We narrowed to 2,377 results for: pet28
-
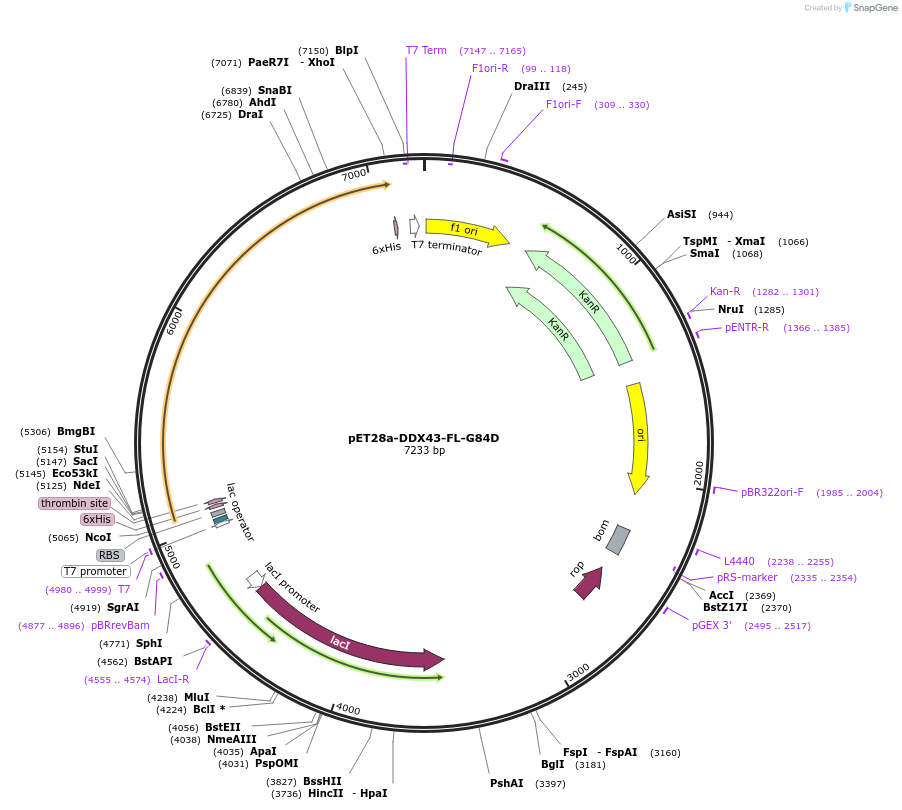
Plasmid#134577PurposeExpress human DDX43 mutant (full-length with G84D mutation) in E. coliDepositorInsertDDX43-G84D (DDX43 Human)
TagsHisExpressionBacterialMutationfull-length (G84D)PromoterT7Available SinceJan. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
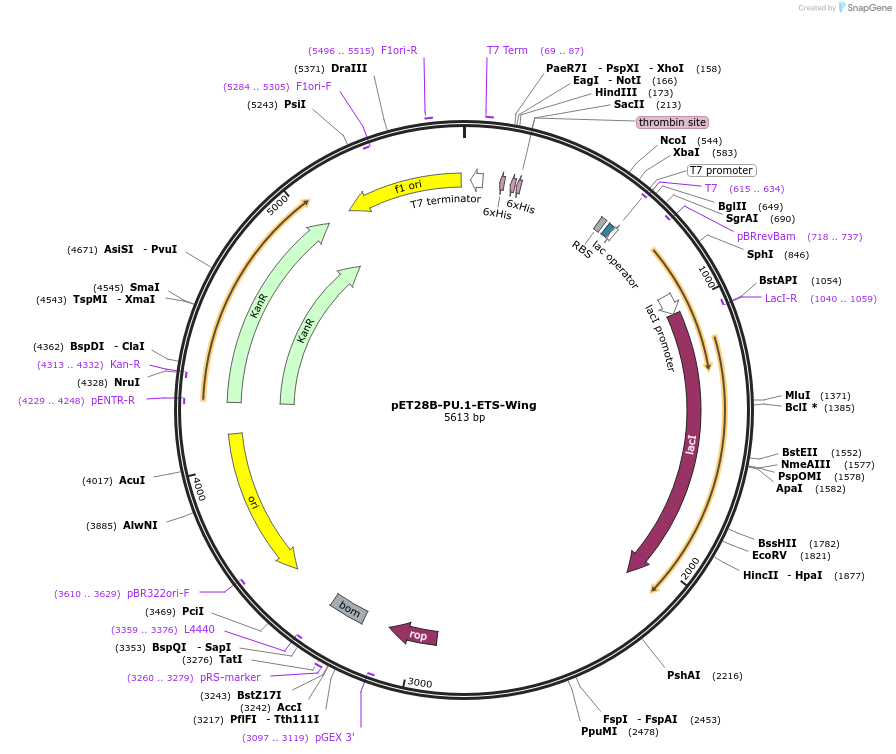
pET28B-PU.1-ETS-Wing
Plasmid#124552PurposeChimeric ETS domain: Wing of Ets-1 in PU.1 scaffoldDepositorTags6xHis tagExpressionBacterialMutationThe 5 residues making up the "wing" in …Available SinceJune 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
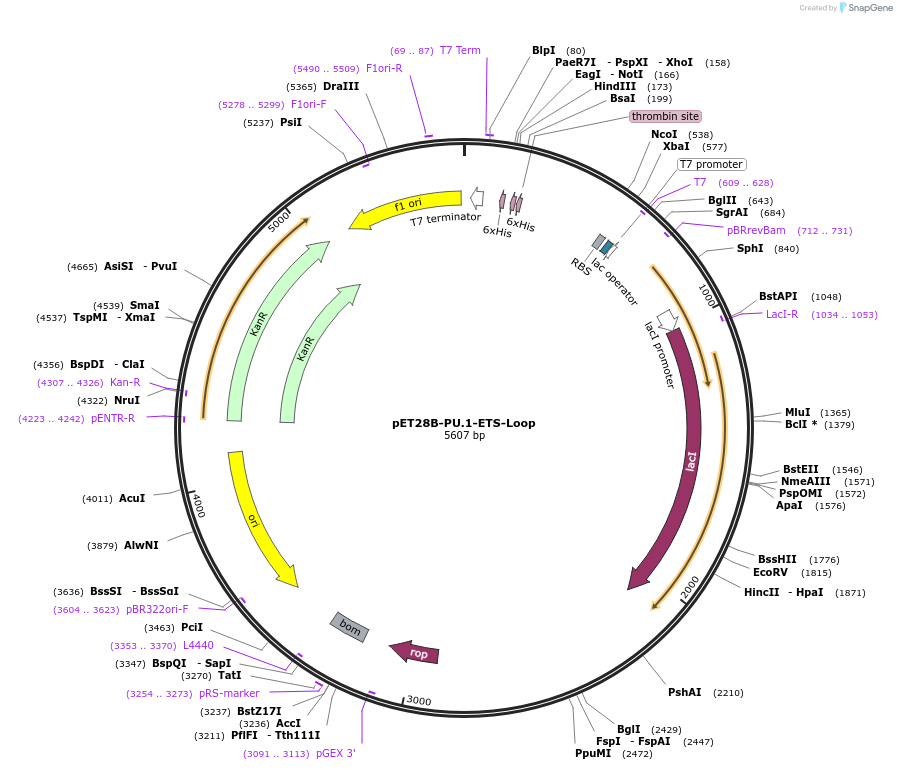
pET28B-PU.1-ETS-Loop
Plasmid#124551PurposeChimeric ETS domain: Loop of Ets-1 in PU.1 scaffoldDepositorTags6xHis tagExpressionBacterialMutationResidues 216 to 223 replaced with corresponding s…Available SinceJune 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
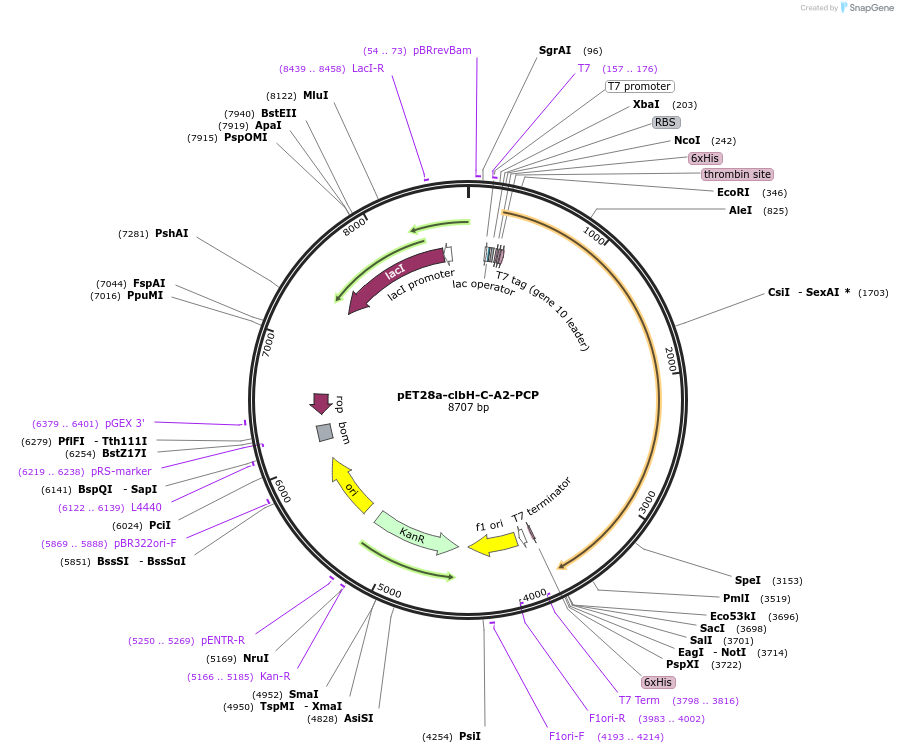
pET28a-clbH-C-A2-PCP
Plasmid#114157PurposeExpresses ClbH-C-A2-PCPDepositorInsertclbH
Tags6xHis-thrombin-T7 tagExpressionBacterialMutationOnly contains the C-A2-PCP domains of ClbHAvailable SinceAug. 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
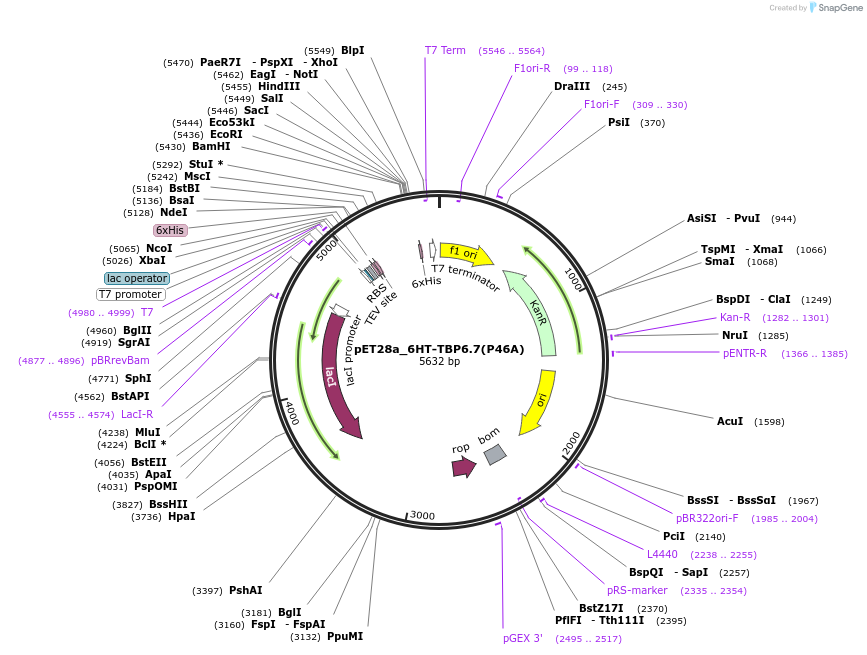
pET28a_6HT-TBP6.7(P46A)
Plasmid#112721PurposeThis mutation (P46A) marks the first residue within the evolved region of U1A.DepositorInsert6His-TEV-TBP6.7(P46A)
Tags6His-TEVExpressionBacterialMutationP46APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
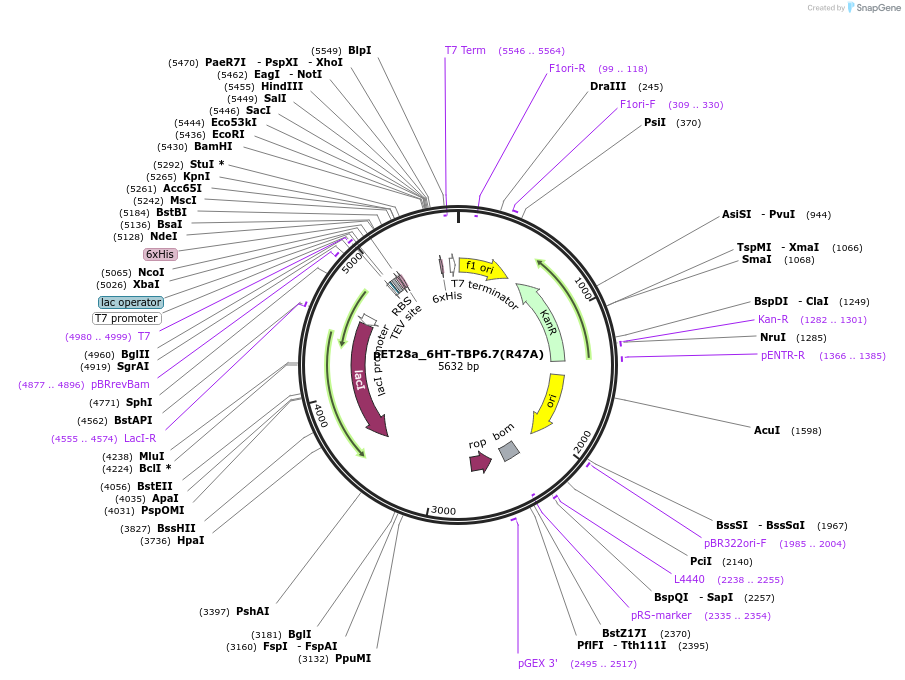
pET28a_6HT-TBP6.7(R47A)
Plasmid#112722PurposeArg47 within TBP6.7 is the most critical residue in terms of forming interactions with WT TAR. Mutating this amino acid to Ala results in ~600-fold reduction in Kd.DepositorInsert6His-TEV-TBP6.7(R47A)
Tags6His-TEVExpressionBacterialMutationR47APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
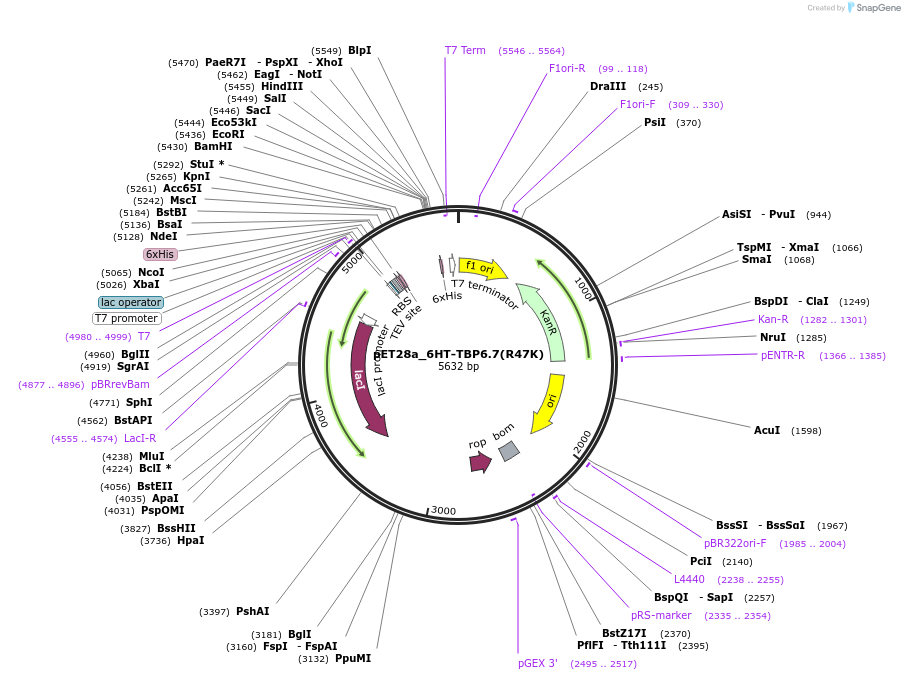
pET28a_6HT-TBP6.7(R47K)
Plasmid#112723PurposeArg47 of TBP6.7 interacts with Hoogsteen edge of TAR's Gua26. Mutating this amino acid to Lys reduces binding to the WT TAR RNA ~330-fold.DepositorInsert6His-TEV-TBP6.7(R47K)
Tags6His-TEVExpressionBacterialMutationR47KPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
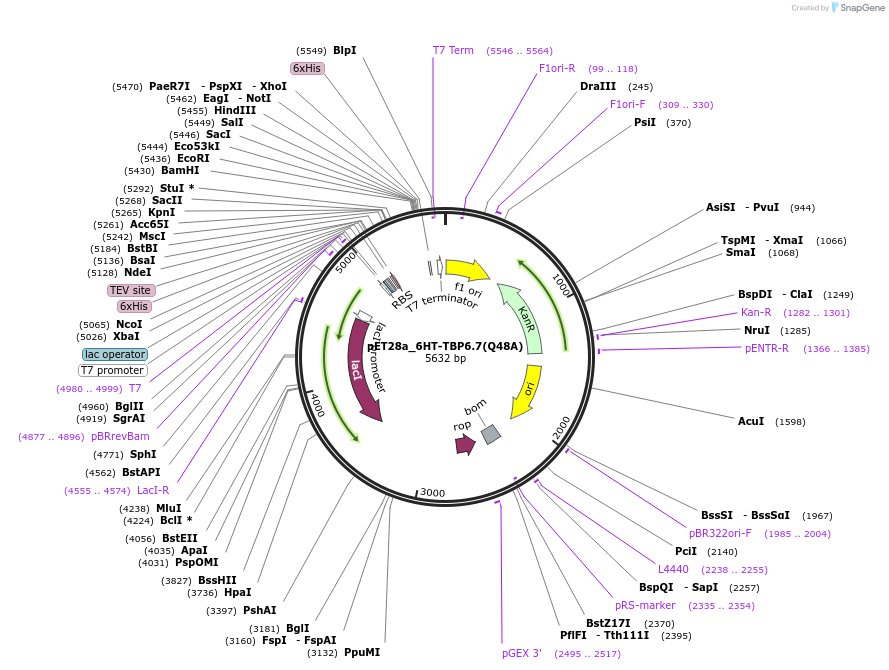
pET28a_6HT-TBP6.7(Q48A)
Plasmid#112724PurposeGln48 contacts the backbone of TAR RNA as well as mediates intramoecular interaction to stabilize the conformation of beta2-beta3 loop within TBP6.7.DepositorInsert6His-TEV-TBP6.7(Q48A)
Tags6His-TEVExpressionBacterialMutationQ48APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
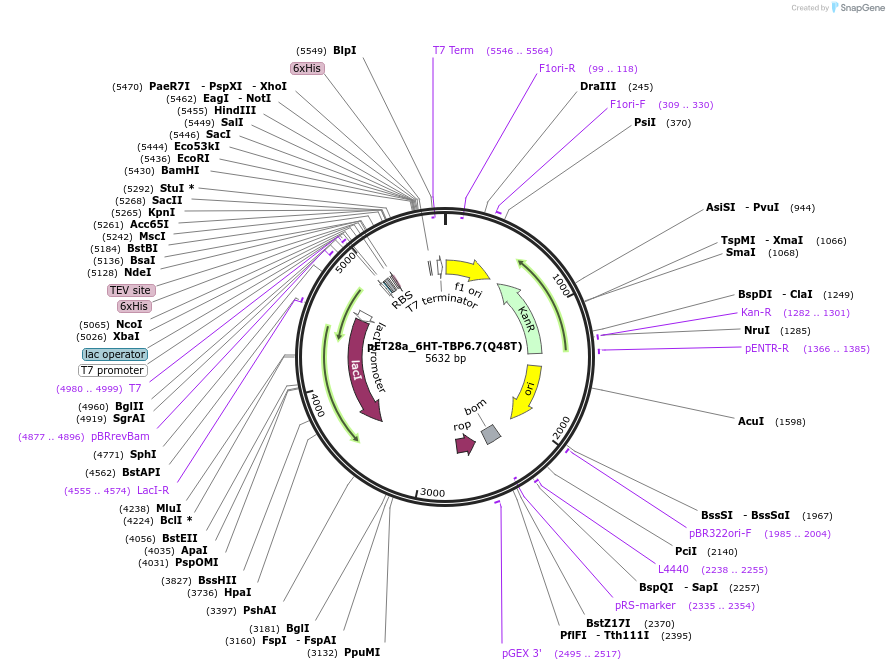
pET28a_6HT-TBP6.7(Q48T)
Plasmid#112725PurposeThis mutant of TBP6.7 binds to TAR very similarly to TBP6.7(WT), with only 1.4-fold reduced Kd.DepositorInsert6His-TEV-TBP6.7(Q48T)
Tags6His-TEVExpressionBacterialMutationQ48TPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
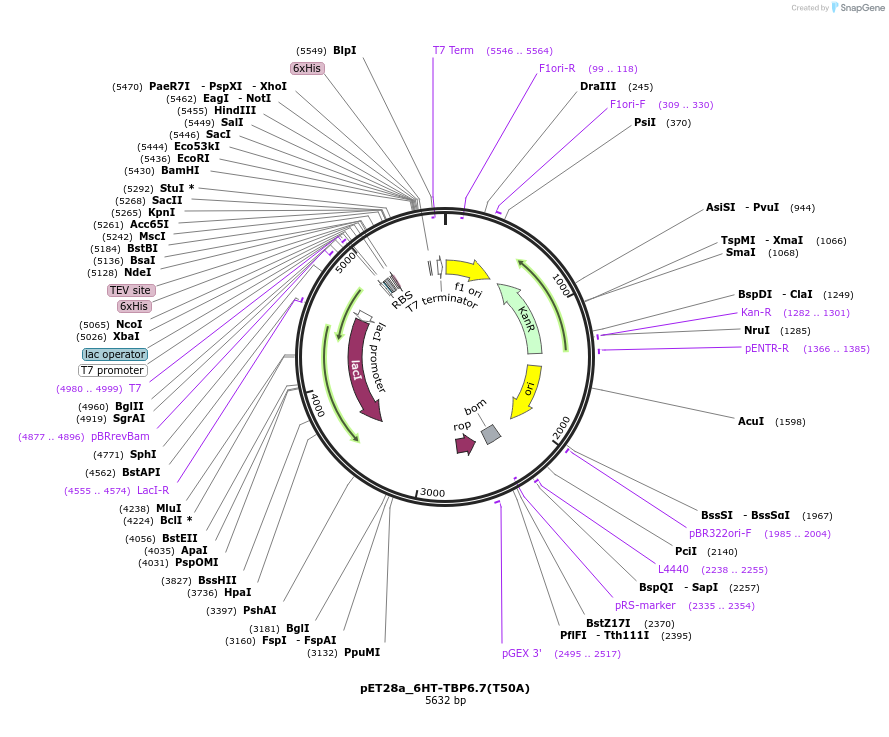
pET28a_6HT-TBP6.7(T50A)
Plasmid#112727PurposeLav-evolved amino acid T50 within TBPs is important in engaging intramolecularly to steer also lab-evolved R49 into conformation compatible binding to RNA.DepositorInsert6His-TEV-TBP6.7(T50A)
Tags6His-TEVExpressionBacterialMutationT50APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
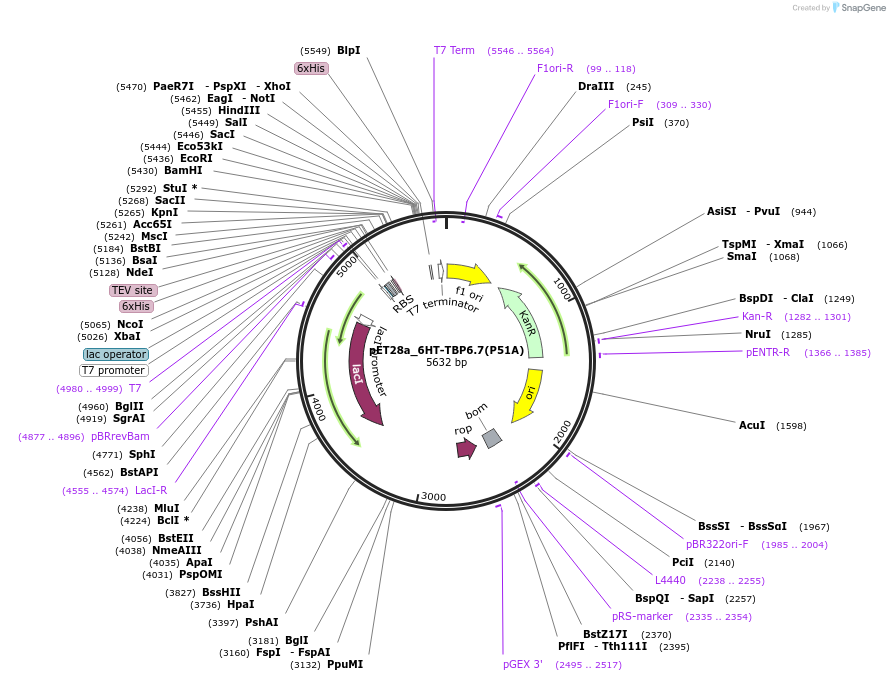
pET28a_6HT-TBP6.7(P51A)
Plasmid#112728PurposeSimilar to P46, P51 was selected and is present in all evolved TBPs. Mutating this position into Ala has a modest effect on binding.DepositorInsert6His-TEV-TBP6.7(P51A)
Tags6His-TEVExpressionBacterialMutationP51APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
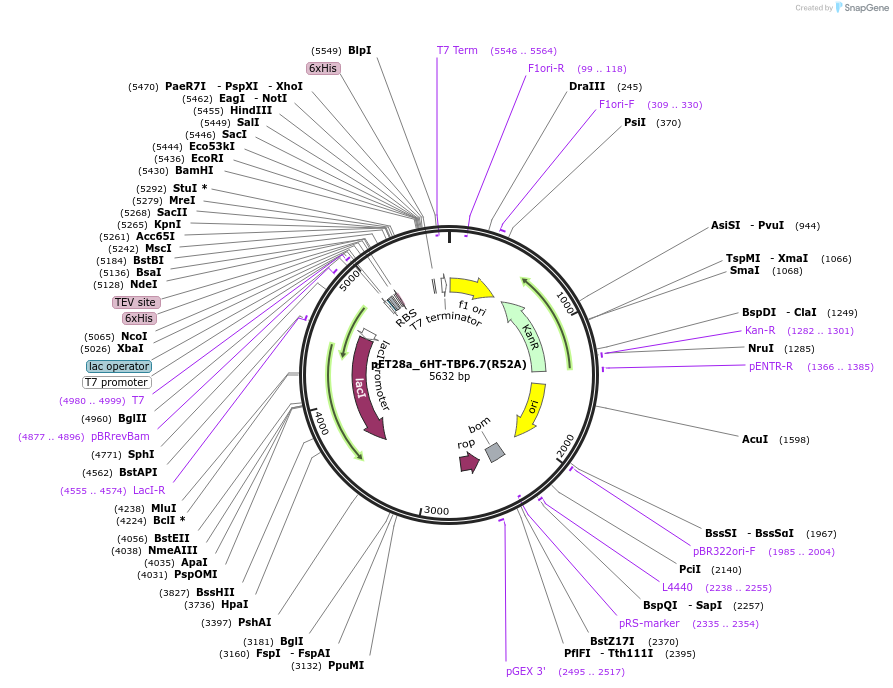
pET28a_6HT-TBP6.7(R52A)
Plasmid#112729Purpose52nd position is Arg in both wild-type U1A and TBPs, but the RNA recognition by this residue in both types of proteins is fundamentally different.DepositorInsert6His-TEV-TBP6.7(R52A)
Tags6His-TEVExpressionBacterialMutationR52APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
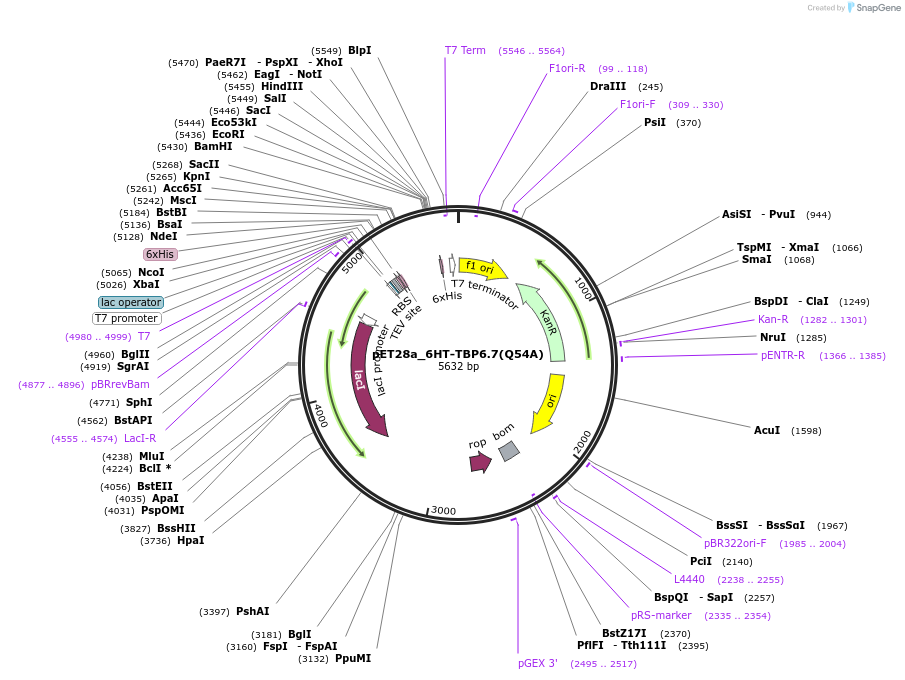
pET28a_6HT-TBP6.7(Q54A)
Plasmid#112730PurposeAlthough not evolved, this residue within TBPs participates in stabilizing the polypeptide loop responsible for RNA recognition.DepositorInsert6His-TEV-TBP6.7(Q54A)
Tags6His-TEVExpressionBacterialMutationQ54APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
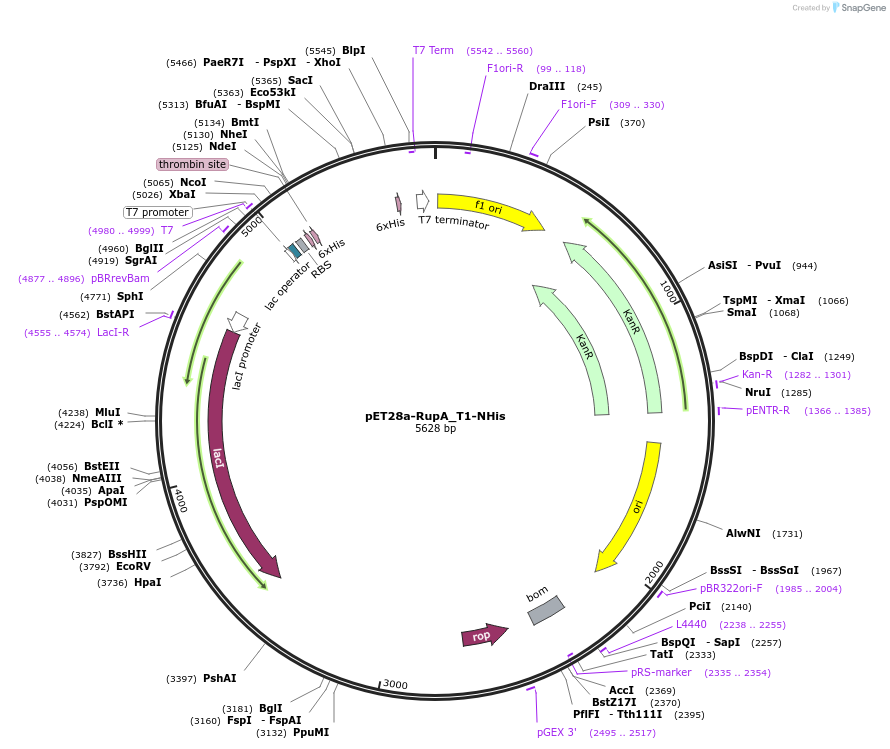
pET28a-RupA_T1-NHis
Plasmid#109259PurposeNRPS T domain with N-terminal 6His tagDepositorArticleInsertRupA_T1
TagsHis tagExpressionBacterialMutationcontains aa 917..1025 of WP_015523502.1PromoterT7Available SinceJuly 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
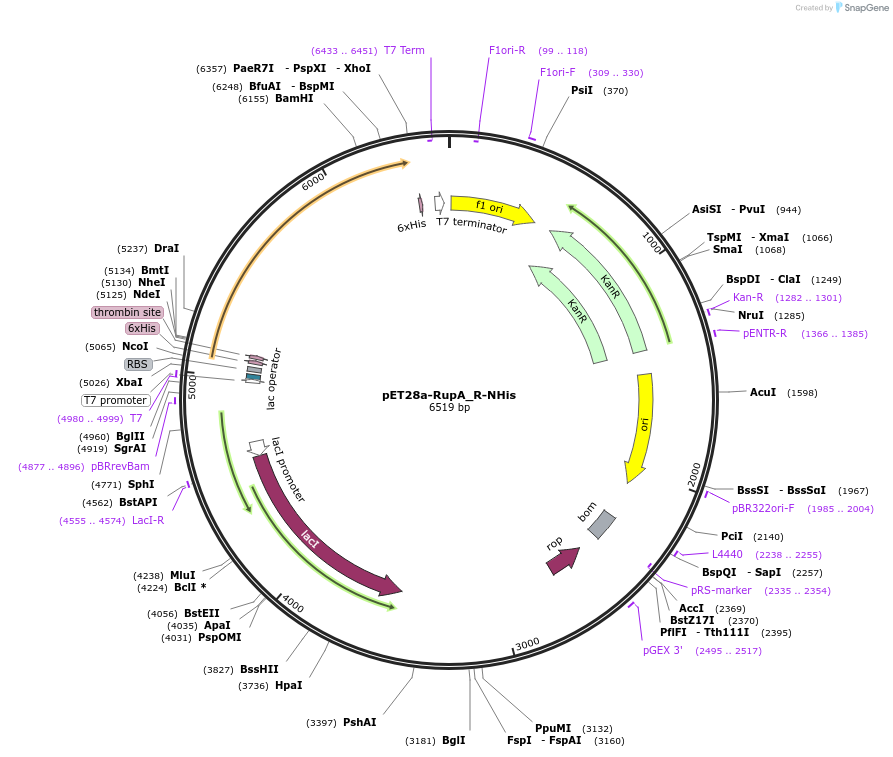
pET28a-RupA_R-NHis
Plasmid#109257PurposeNRPS R domain with N-terminal 6His tagDepositorArticleInsertRupA_R
TagsHis tagExpressionBacterialMutationcontains aa 2038..2443 of WP_015523502.1PromoterT7Available SinceJuly 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
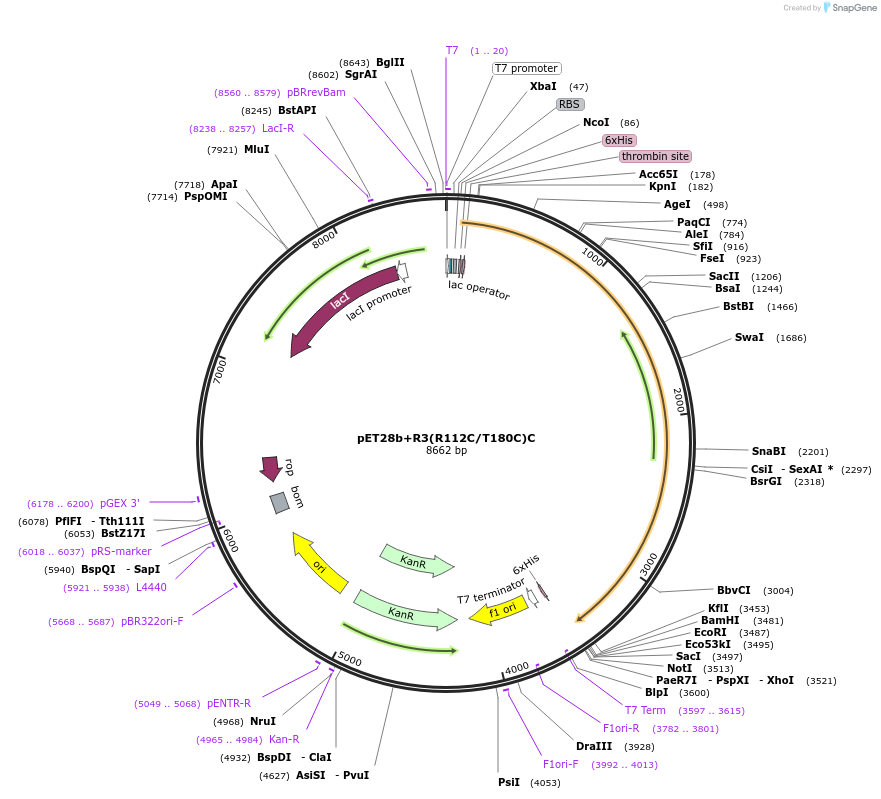
pET28b+R3(R112C/T180C)C
Plasmid#108194PurposeExpresses PdR-PCNA3(R112C/T180C)-P450cam fusion protein in E. coliDepositorInsertPdR-PCNA3(R112C/T180C)-P450cam fusion protein
TagsHis6 tagExpressionBacterialMutationR112C/T180C(PCNA3)PromoterT7 promoterAvailable SinceApril 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
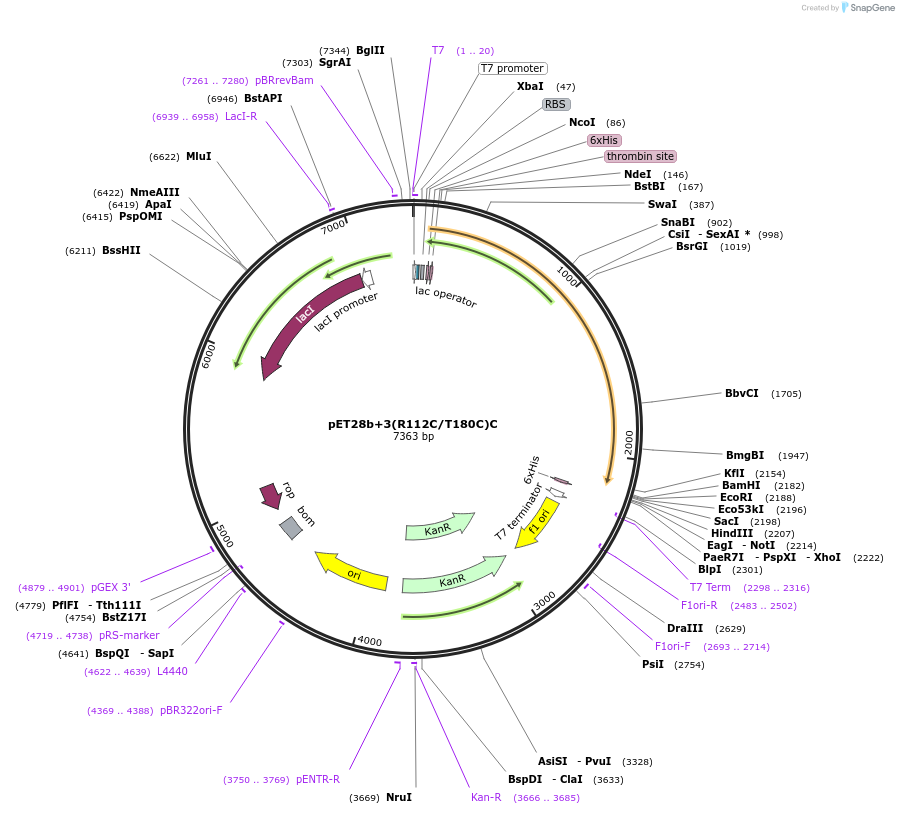
pET28b+3(R112C/T180C)C
Plasmid#108193PurposeExpresses PCNA3(R112C/T180C)-P450cam fusion protein in E. coliDepositorInsertPCNA3(R112C/T180C)-P450cam fusion protein
TagsHis6 tagExpressionBacterialMutationR112C/T180C(PCNA3)PromoterT7 promoterAvailable SinceApril 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
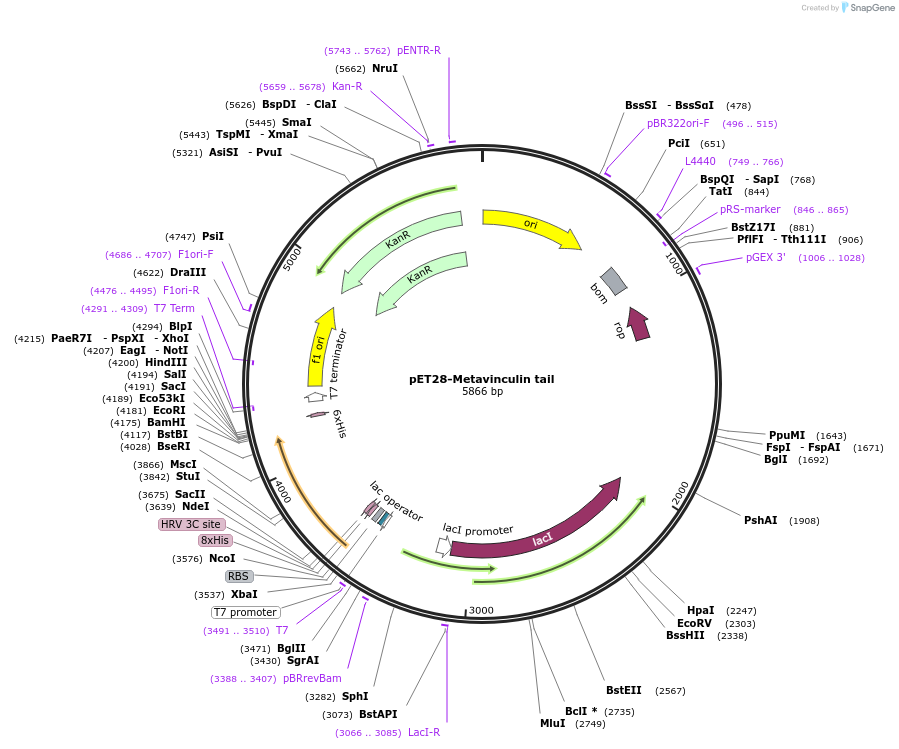
pET28-Metavinculin tail
Plasmid#107152Purposebacterial expressionDepositorAvailable SinceMarch 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
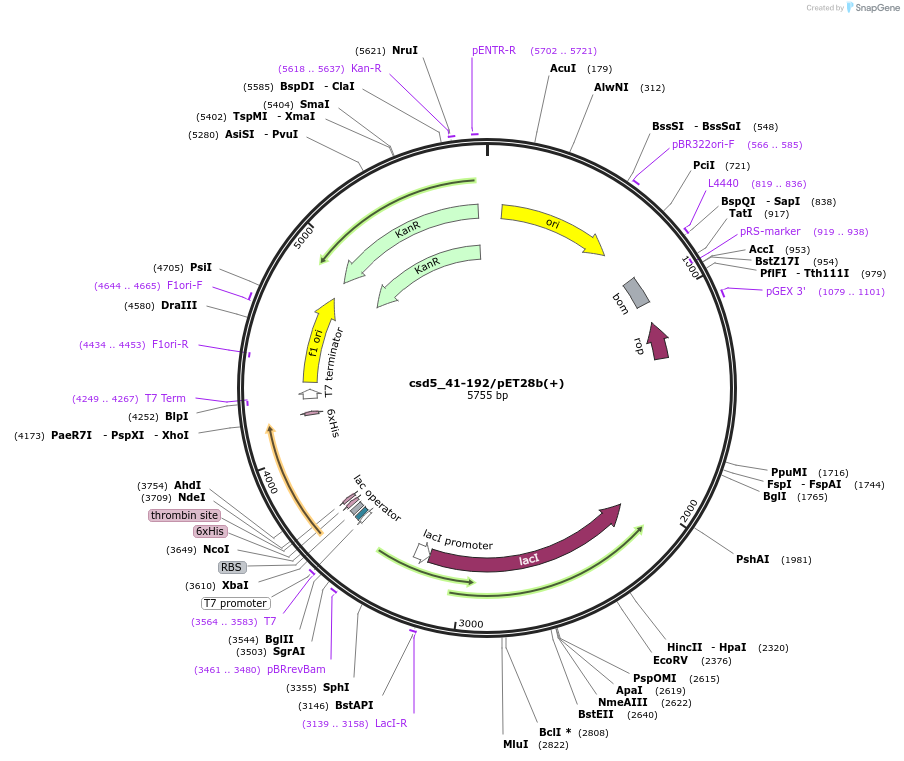
csd5_41-192/pET28b(+)
Plasmid#83296Purposecsd5, construct: 41-192, pET28b(+) N-tagDepositorInserthp1250
TagsHis tagExpressionBacterialMutationdeleted amino acid 1-40Available SinceNov. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
csd4_22-438/pET28b(+)
Plasmid#83295Purposecsd4, construct: 22-438, pET28b(+) N-tagDepositorInserthp1075
TagsHis tagExpressionBacterialMutationdeleted amino acid 1-21Available SinceNov. 3, 2016AvailabilityAcademic Institutions and Nonprofits only