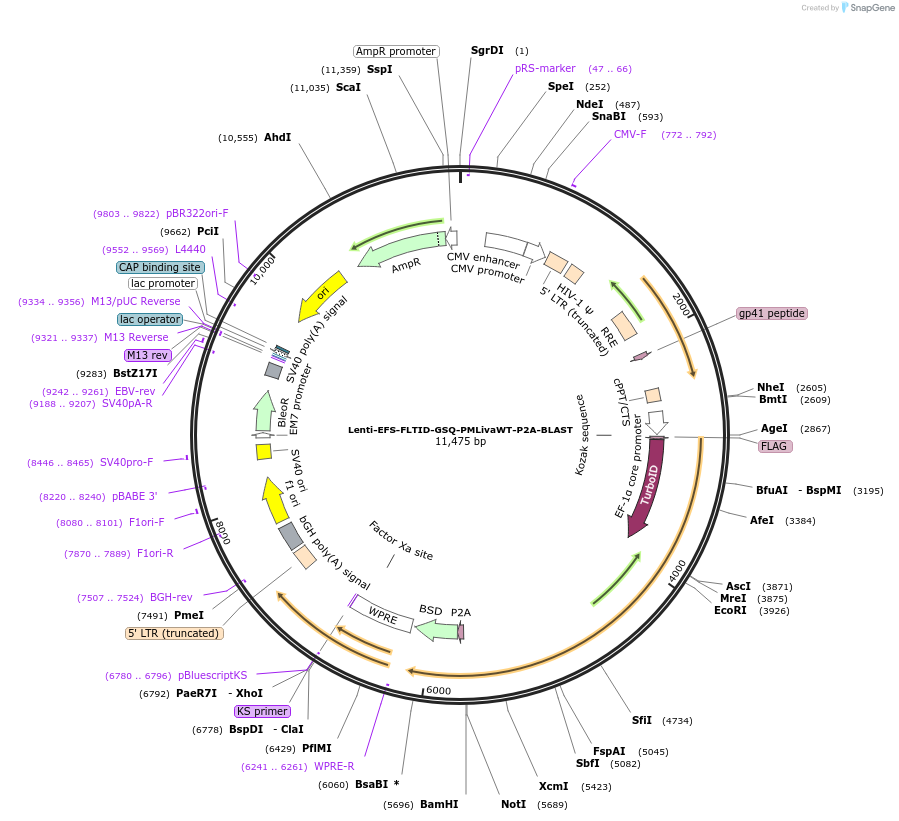
Lenti-EFS-FLTID-GSQ-PMLivaWT-P2A-BLAST
Plasmid#208040PurposeEnables constitutive expression of N-terminal TurboID-fused PML (isoform IVa; WT); selection with blasticidinDepositorAvailable SinceOct. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
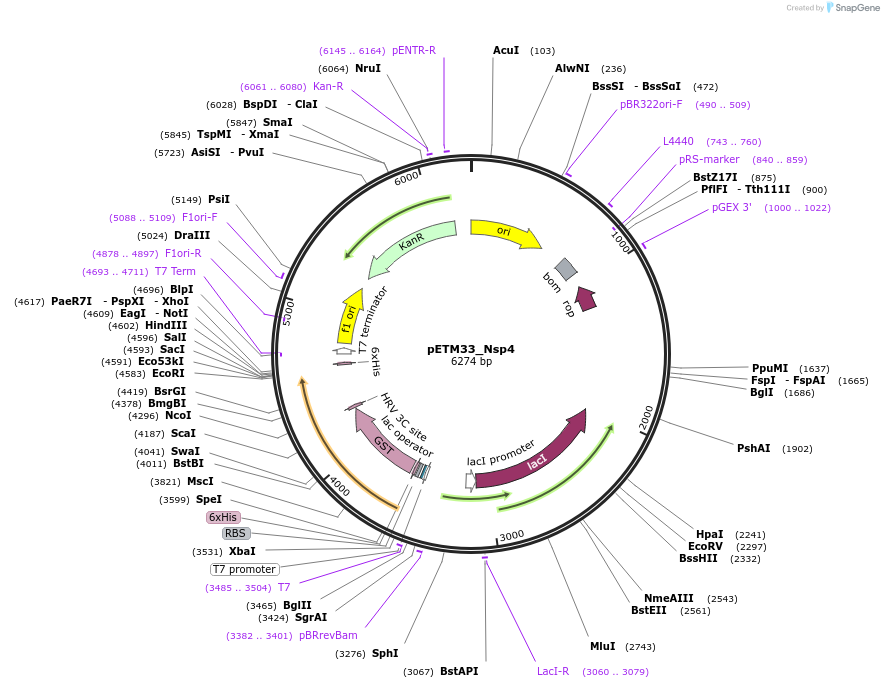
pETM33_Nsp4
Plasmid#156474PurposeBacterial expression of Sars-CoV2 Nsp4 protein with His-tag and GST-tagDepositorInsertNsp4 (ORF1ab Severe acute respiratory syndrome coronavirus 2, Synthetic)
TagsHis-GSTExpressionBacterialMutationGene insert is codon optimized for Ecoli.PromoterT7/LacOAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
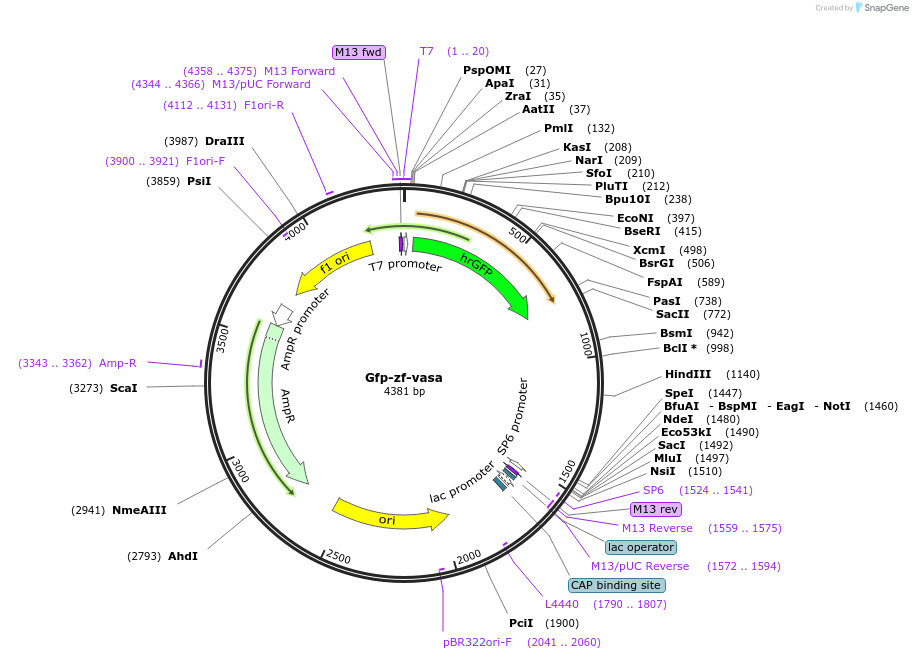
Gfp-zf-vasa
Plasmid#167323PurposePlasmid for synthesis of GFP chimeric RNA that contained vasa-3'UTRs of zebrafish by in vitro transcription.DepositorInsertzf vasa 3'UTRs (ddx4 Zebrafish)
ExpressionBacterialAvailable SinceJune 28, 2021AvailabilityAcademic Institutions and Nonprofits only