We narrowed to 9,393 results for: sacs
-
Plasmid#205071PurposeMTF1 protein expressionDepositorAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only
-
pJAT58
Plasmid#204304PurposeExpresses jGCaMP7s-T2A-tdTomato under UAS control. HDR event marked with removable 3XP3-DsRed cassette expressed in eyes.DepositorInsert3XP3-DsRed
UseCRISPRPromoter3XP3Available SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
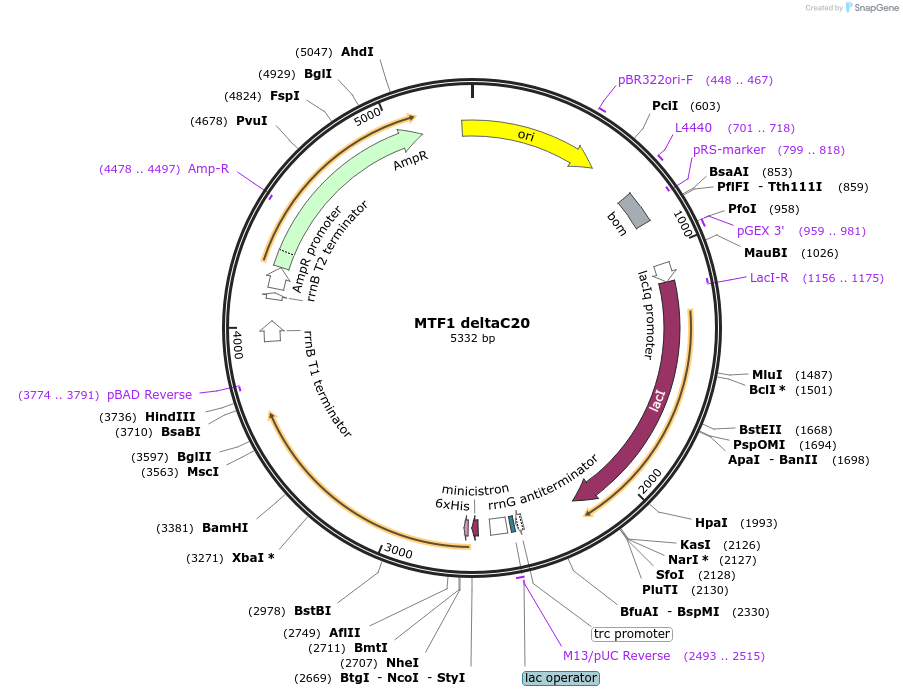
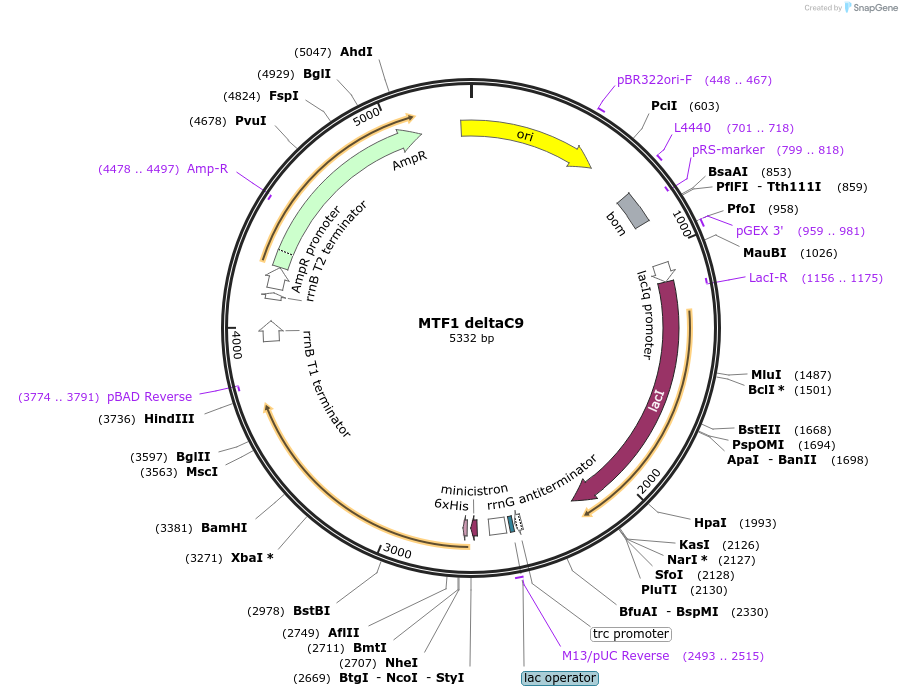
MTF1 deltaC9
Plasmid#205067PurposeMTF1 protein expressionDepositorAvailable SinceAug. 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
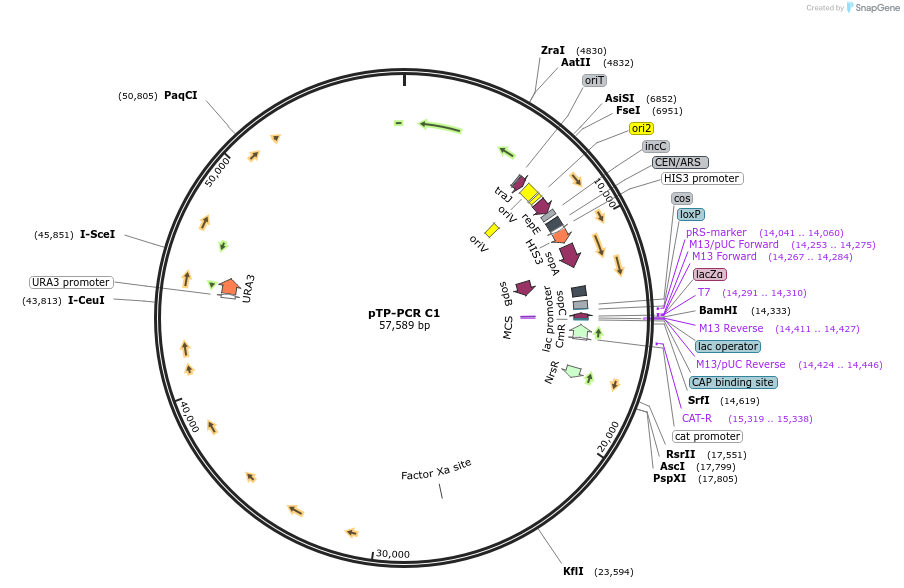
pTP-PCR C1
Plasmid#189618PurposeCloned Mitochondrial Genome of the diatom Thalassiosira pseudonana in yeast and bacteriaDepositorInsertMitochondrial Genome of Thalassiosira pseudonana
UseSynthetic Biology; Diatom expressionExpressionBacterial and YeastMutationMinor additional mutations described in manuscriptAvailable SinceJuly 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
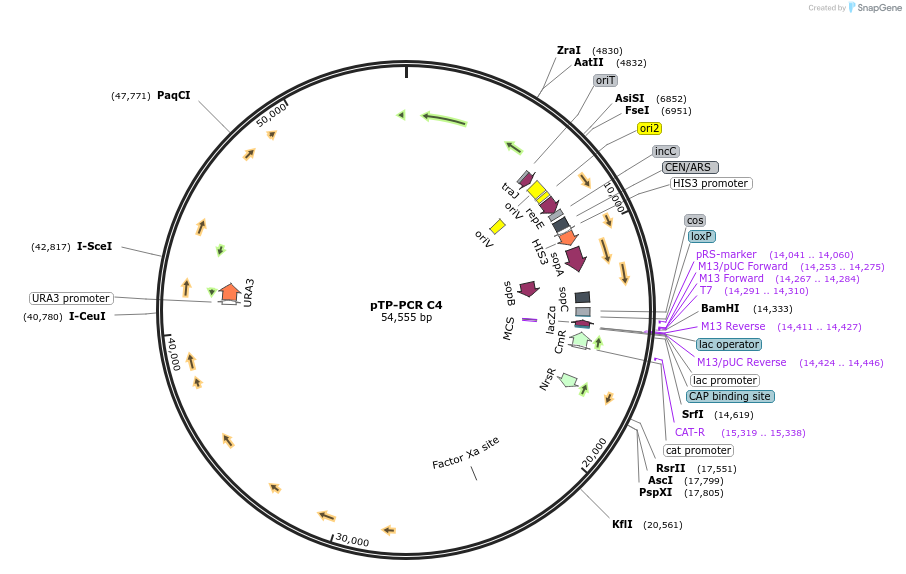
pTP-PCR C4
Plasmid#189621PurposeCloned Reduced Mitochondrial Genome of the diatom Thalassiosira pseudonana in yeast and bacteriaDepositorInsertReduced Mitochondrial Genome of Thalassiosira pseudonana
UseSynthetic Biology; Diatom expressionExpressionBacterial and YeastMutationRepeat region removed, and minor additional mutat…Available SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
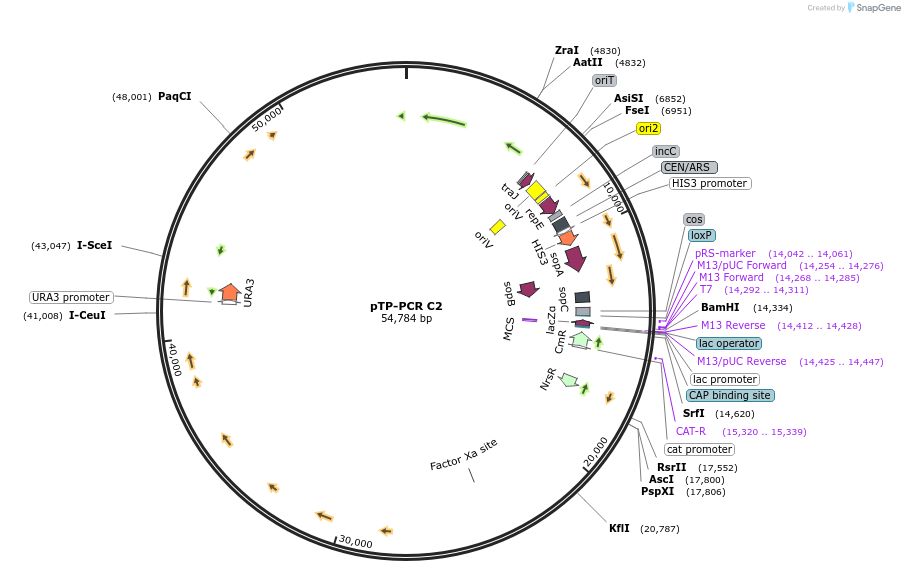
pTP-PCR C2
Plasmid#189619PurposeCloned Mitochondrial Genome of the diatom Thalassiosira pseudonana in yeast and bacteriaDepositorInsertMitochondrial Genome of Thalassiosira pseudonana
UseSynthetic Biology; Diatom expressionExpressionBacterial and YeastMutationMinor additional mutations described in manuscriptAvailable SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
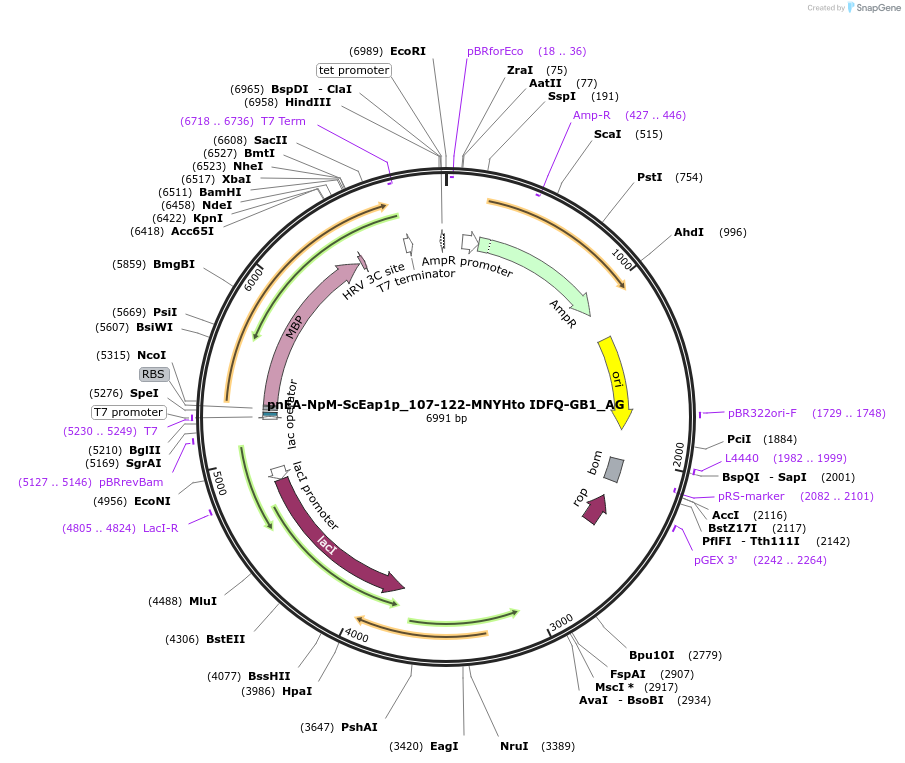
pnEA-NpM-ScEap1p_107-122-MNYHto IDFQ-GB1_AG
Plasmid#148812PurposeBacterial Expression of ScEap1p_107-122-MNYHtoIDFQDepositorInsertScEap1p_107-122-MNYHtoIDFQ (EAP1 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
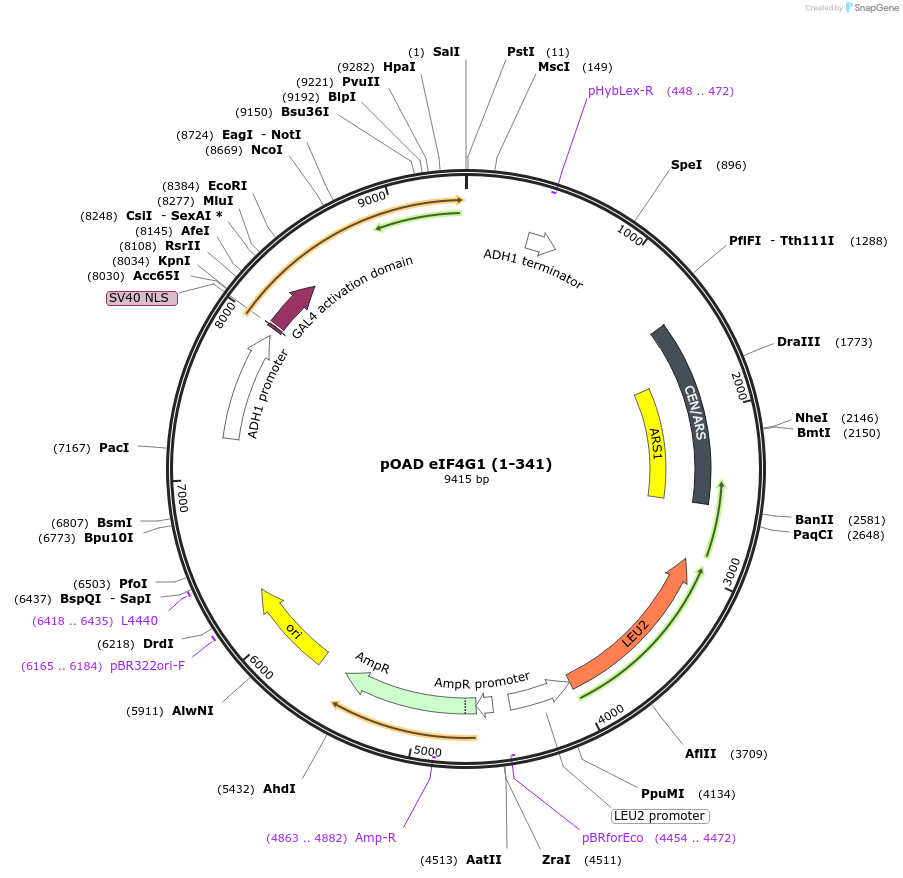
pOAD eIF4G1 (1-341)
Plasmid#196833PurposeExpesses S.cerevisiae eIF4G1 amino acids 1-341 as Gal4 activation domain fusion.DepositorInserteIF4G1 (TIF4631 Budding Yeast)
TagsGal4ExpressionYeastMutationamino acids 1-341PromoterADHAvailable SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
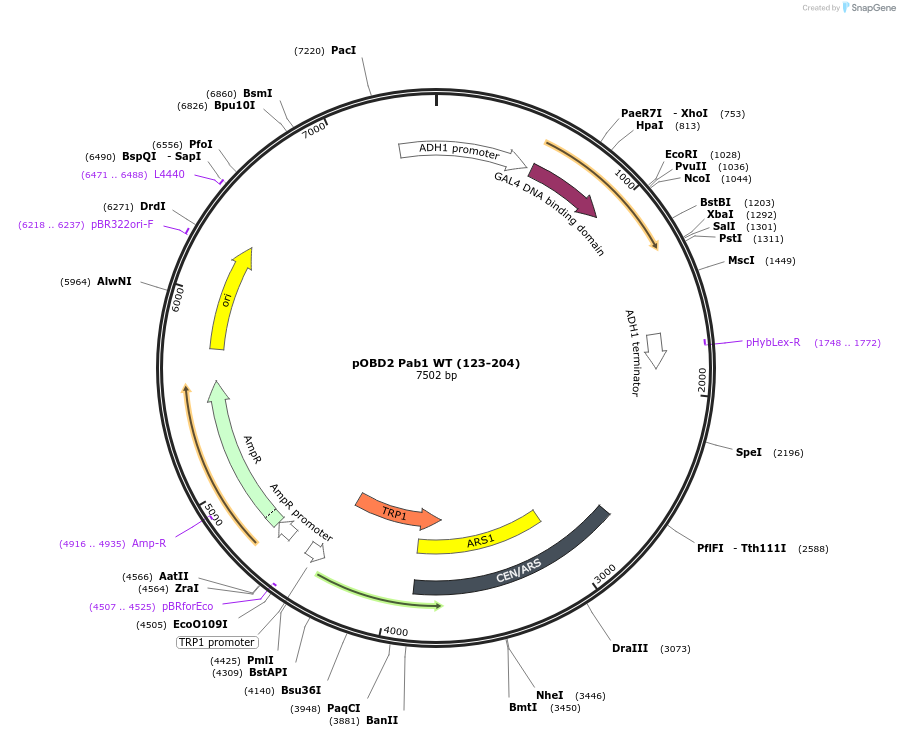
pOBD2 Pab1 WT (123-204)
Plasmid#196834PurposeExpresses Pab1 RRM2 fragment fused to GAL4 DNA binding domainDepositorInsertPab1 (PAB1 Budding Yeast)
TagsGal4ExpressionYeastMutation123-204 amino acidsPromoterADHAvailable SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
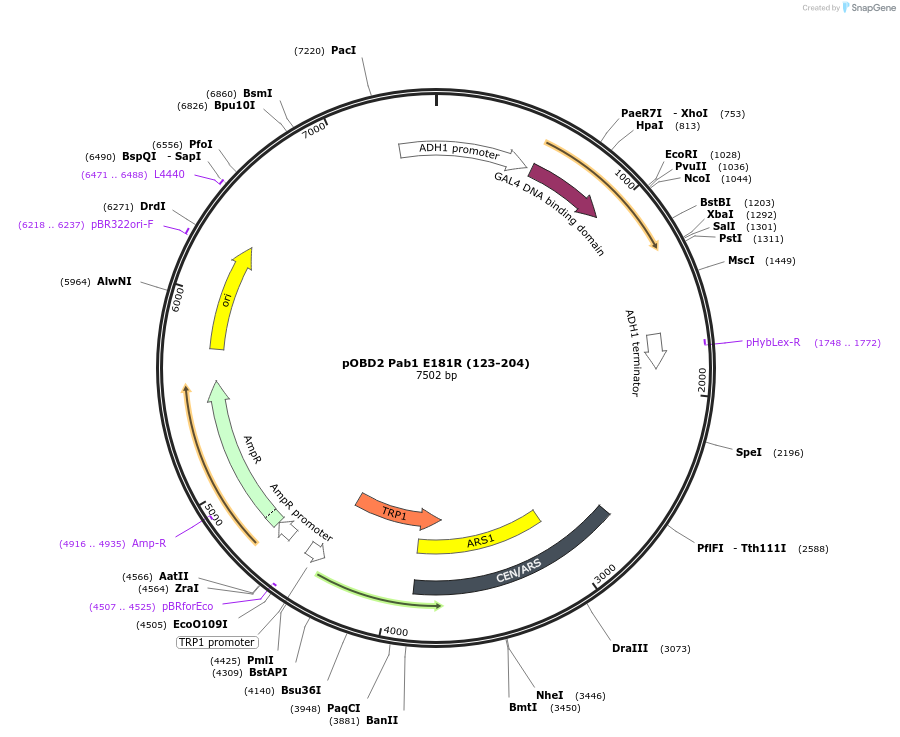
pOBD2 Pab1 E181R (123-204)
Plasmid#196835PurposeExpresses Pab1 RRM2 fragment E181R fused to GAL4 DNA binding domainDepositorInsertPab1 (PAB1 Budding Yeast)
TagsGal4ExpressionYeastMutation123-204 amino acids & E181RPromoterADHAvailable SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
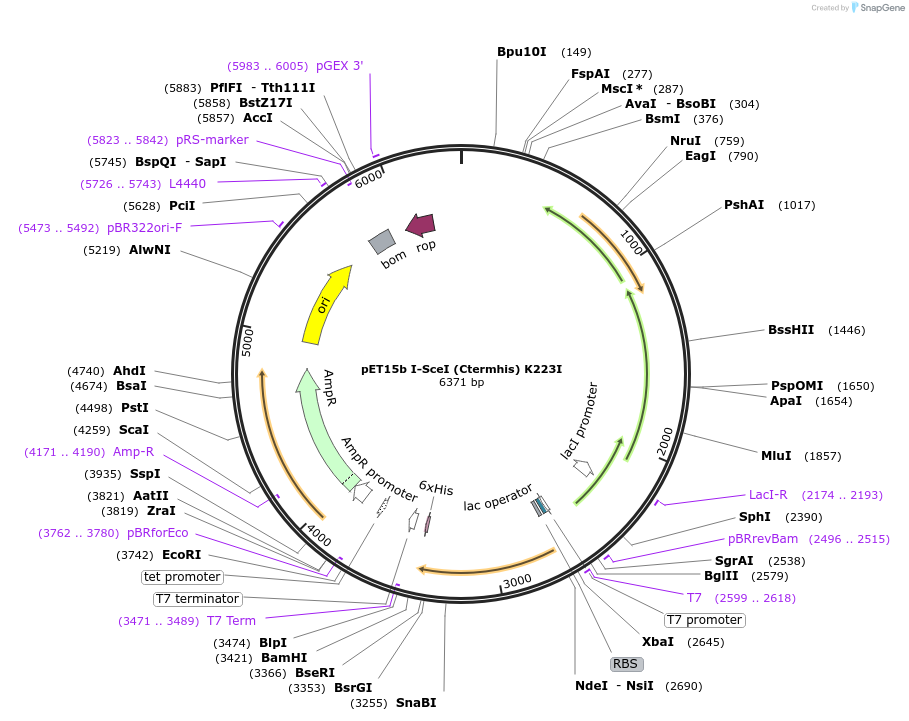
pET15b I-SceI (Ctermhis) K223I
Plasmid#195615PurposeExpresses I-SceI homing endonuclease from the T7 promoter containing an additional methionine-histidine sequence at the amino terminus and a K223I mutation that results in nicking activityDepositorInsertI-SceI
ExpressionBacterialMutationC2694G, G2345C, G2351C, A3365T, A3366TPromoterT7Available SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
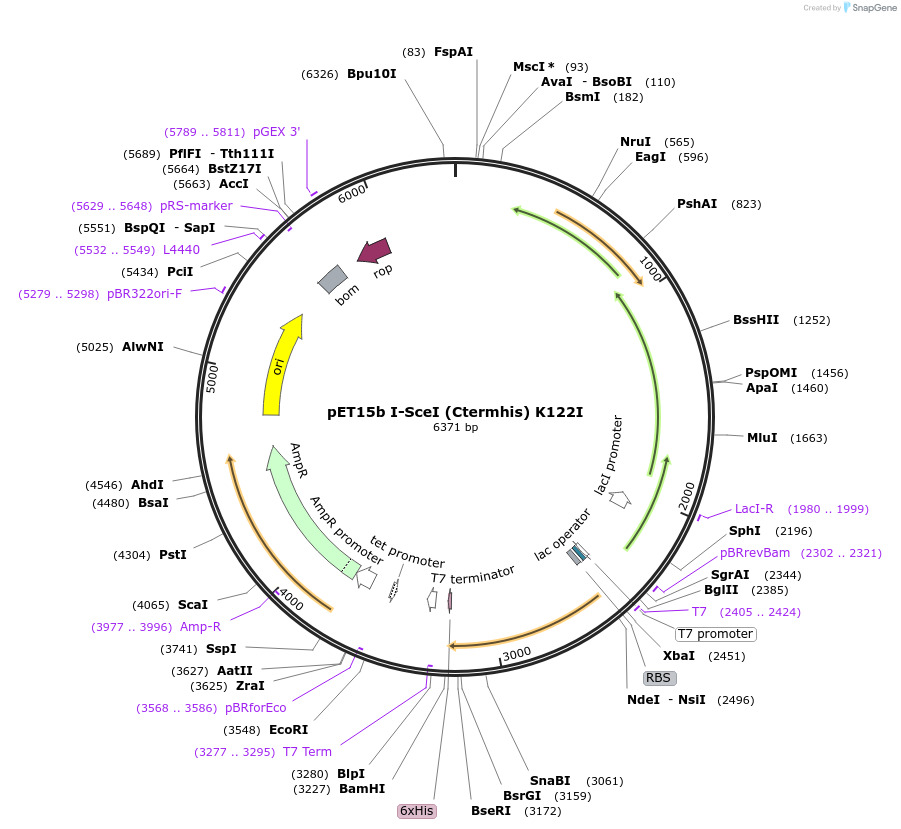
pET15b I-SceI (Ctermhis) K122I
Plasmid#195614PurposeExpresses I-SceI homing endonuclease from the T7 promoter containing an additional methionine-histidine sequence at the amino terminus and a K122I mutation that results in nicking activityDepositorInsertI-SceI
ExpressionBacterialMutationC2458G, A2826T, A2827TPromoterT7Available SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
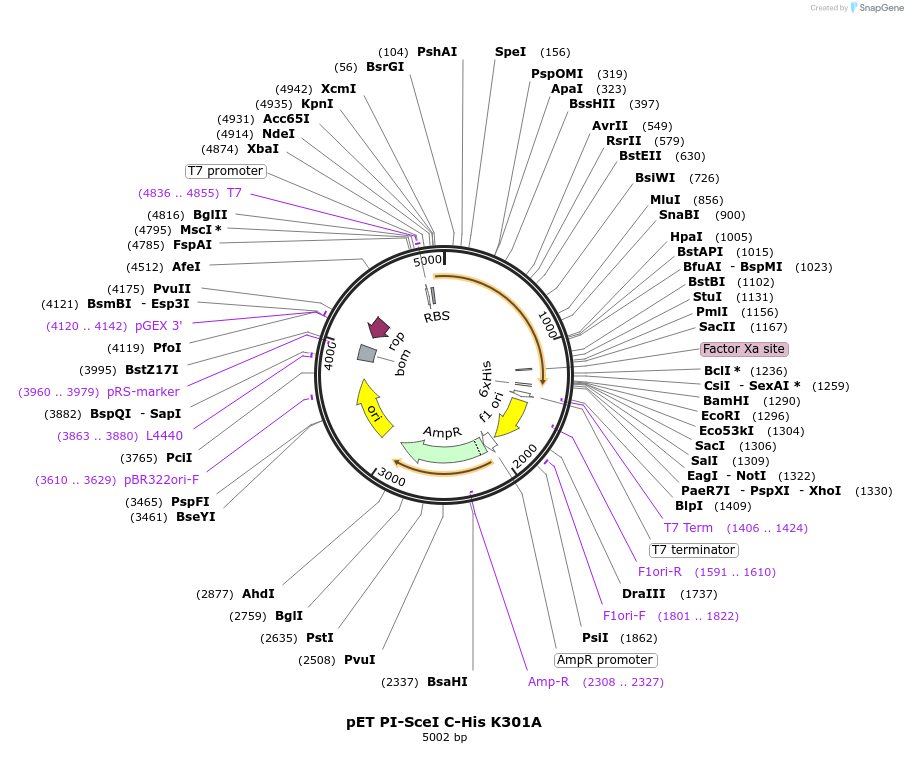
pET PI-SceI C-His K301A
Plasmid#195611PurposeExpresses PI-SceI (VDE) homing endonuclease/intein from the T7 promoter that contains the K301A active site mutationDepositorInsertPI-SceI
ExpressionBacterialMutationA822G, A823C, A824TPromoterT7Available SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
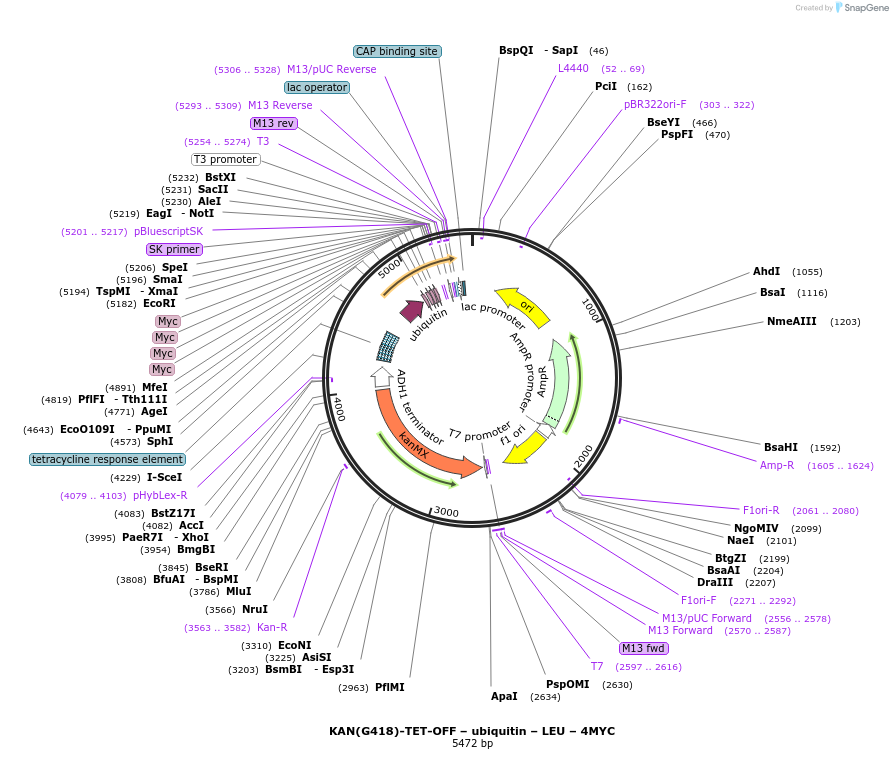
KAN(G418)-TET-OFF – ubiquitin – LEU – 4MYC
Plasmid#193318PurposeDoxycycline-induced transcriptional regulation and Ubiquitin - LEU - 4xMYC N-terminal tagging. Ubiquitin is cleaved off, resulting in an unstable protein with an N-terminal leucine.DepositorInsertUbiquitin - Leucine - 4MYC N-terminal tagging
Tags4MYCExpressionBacterialAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
KAN(G418)-TET-OFF – ubiquitin – LEU – 4FLAG
Plasmid#193317PurposeDoxycycline-induced transcriptional regulation and Ubiquitin - LEU - 4xFLAG N-terminal tagging. Ubiquitin is cleaved off, resulting in an unstable protein with an N-terminal leucine.DepositorInsertUbiquitin - Leucine - 4FLAG N-terminal tagging
Tags4FLAGExpressionBacterialAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
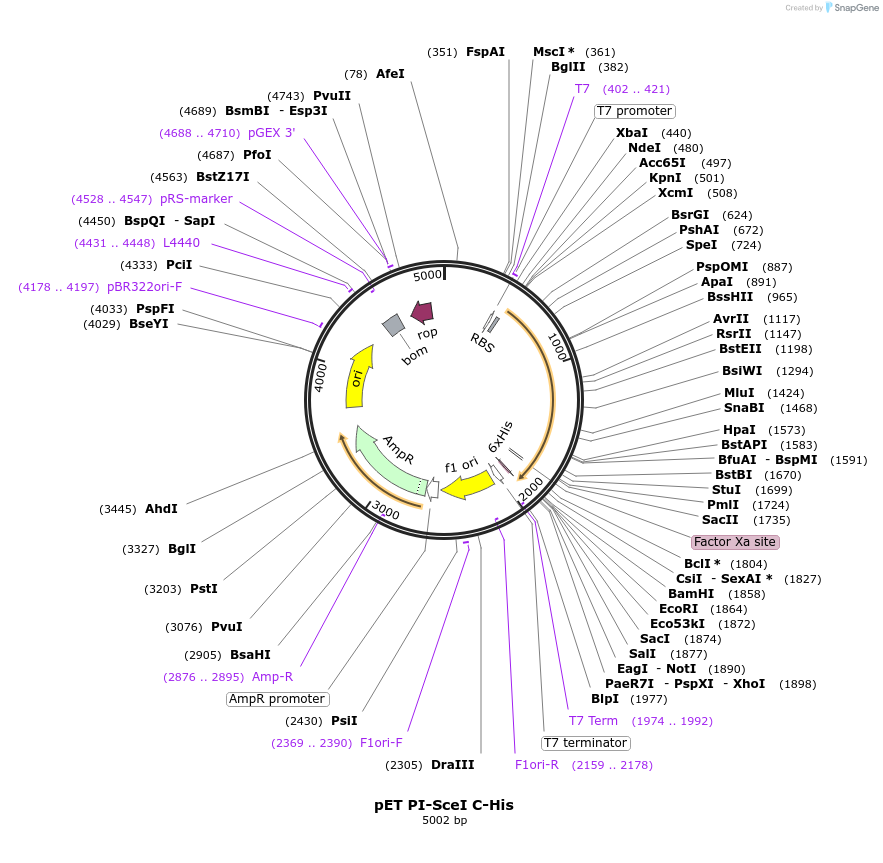
pET PI-SceI C-His
Plasmid#195610PurposeExpresses PI-SceI (VDE) homing endonuclease/intein from the T7 promoter with a carboxyl-terminal histidine tagDepositorInsertPI-SceI
ExpressionBacterialMutationT825C, C900G, A901C, A903C, T1131G, T1137C, T1227…PromoterT7Available SinceJan. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
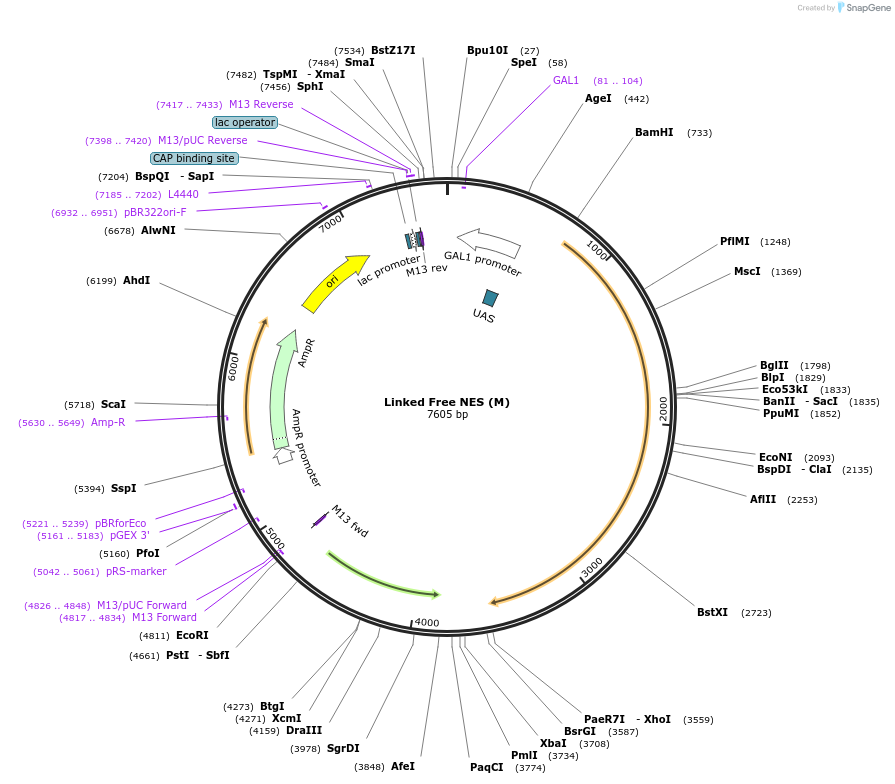
Linked Free NES (M)
Plasmid#182436PurposeYeast integrative plasmid for expressing fusion protein ERG20(F96W-N127W)-AcNES1 (GAL10 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertFPPS(M)-NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10Available SinceDec. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
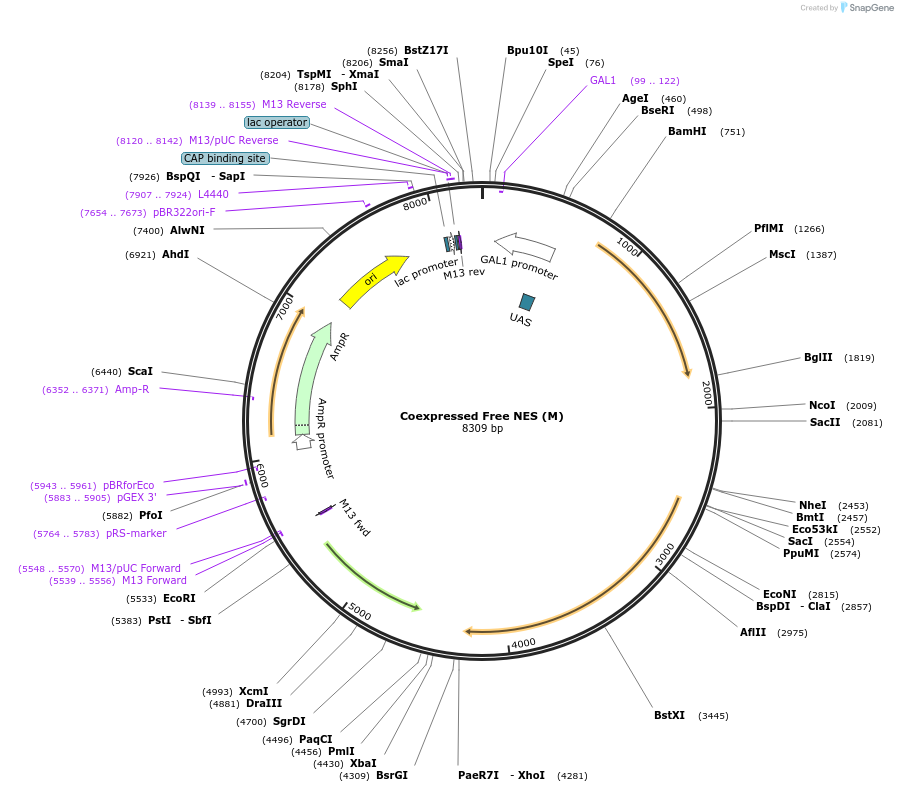
Coexpressed Free NES (M)
Plasmid#182437PurposeYeast integrative plasmid for expressing ERG20(F96W-N127W) (GAL10 promoter) and AcNES1 (GAL7 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertsFPPS(M)
NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10 and GAL7Available SinceDec. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
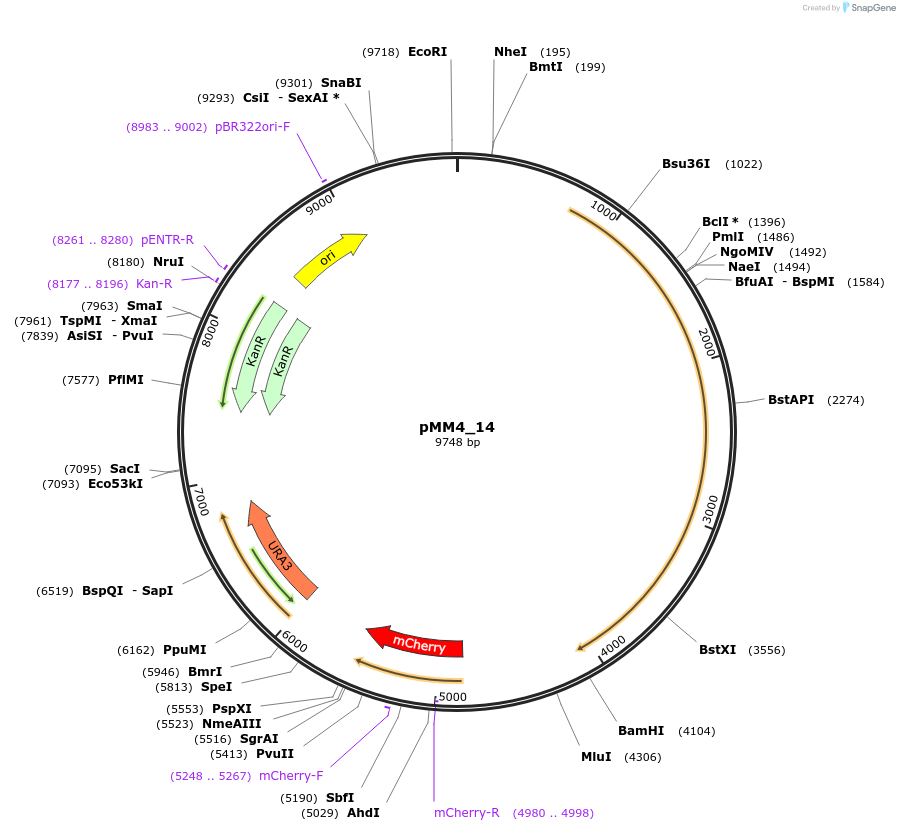
pMM4_14
Plasmid#183062PurposePlasmid expressing a biosensor for acetic acid: BM3R1-Haa1-mTurquoise2 under RET2p and mCherry under HREsYGP1p with flanks for genome integration into HO locusDepositorInsertsmCherry
BM3R1-HAA1-mTurquoise2
UseSynthetic BiologyTagsmCherry and mTurquoise2ExpressionYeastPromoterHREsYGP1p and RET2Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
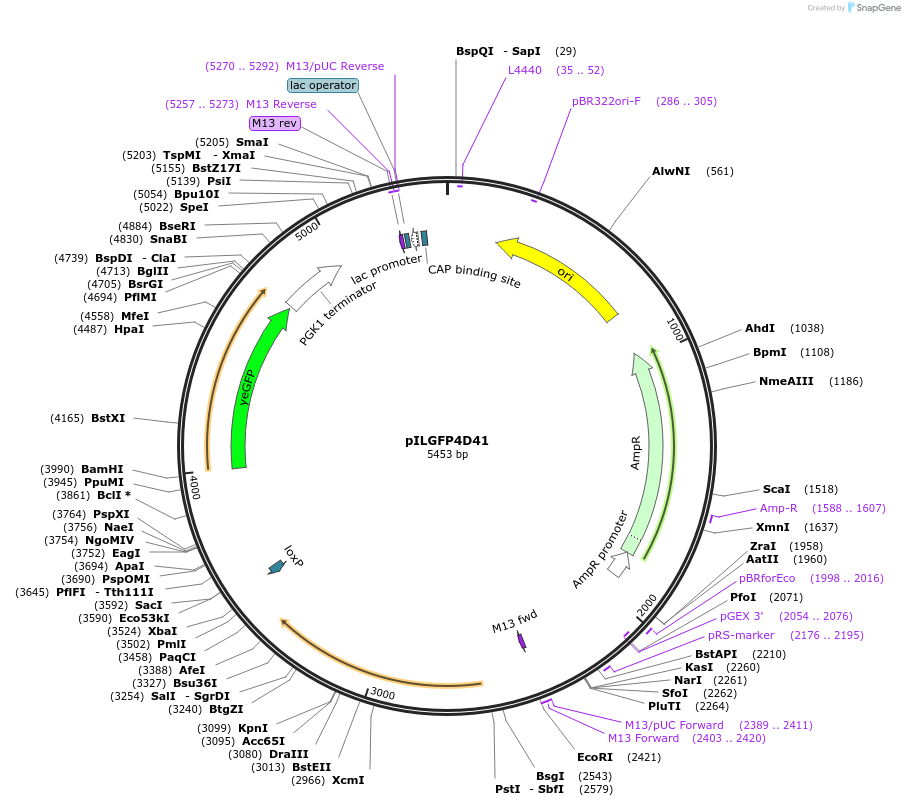
pILGFP4D41
Plasmid#185841PurposeTesting the CYC1 core promoter inserted with 4 Z268 elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PCYC1+[Z268]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only