We narrowed to 169,429 results for: HTT
-
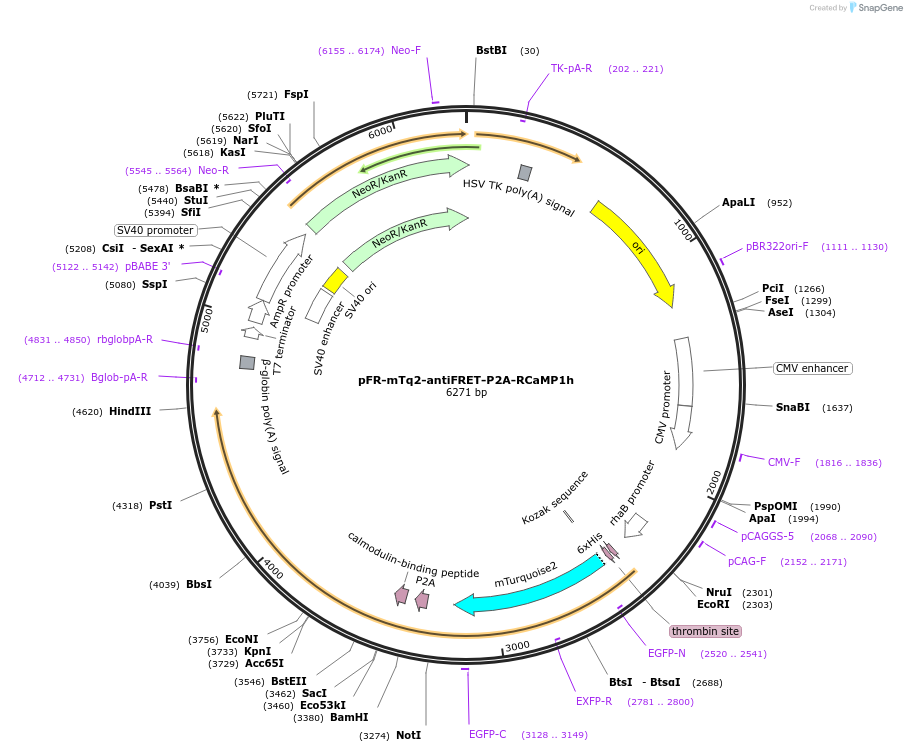
Plasmid#191467PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) RCaMP1h to mTurquoise2 fluorescent proteinDepositorInsertRCaMP1h
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
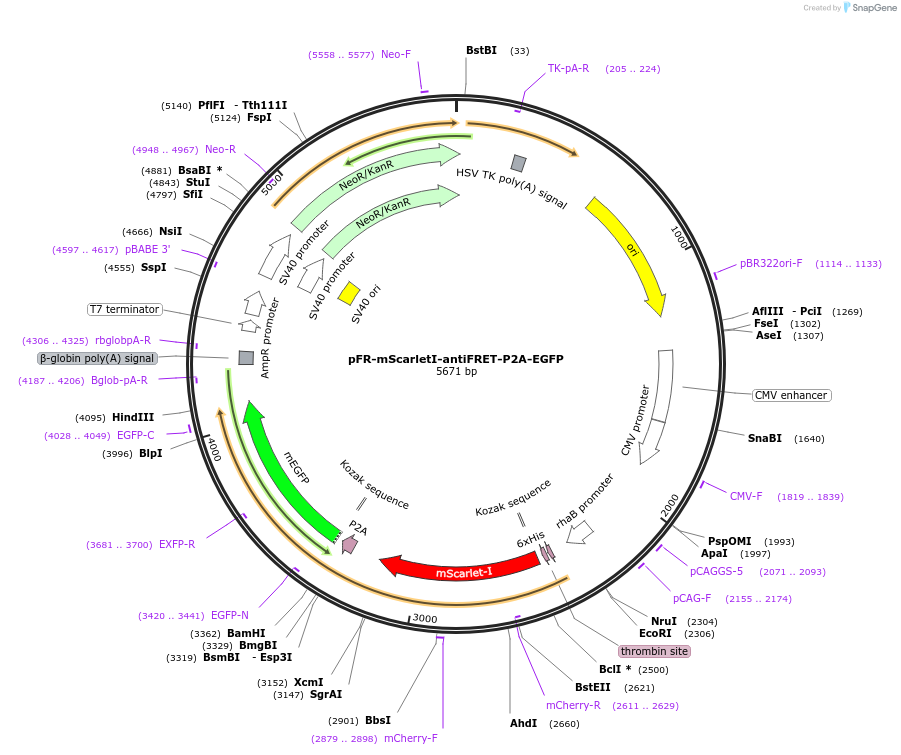
pFR-mScarletI-antiFRET-P2A-EGFP
Plasmid#191458PurposeCompare the intensity of EGFP to mScarlet-I fluorescent proteinDepositorInsertEGFP
TagsHis-mScarletI-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
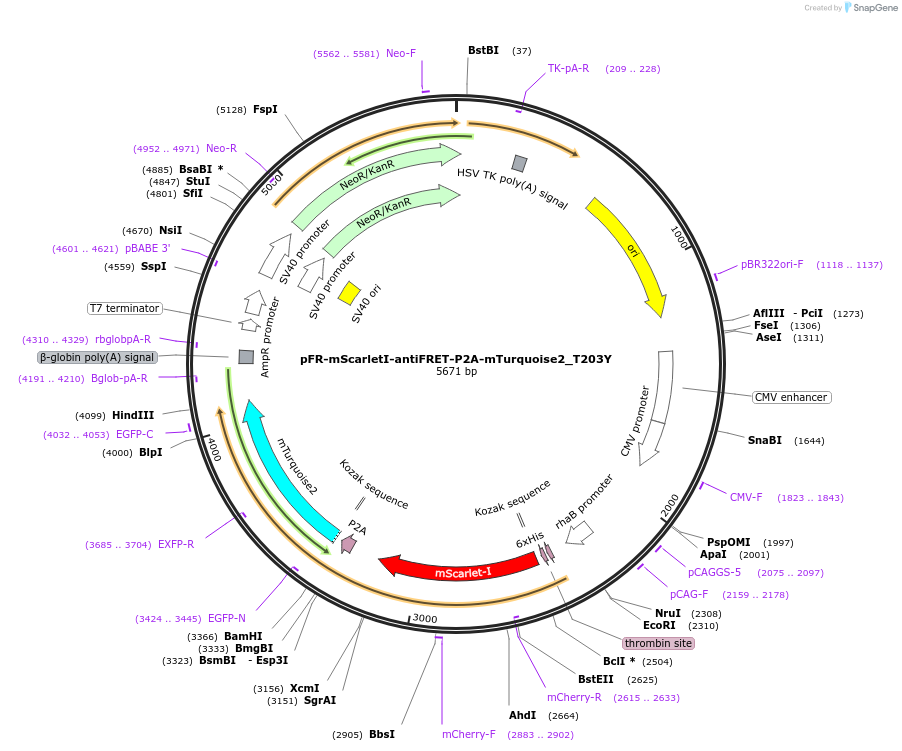
pFR-mScarletI-antiFRET-P2A-mTurquoise2_T203Y
Plasmid#191459PurposeCompare the intensity of mTurquoise2_T203Y to mScarlet-I fluorescent proteinDepositorInsertmTurquoise2_T203Y
TagsHis-mScarletI-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
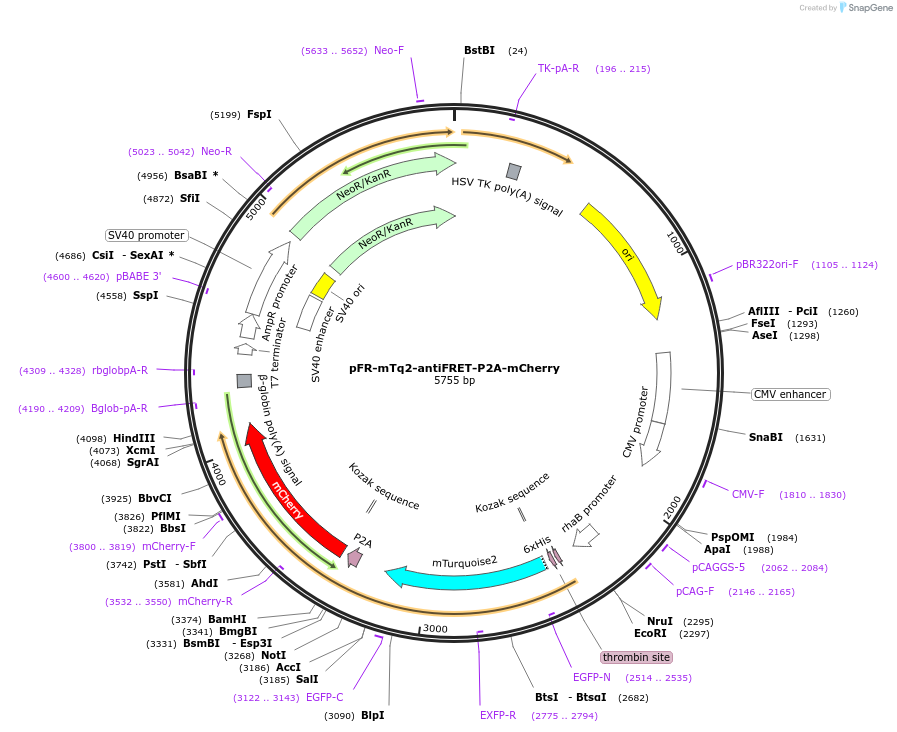
pFR-mTq2-antiFRET-P2A-mCherry
Plasmid#191466PurposeCompare the intensity of mCherry to mTurquoise2 fluorescent proteinDepositorInsertmCherry
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
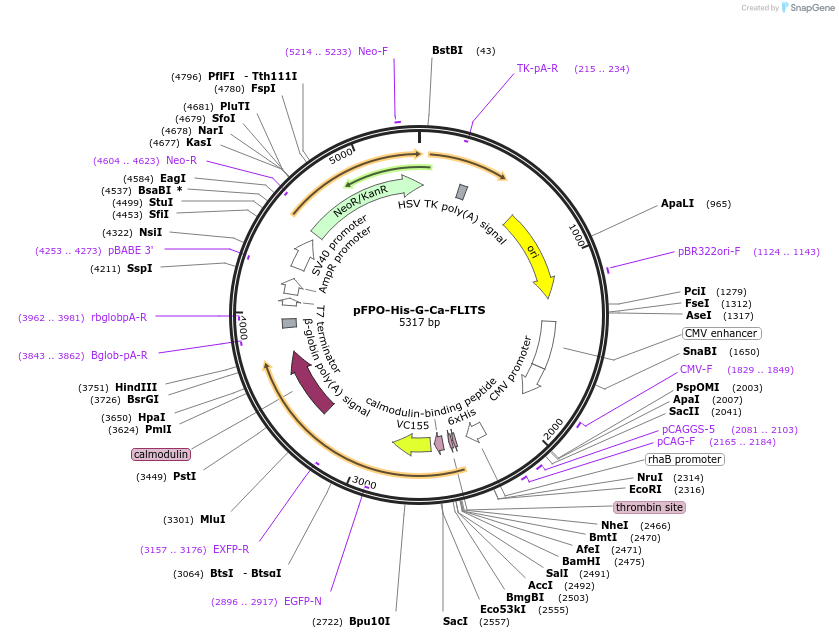
pFPO-His-G-Ca-FLITS
Plasmid#191455PurposeBacterial isolation of genetically encoded calcium sensor (GECI) G-Ca-FLITS that reports with a lifetime changeDepositorInsertG-Ca-FLITS
TagsHisExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
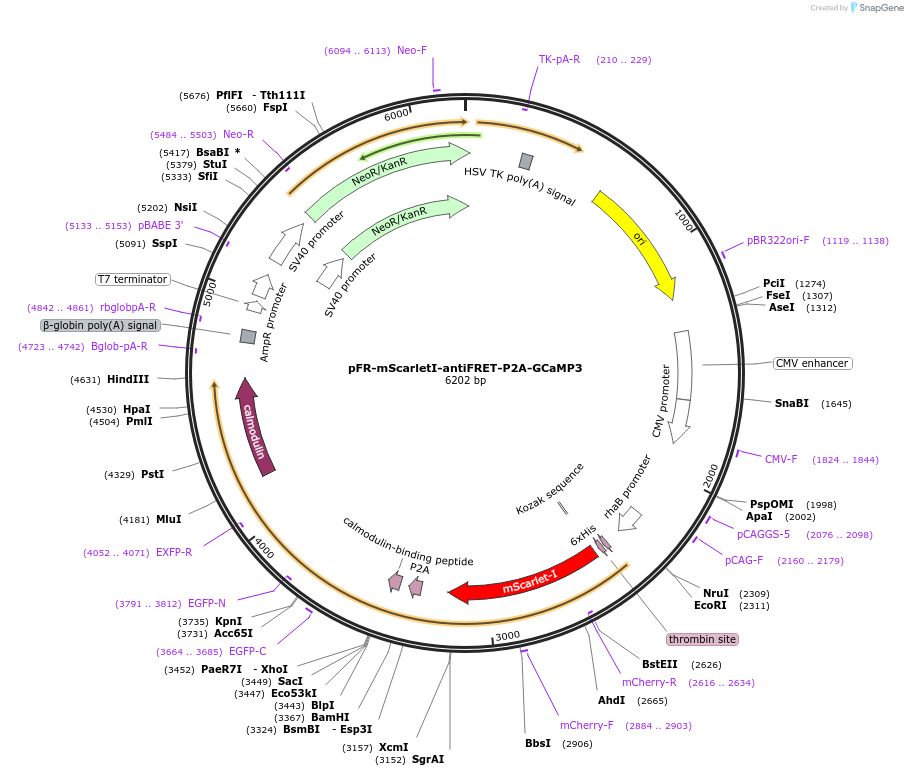
pFR-mScarletI-antiFRET-P2A-GCaMP3
Plasmid#191460PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) GCaMP3 to mScarlet-I fluorescent proteinDepositorInsertGCaMP3
TagsHis-mScarletI-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
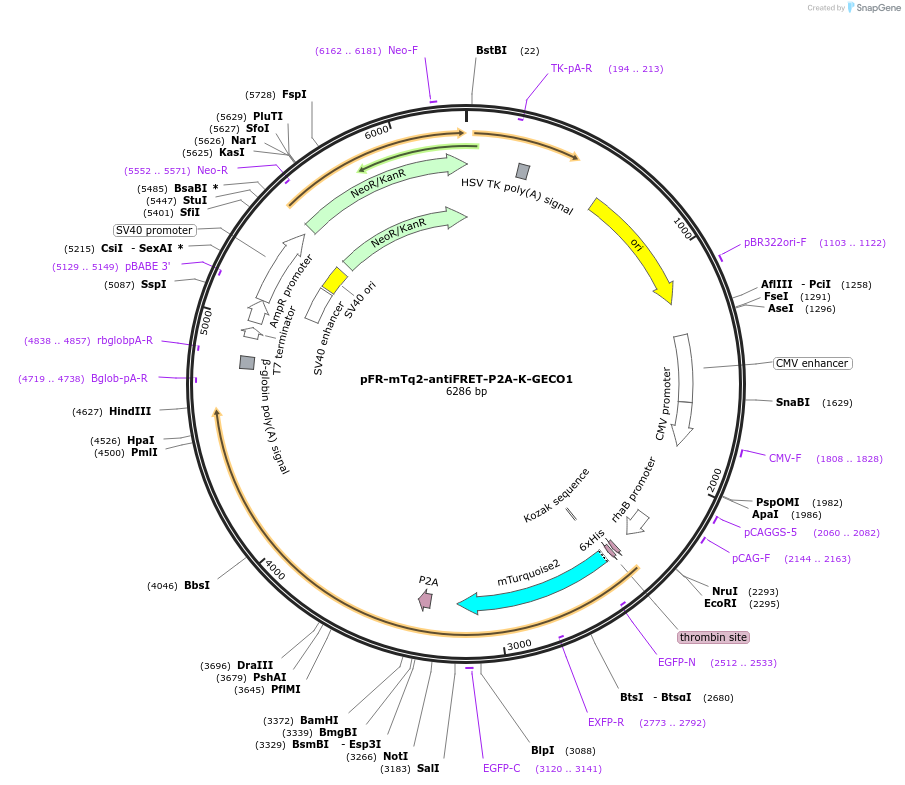
pFR-mTq2-antiFRET-P2A-K-GECO1
Plasmid#191469PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) K-GECO1 to mTurquoise2 fluorescent proteinDepositorInsertK-GECO1
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
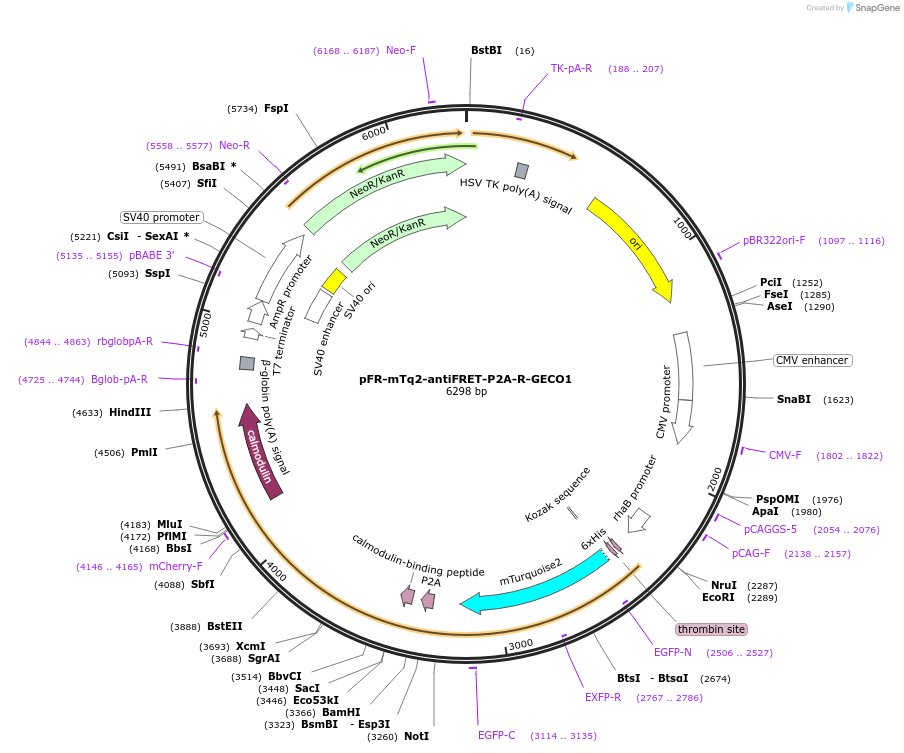
pFR-mTq2-antiFRET-P2A-R-GECO1
Plasmid#191471PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) R-GECO1 to mTurquoise2 fluorescent proteinDepositorInsertR-GECO1
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceDec. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
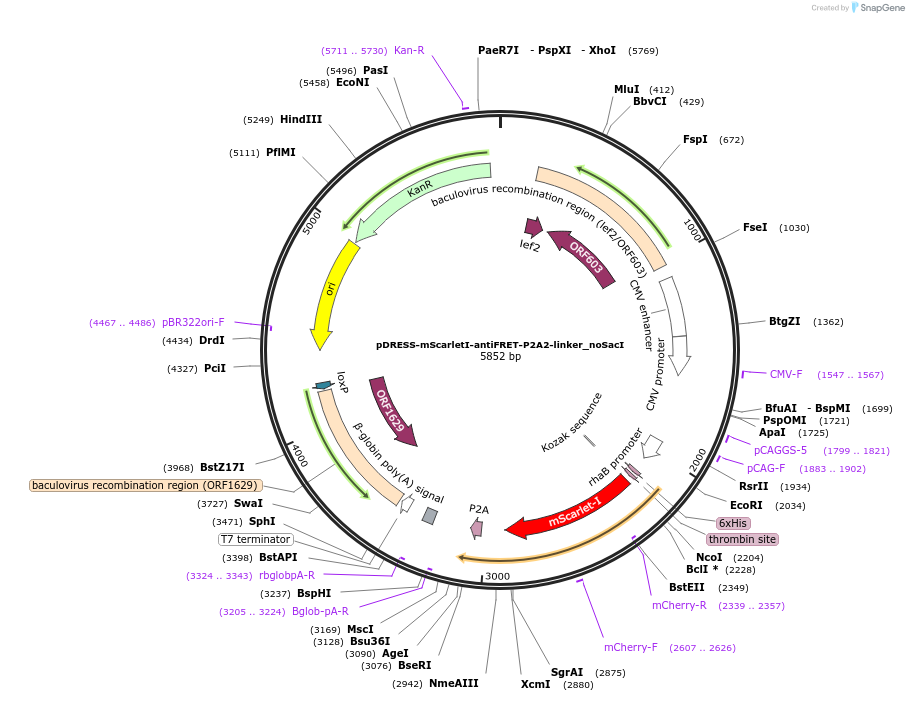
pDRESS-mScarletI-antiFRET-P2A2-linker_noSacI
Plasmid#191473PurposeNegative control for ratiometric comparison between green fluorescent proteins and sensorsDepositorInsertnone
TagsHis-mScarletI-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
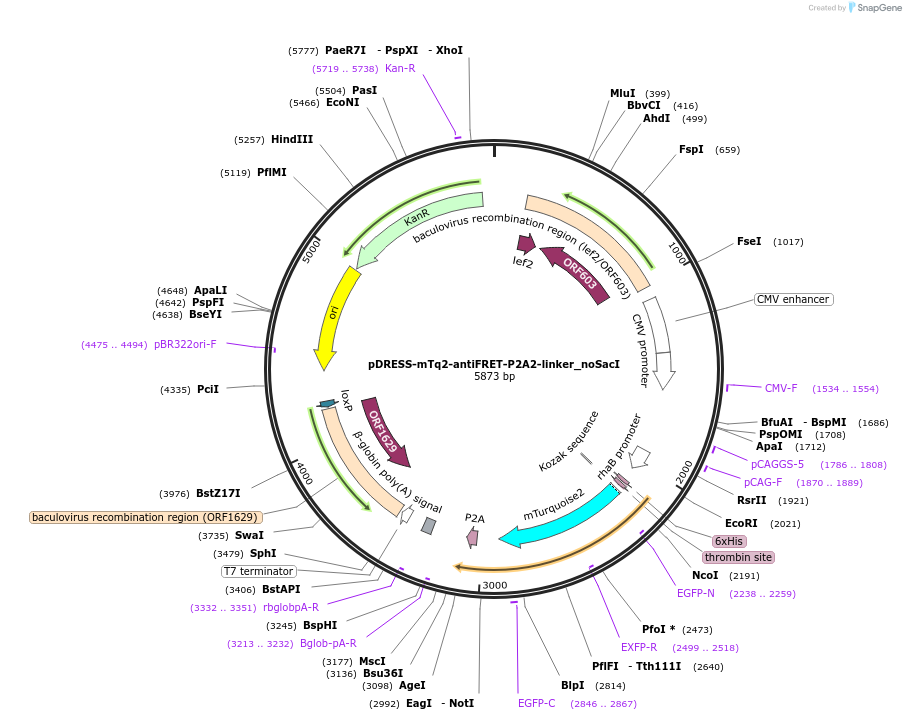
pDRESS-mTq2-antiFRET-P2A2-linker_noSacI
Plasmid#191472PurposeNegative control for ratiometric comparison between red fluorescent proteins and sensorsDepositorInsertnone
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
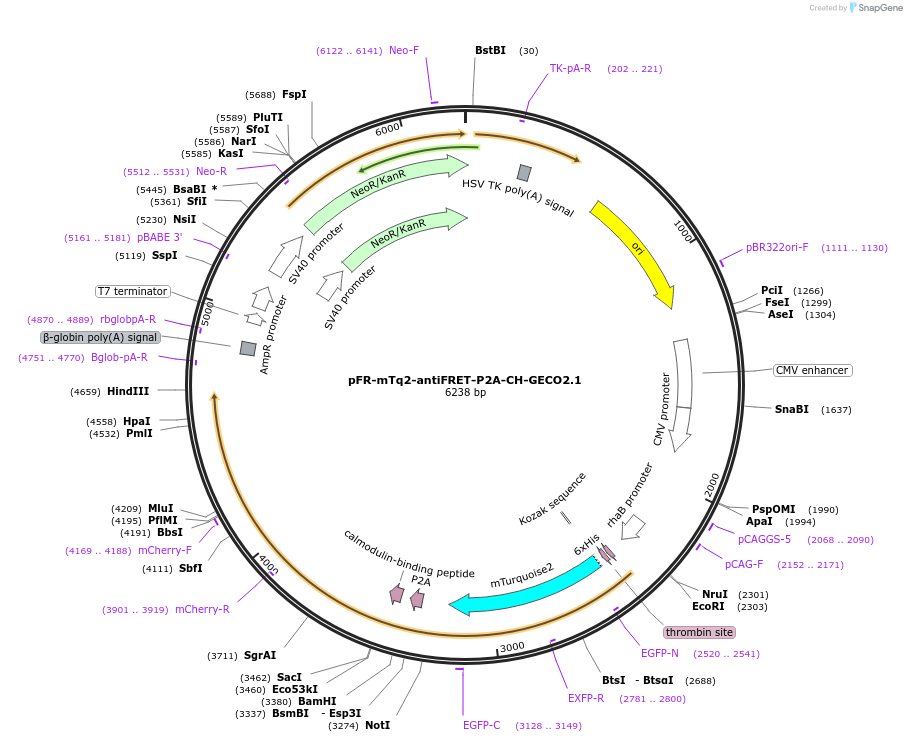
pFR-mTq2-antiFRET-P2A-CH-GECO2.1
Plasmid#191470PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) CH-GECO2.1 to mTurquoise2 fluorescent proteinDepositorInsertCH-GECO2.1
TagsHis-mTurquoise2-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
pFR-mScarletI-antiFRET-P2A-jGCaMP7c
Plasmid#191461PurposeCompare the intensity of the genetically encoded calcium sensor (GECI) jGCaMP7c to mScarlet-I fluorescent proteinDepositorInsertjGCaMP7c
TagsHis-mScarletI-antiFRETlinker-P2AExpressionBacterial and MammalianAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
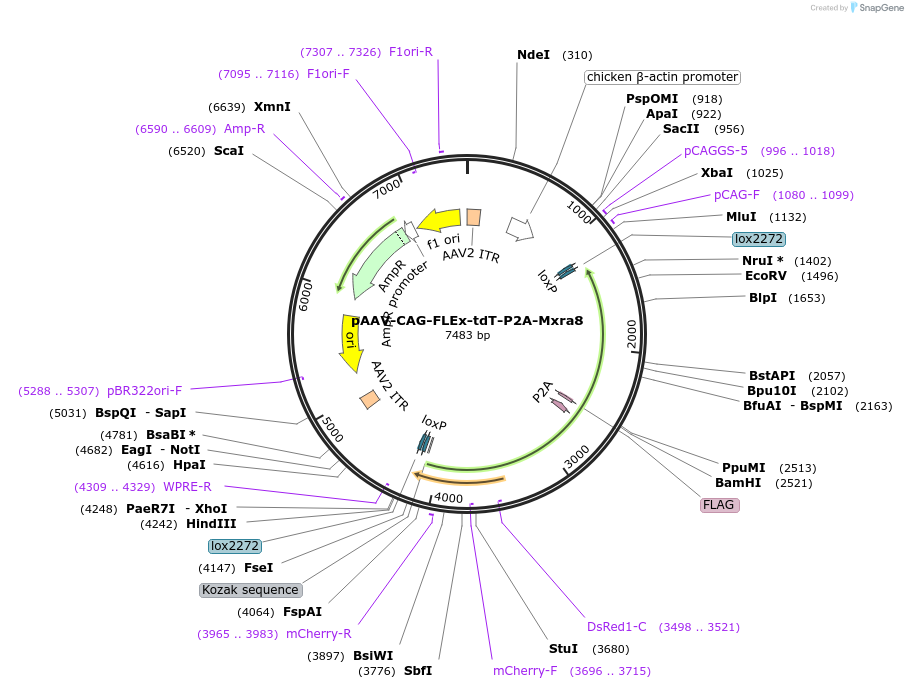
pAAV-CAG-FLEx-tdT-P2A-Mxra8
Plasmid#242478PurposeAAV vector expressing Mxra8 and tdTomato under cre-recombinase controlDepositorAvailable SinceSept. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
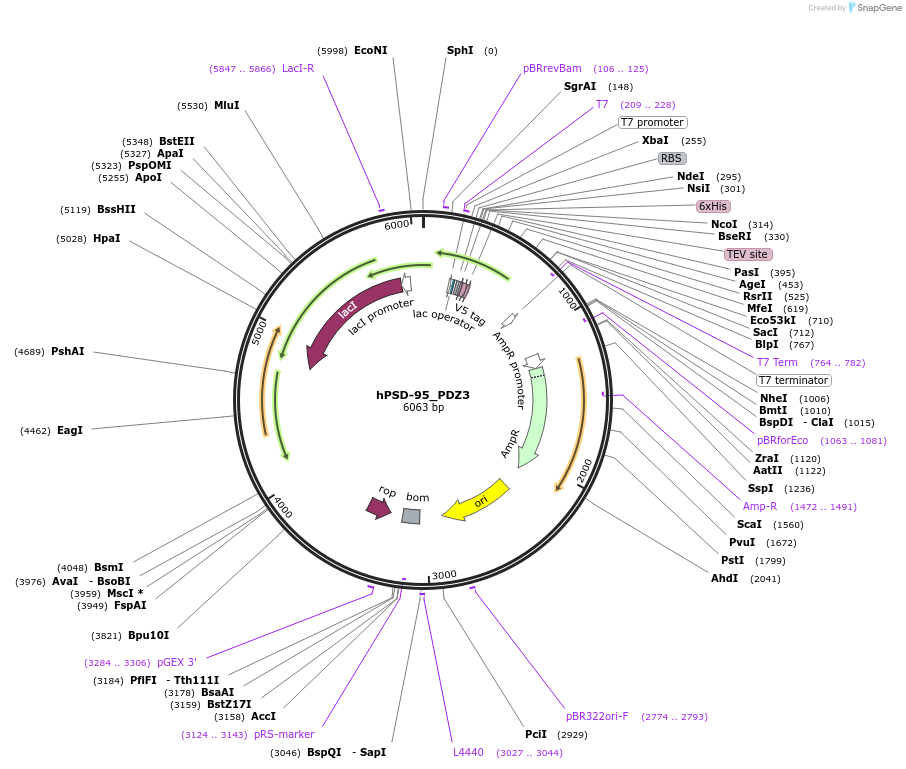
hPSD-95_PDZ3
Plasmid#245905PurposeBacterial expression of PSD-95 PDZ3 domain (302-402)DepositorAvailable SinceOct. 23, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
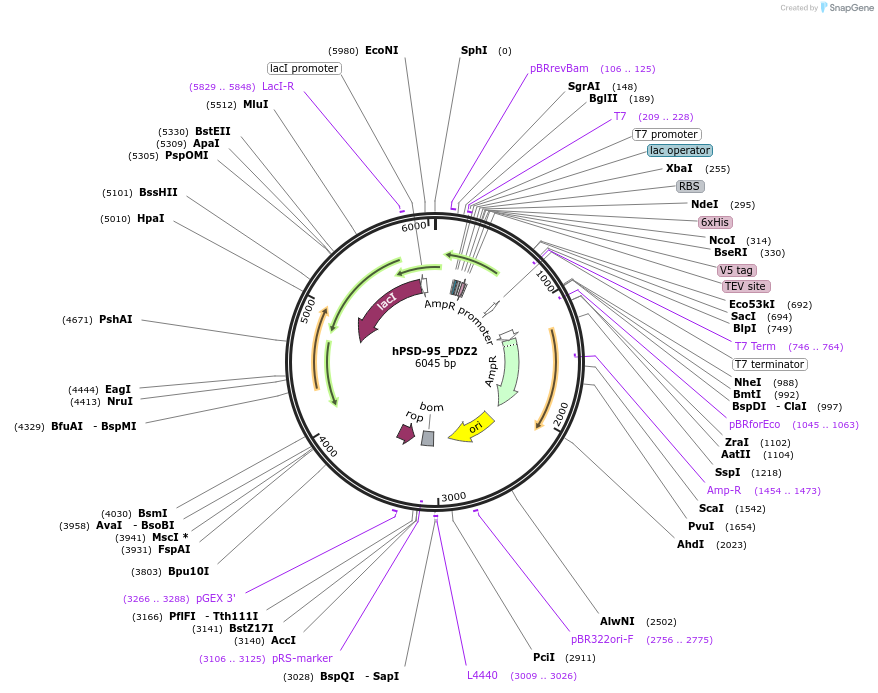
hPSD-95_PDZ2
Plasmid#245904PurposeBacterial expression of PSD-95 PDZ2 domain (155-249)DepositorAvailable SinceOct. 23, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
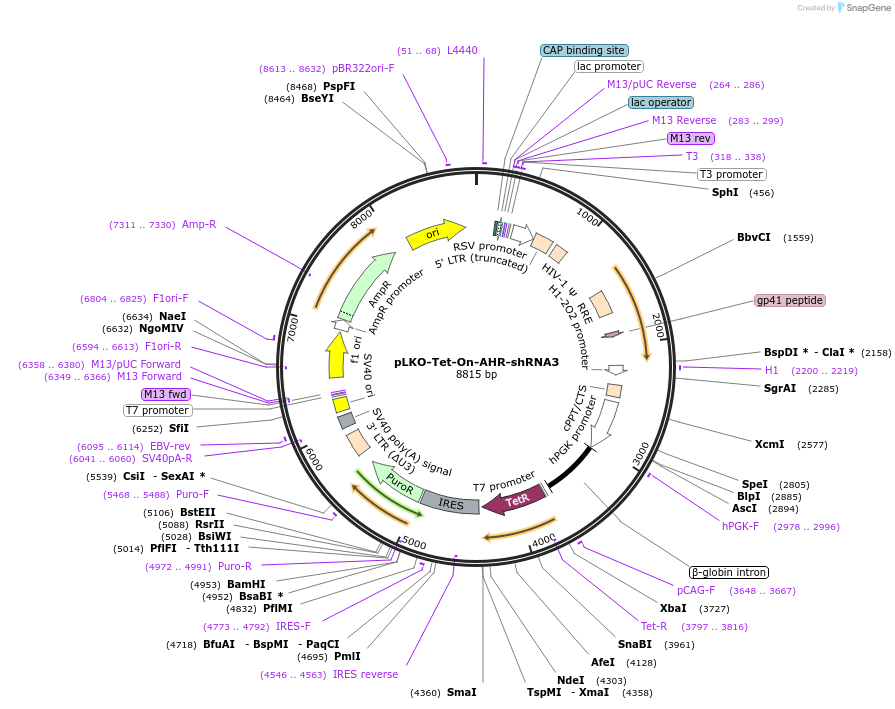
pLKO-Tet-On-AHR-shRNA3
Plasmid#166909PurposeLentivirus for inducible knockdown of human AHRDepositorAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
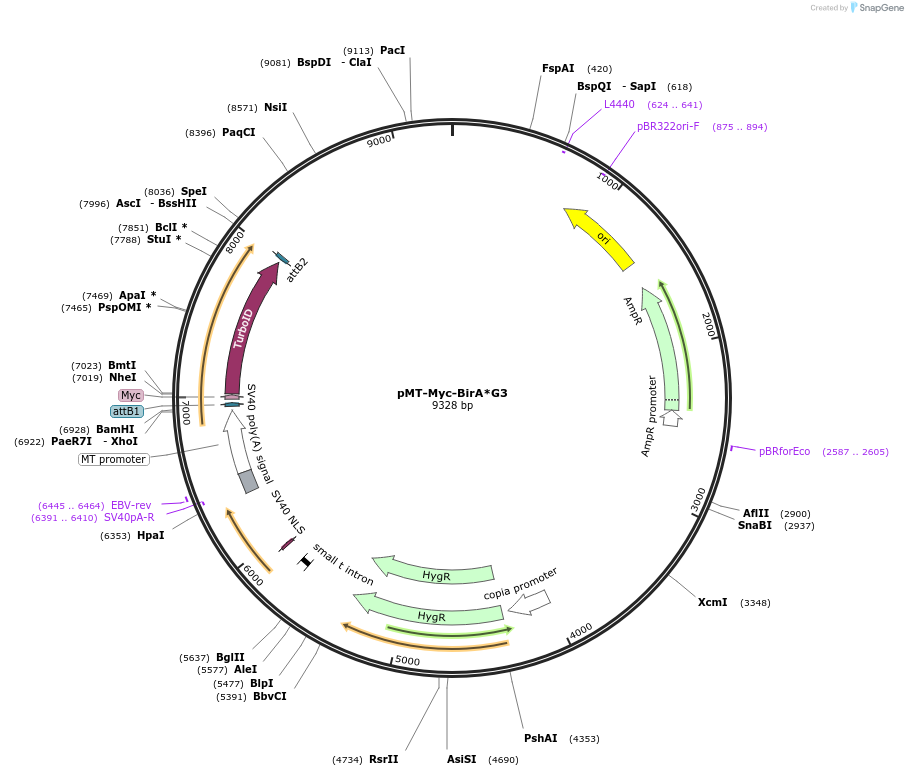
pMT-Myc-BirA*G3
Plasmid#240233PurposeExpression of Myc-BirA*-G3 under control of copper-inducible promoter. Can be used to generate stable cell lines by Hygromycin selection.DepositorInsertMyc-BirA*G3
ExpressionInsectPromoterMTAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
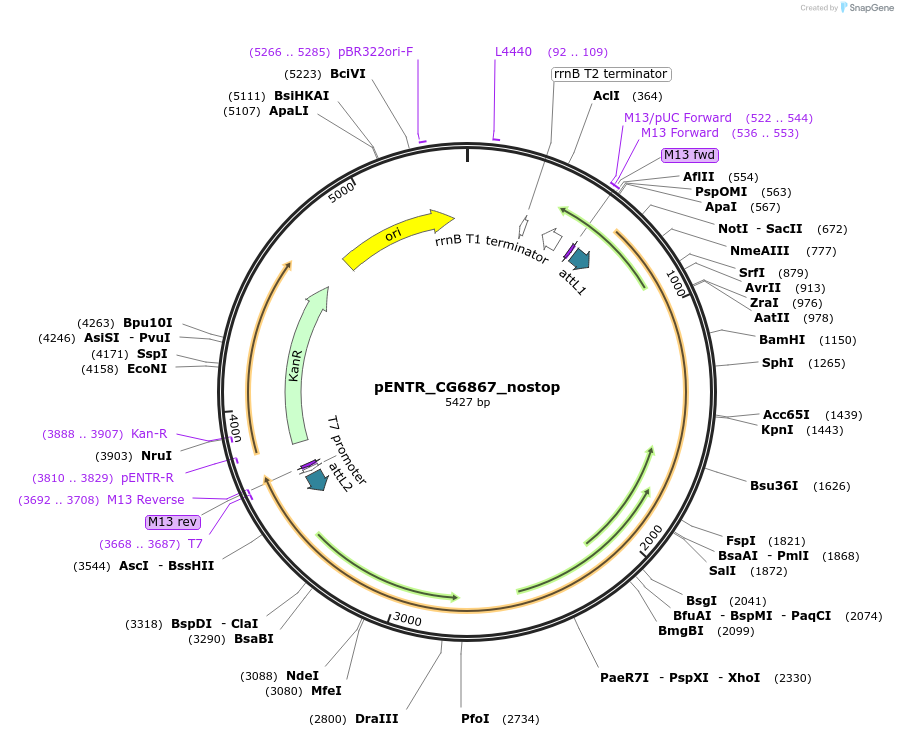
pENTR_CG6867_nostop
Plasmid#240238PurposeGateway entry clone with CG6867 without stop codonDepositorInsertCG6867 (CG6867 Fly)
UseGateway shuttle vectorAvailable SinceJuly 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
pBS-GMR-eya(shRNA)_EcoRImut
Plasmid#240222PurposepBS-GMR-eya(shRNA) vector with one of two EcoRI sites mutated (T->A)DepositorTypeEmpty backboneExpressionInsectAvailable SinceJuly 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
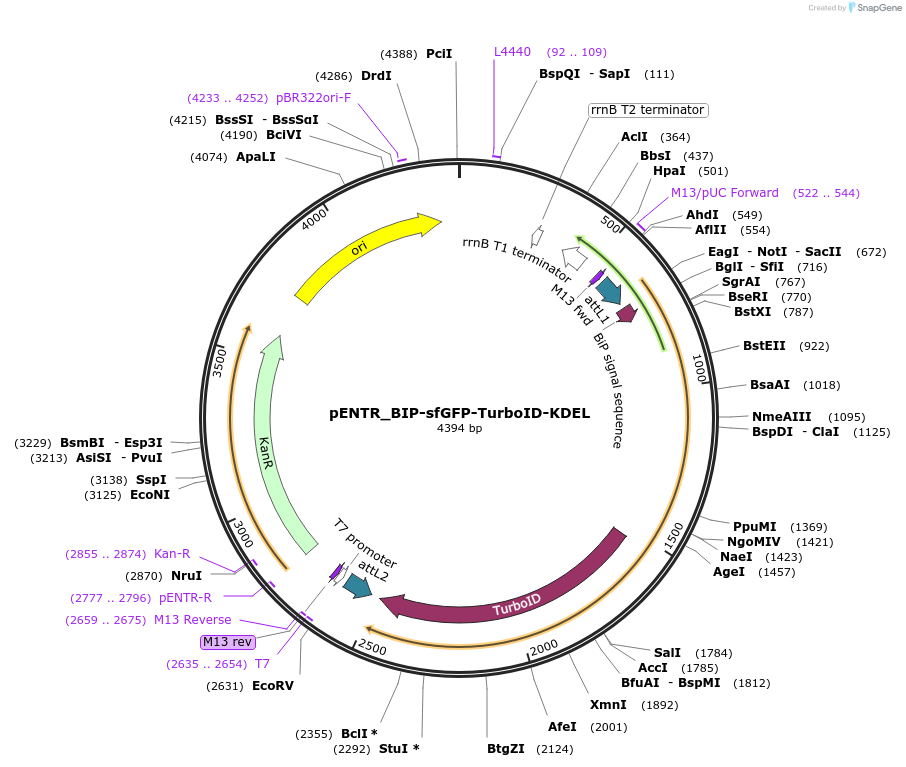
pENTR_BIP-sfGFP-TurboID-KDEL
Plasmid#240223PurposeGateway entry clone with ER-localized TurboID tagged with superfolder GFP (contains stop codon)DepositorInsertBIP-sfGFP-TurboID-KDEL
UseGateway shuttle vectorAvailable SinceJuly 11, 2025AvailabilityAcademic Institutions and Nonprofits only