We narrowed to 14,019 results for: OVA
-
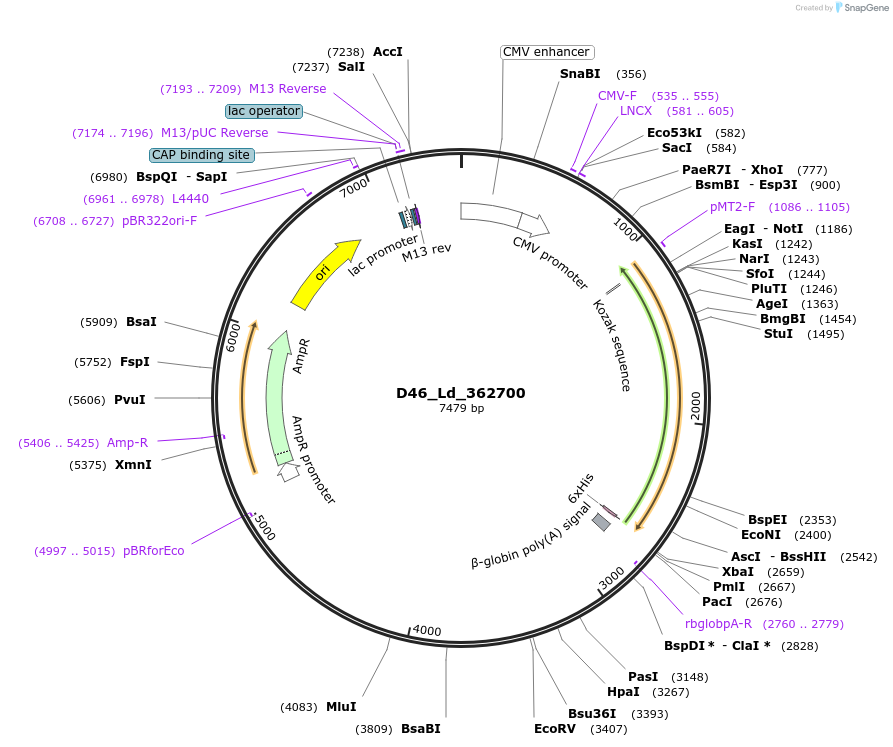
Plasmid#135860PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertmembrane-bound acid phosphatase precursor
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
D64_Ld_010010
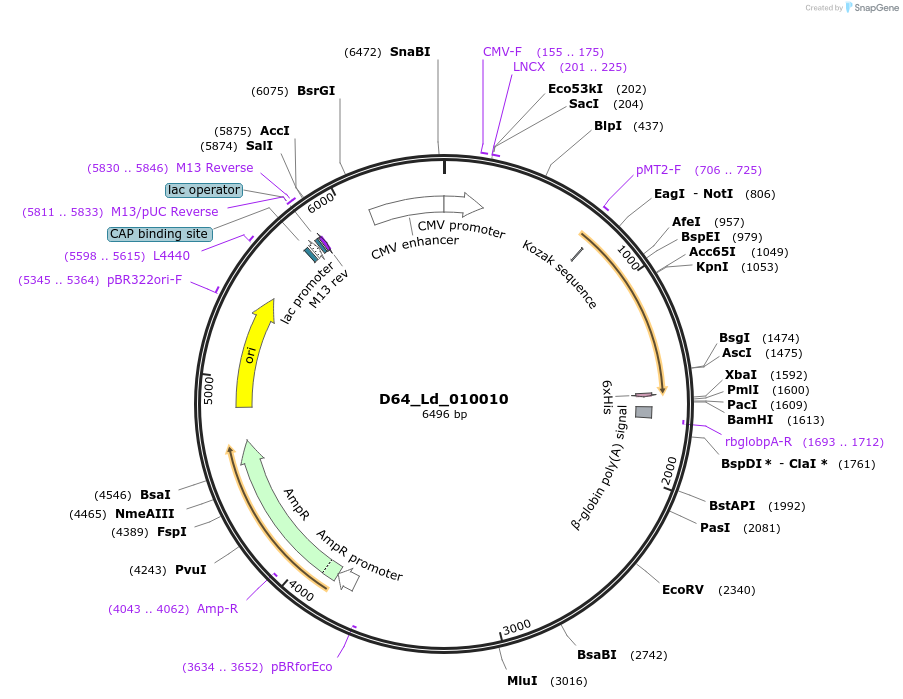
Plasmid#135877PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertProtein of unknown function (DUF2946), putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
D74_Ld_291730
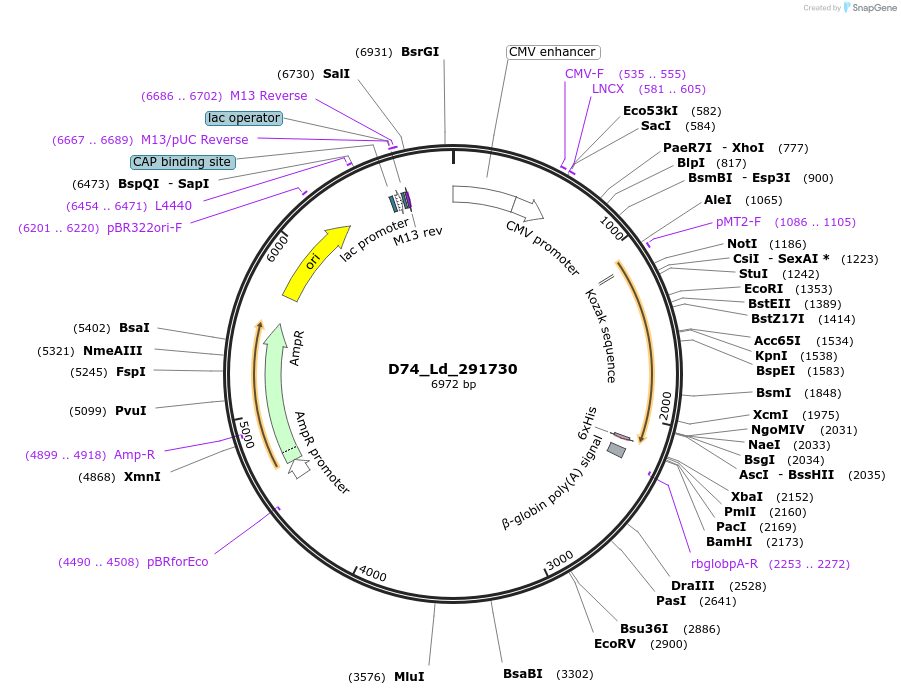
Plasmid#135887PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertendosomal integral membrane protein, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
D79_Ld_366740
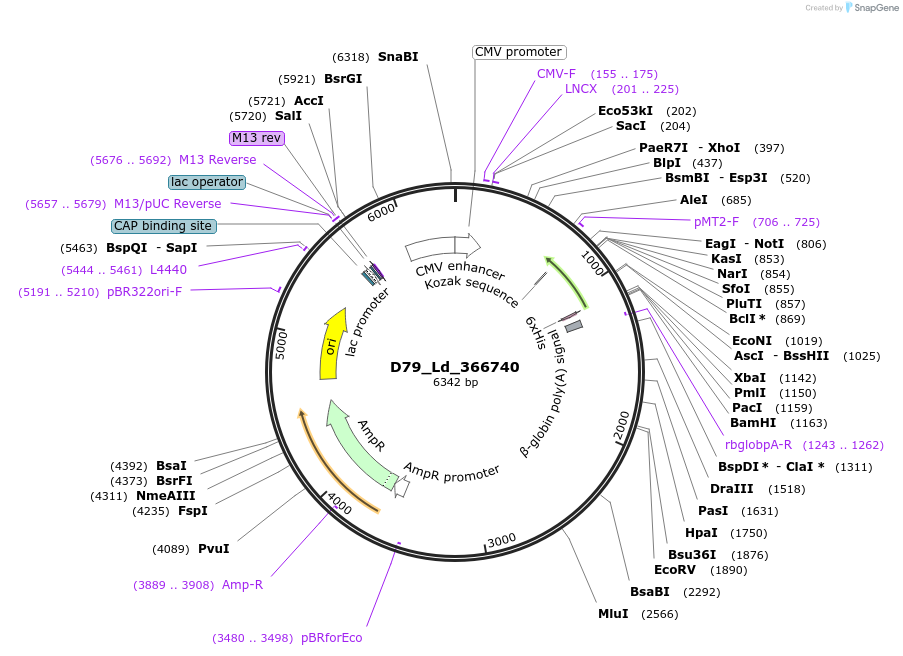
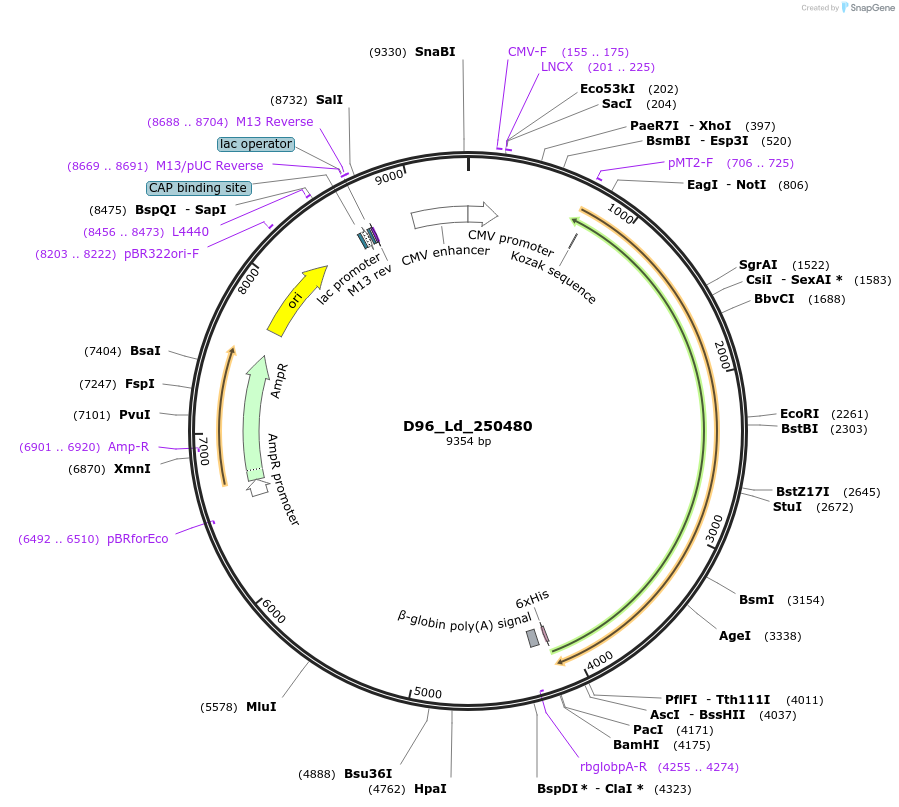
Plasmid#135892PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInserthistidine secretory acid phosphatase, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
D96_Ld_250480
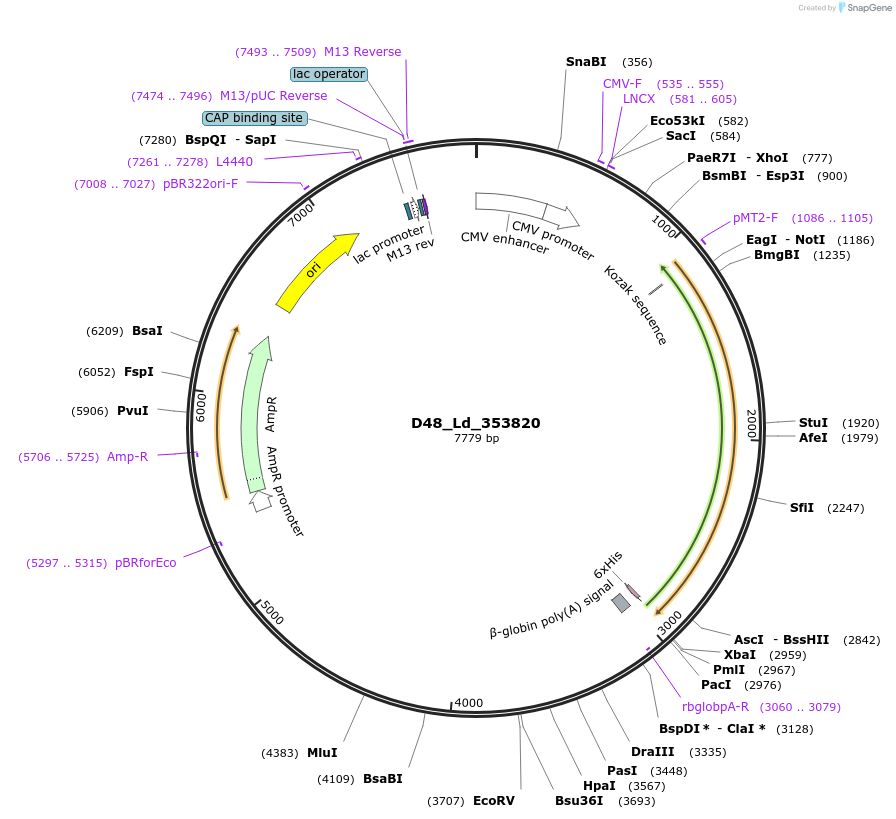
Plasmid#135909PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertKringle domain containing protein, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
D48_Ld_353820
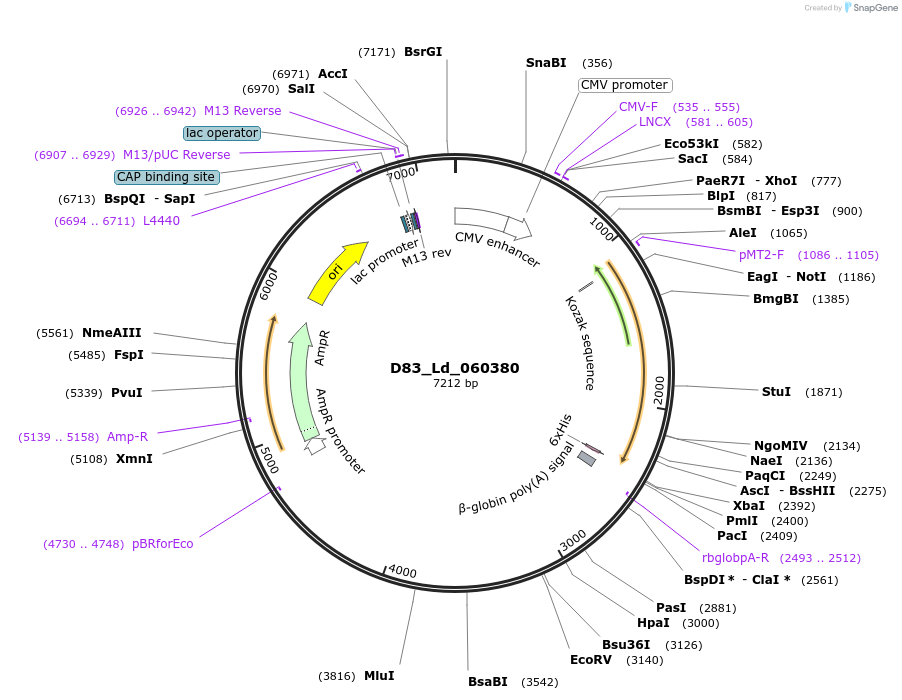
Plasmid#135861PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertEnriched in surface-labeled proteome protein 9, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
D83_Ld_060380
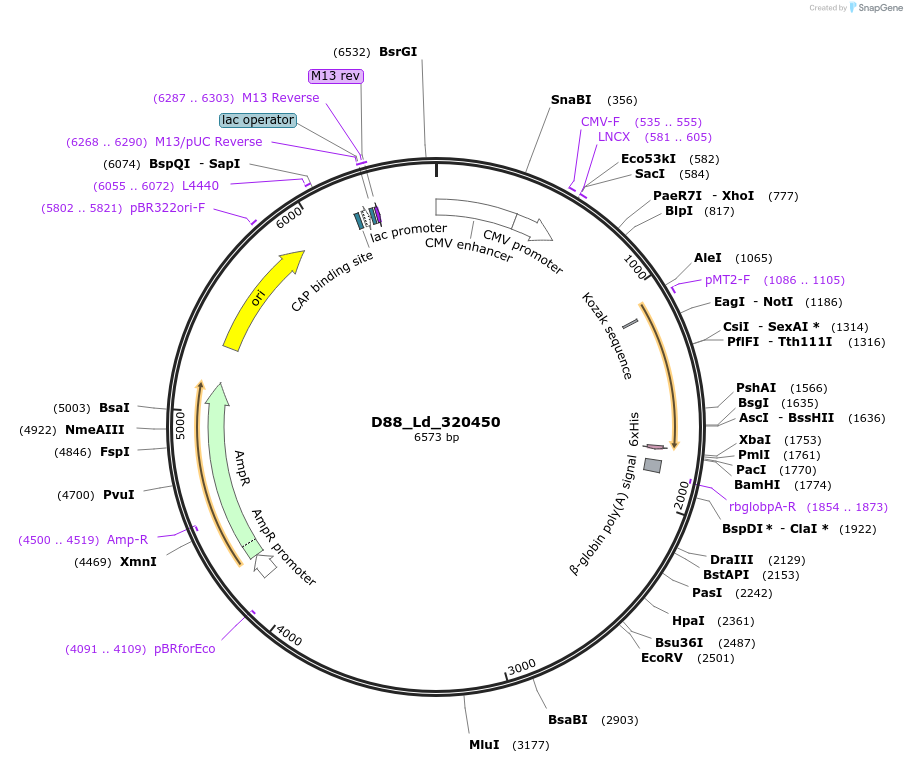
Plasmid#135896PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertPresent in the outer mitochondrial membrane proteome 35
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
D88_Ld_320450
Plasmid#135901PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertProtein of unknown function (DUF1279), putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
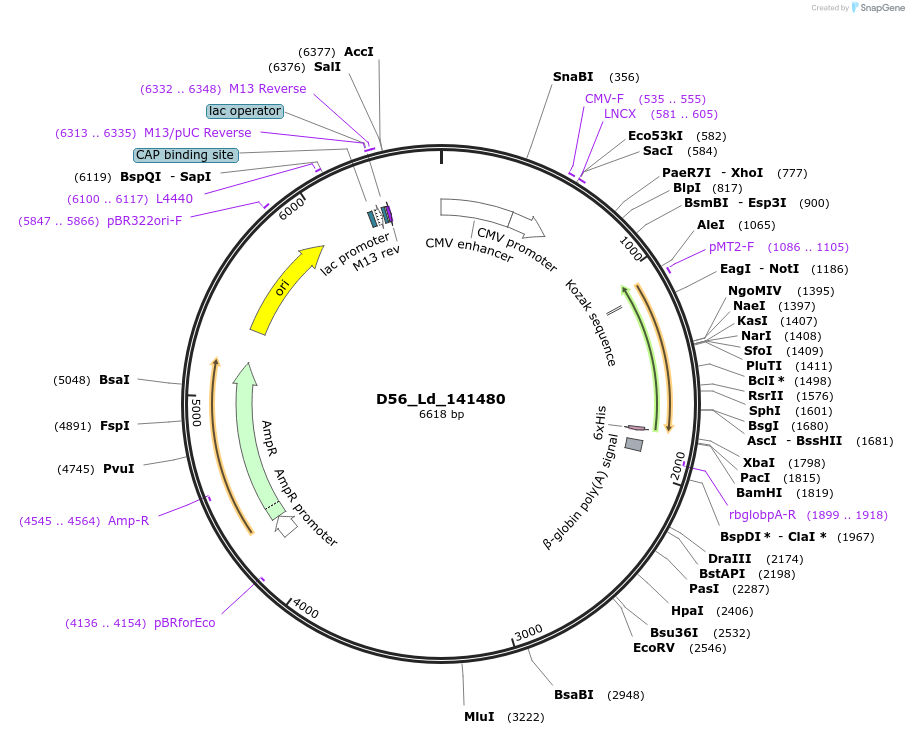
D56_Ld_141480
Plasmid#135869PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertemp24/gp25L/p24 family/GOLD, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
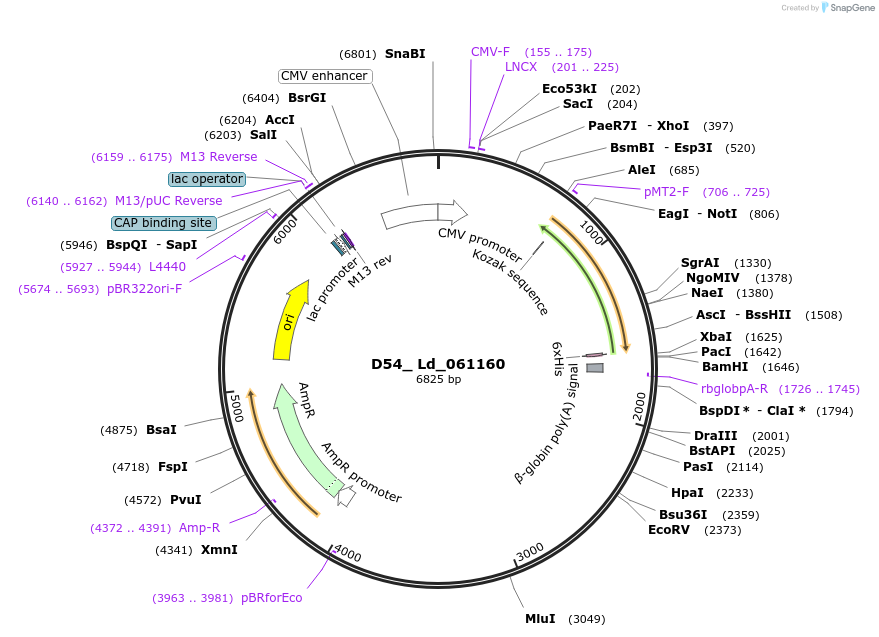
D54_ Ld_061160
Plasmid#135867PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInserthypothetical protein, conserved
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
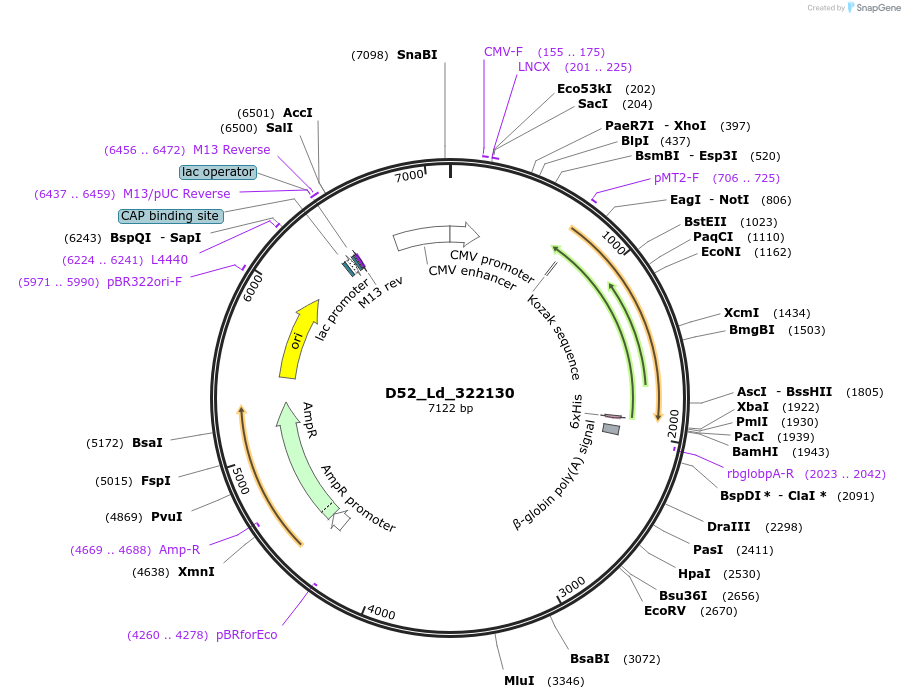
D52_Ld_322130
Plasmid#135865PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertTLR4 regulator and MIR-interacting MSAP, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
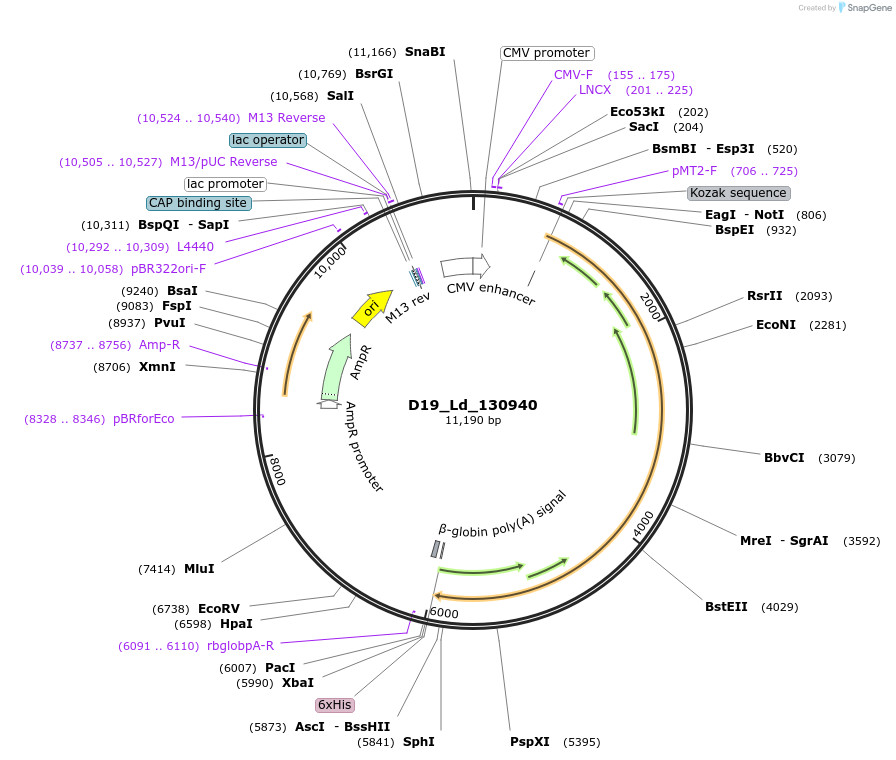
D19_Ld_130940
Plasmid#135835PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertsubtilisin-like serine peptidase,serine peptidase, clan SB, family S8-like protein
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
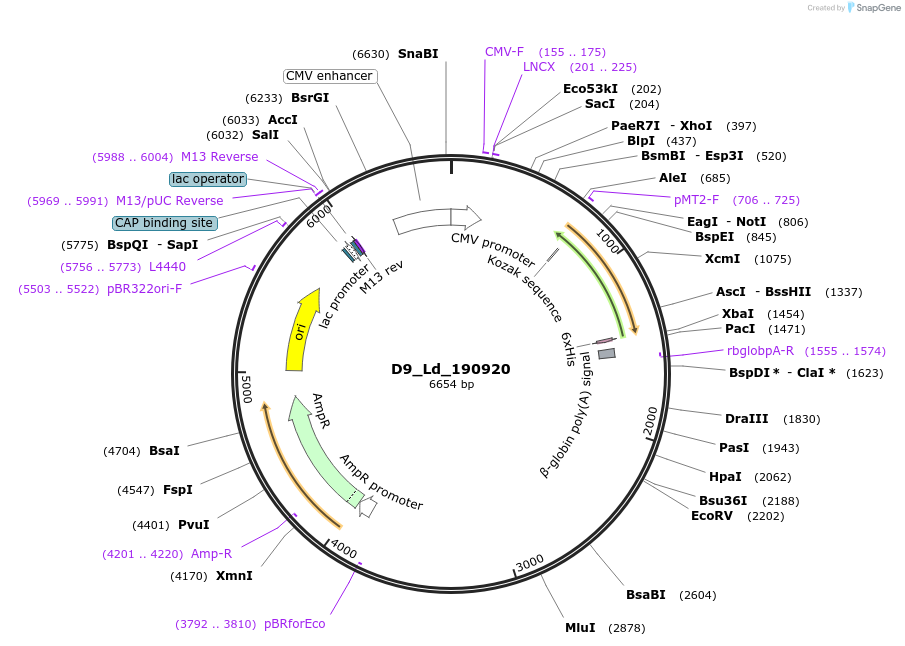
D9_Ld_190920
Plasmid#135825PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertpeptidyl-prolyl cis-trans isomerase, macrophage infectivity potentiator precursor, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
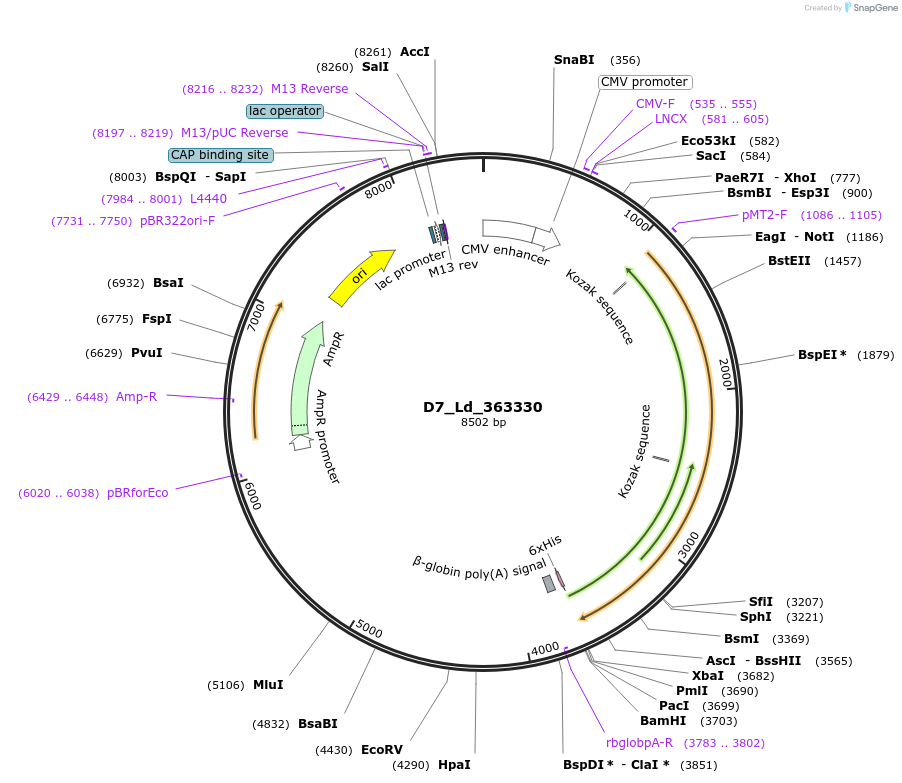
D7_Ld_363330
Plasmid#135823PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertreceptor-type adenylate cyclase a-like protein
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
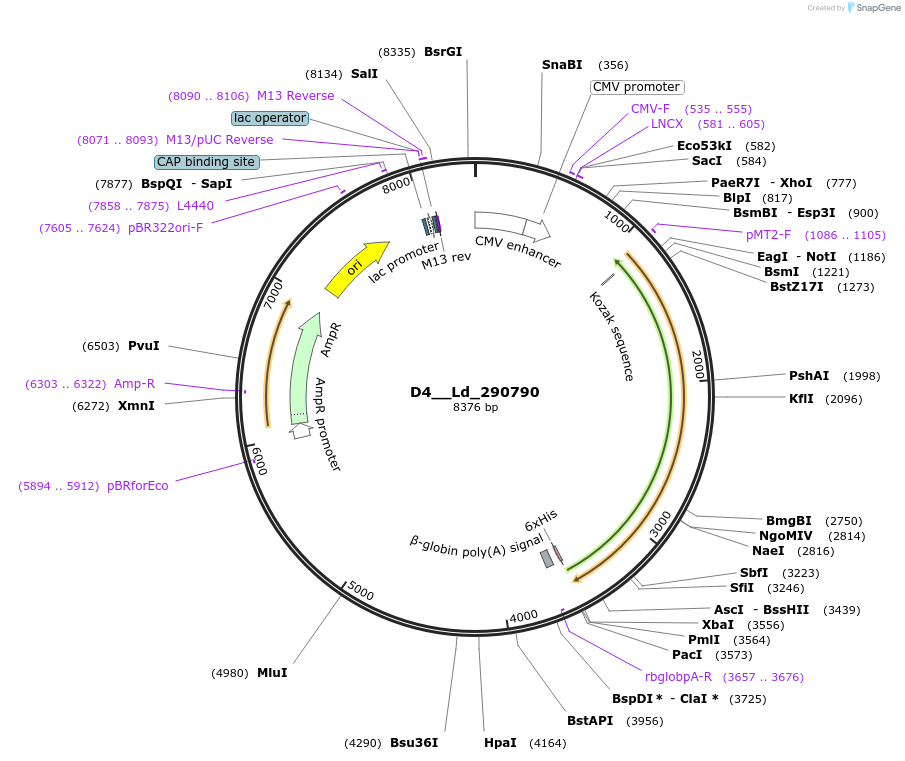
D4__Ld_290790
Plasmid#135820PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertheat shock protein 90, putative,lipophosphoglycan biosynthetic protein, putative,glucose regulated p...
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
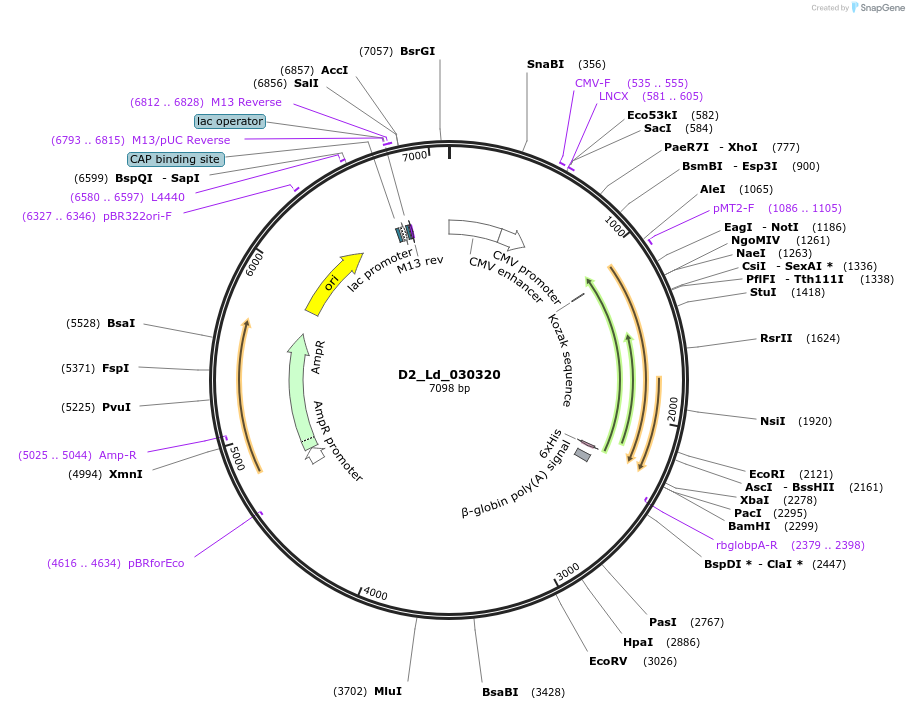
D2_Ld_030320
Plasmid#135818PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertDDRGK domain containing protein, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
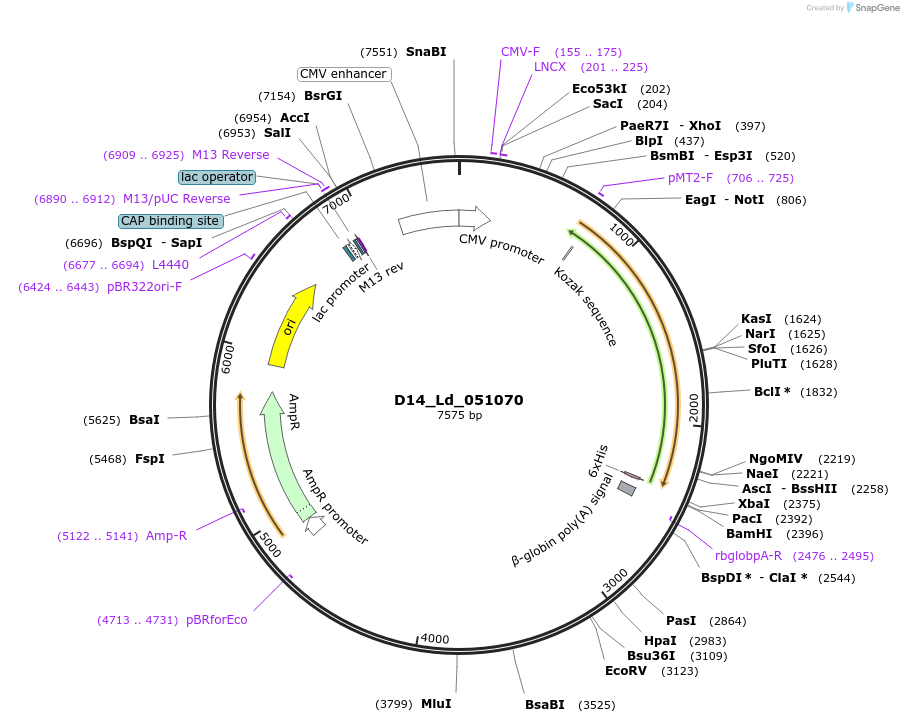
D14_Ld_051070
Plasmid#135830PurposeExpression of Leishmania donovani ectodomain in mammalian cellsDepositorInsertLegume-like lectin family, putative
Tagsbiotinylation sequence and His tagExpressionMammalianAvailable SinceMarch 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
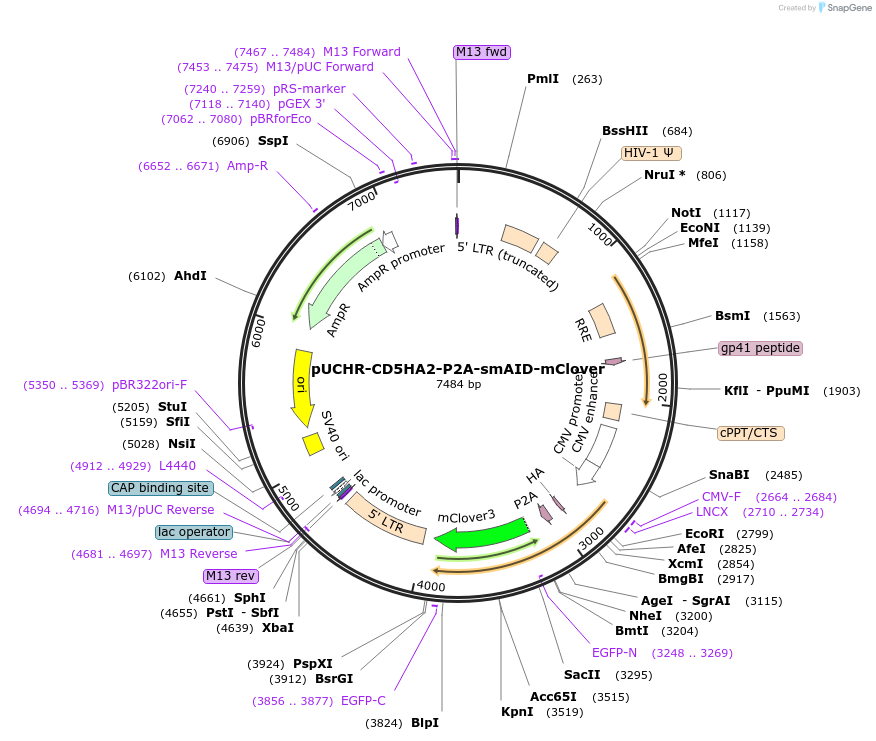
pUCHR-CD5HA2-P2A-smAID-mClover
Plasmid#110658PurposeLentiviral vector expressing cell surface HA-tag and monomeric GFP N-terminaly fused with auxin inducible degronDepositorInsertCD5HA2-P2A-AID-mClover
UseLentiviralPromoterCMVAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
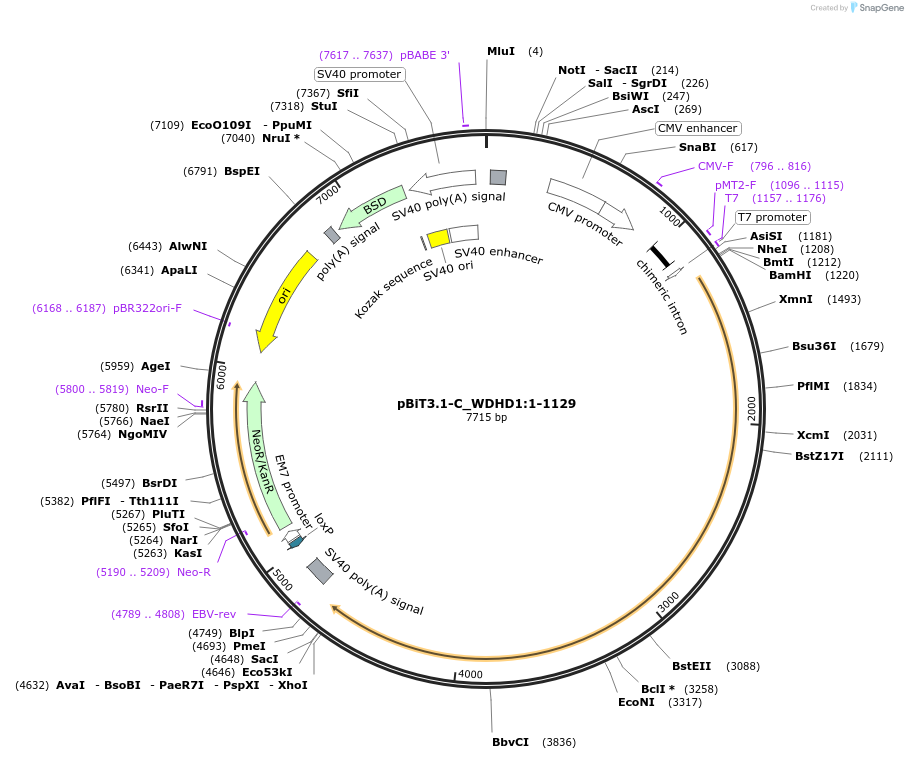
pBiT3.1-C_WDHD1:1-1129
Plasmid#211175PurposeMammalian expression of full-length human WDHD1. C-terminal HiBiT tag.DepositorInsertWDHD1:M1-E1129 (WDHD1 Human)
TagsGSSGGSSGVSGWRLFKKISExpressionMammalianMutationwild typePromoterCMVAvailable SinceMarch 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pBiT3.1-C_WDR77:1-342
Plasmid#211169PurposeMammalian expression of full-length human WDR77. C-terminal HiBiT tag.DepositorInsertWDR77:M1-E342 (WDR77 Human)
TagsGSSGGSSGVSGWRLFKKISExpressionMammalianMutationwild typePromoterCMVAvailable SinceMarch 19, 2024AvailabilityAcademic Institutions and Nonprofits only