We narrowed to 2,087 results for: TRIM
-
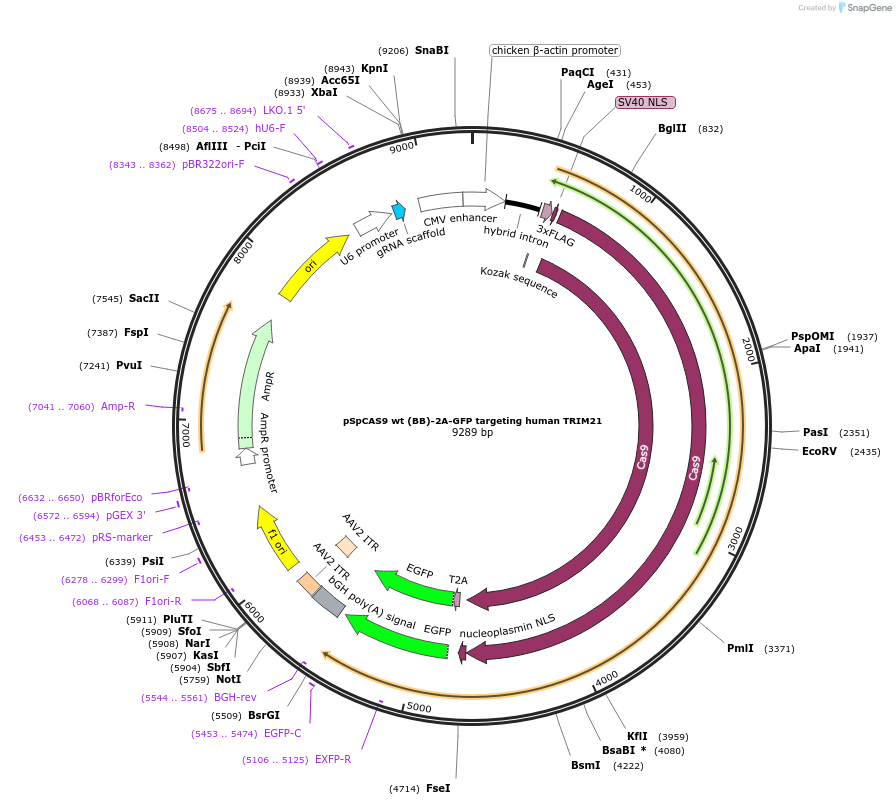
Plasmid#138295PurposegRNA for CRISPR/Cas9 knockout of human TRIM21DepositorAvailable SinceJune 22, 2022AvailabilityAcademic Institutions and Nonprofits only
-
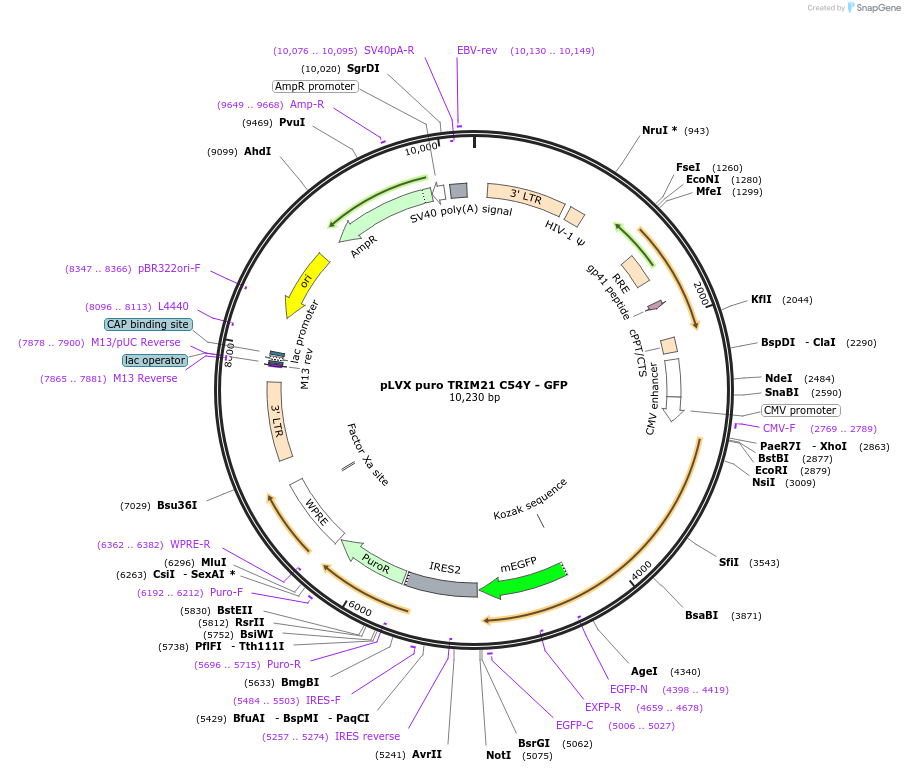
pLVX puro TRIM21 C54Y - GFP
Plasmid#116942PurposeExpression of TRIM21 C54Y -GFP fusionDepositorAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
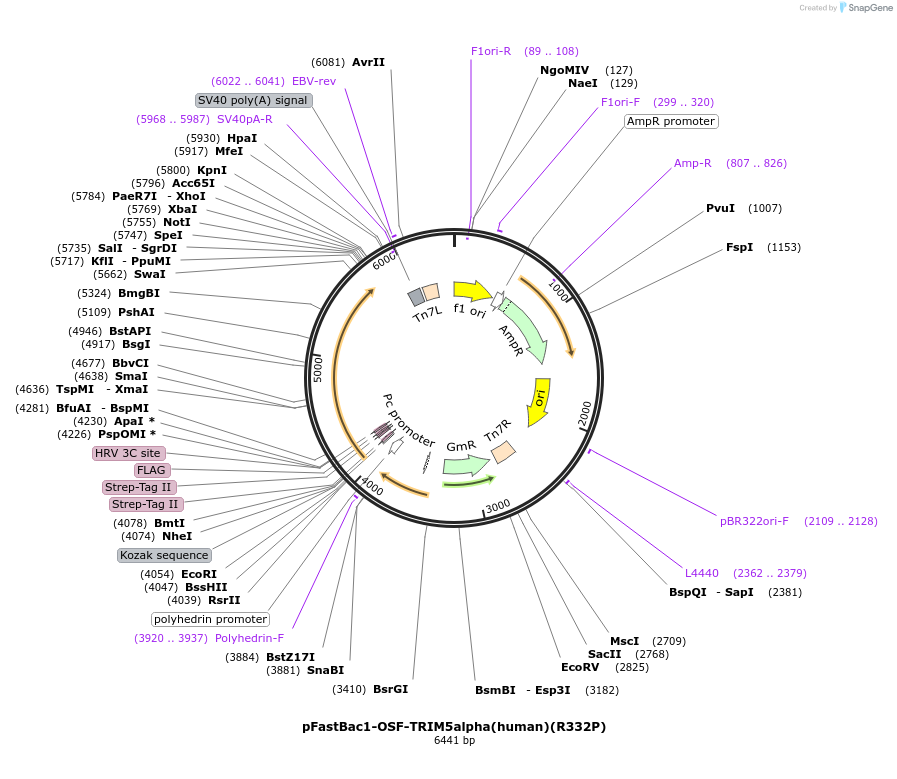
pFastBac1-OSF-TRIM5alpha(human)(R332P)
Plasmid#79037PurposeBaculoviral entry vector to produce Bac-to-bac baculovirusesDepositorInsertOSF-TRIM5alpha(human R332P) (TRIM5 Human)
TagsOneSTrEP-FLAGExpressionInsectMutationChanged Arg 332 to ProPromoterpolyhedrinAvailable SinceMarch 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
Flag-TRIM24-F979A/N980A shRescue
Plasmid#28143DepositorInsertTRIM24 (TRIM24 Human)
TagsFLAGX2ExpressionMammalianMutationA489V, F979A and N980A; aa 308 and 309 mutated fo…Available SinceFeb. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
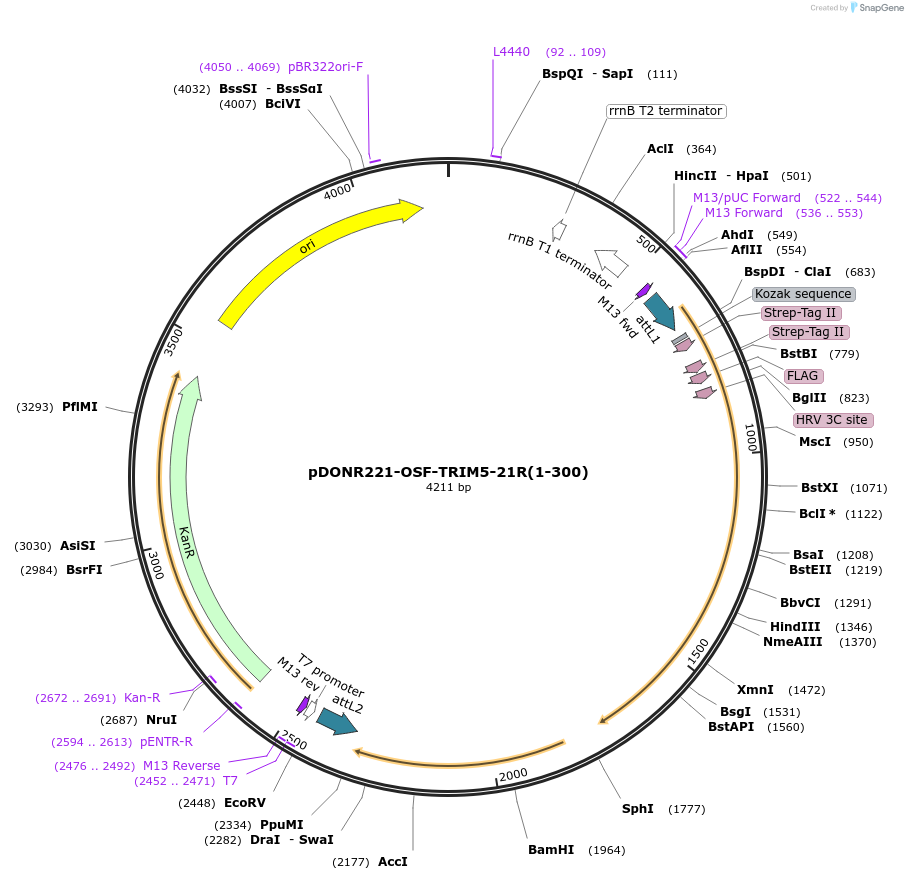
pDONR221-OSF-TRIM5-21R(1-300)
Plasmid#79029PurposeGateway entry vector for cloning and propagation in DH5alphaDepositorInsertTRIM5-21R (1-300)
TagsOneSTrEP-FLAGExpressionInsectPromoterT7Available SinceMarch 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
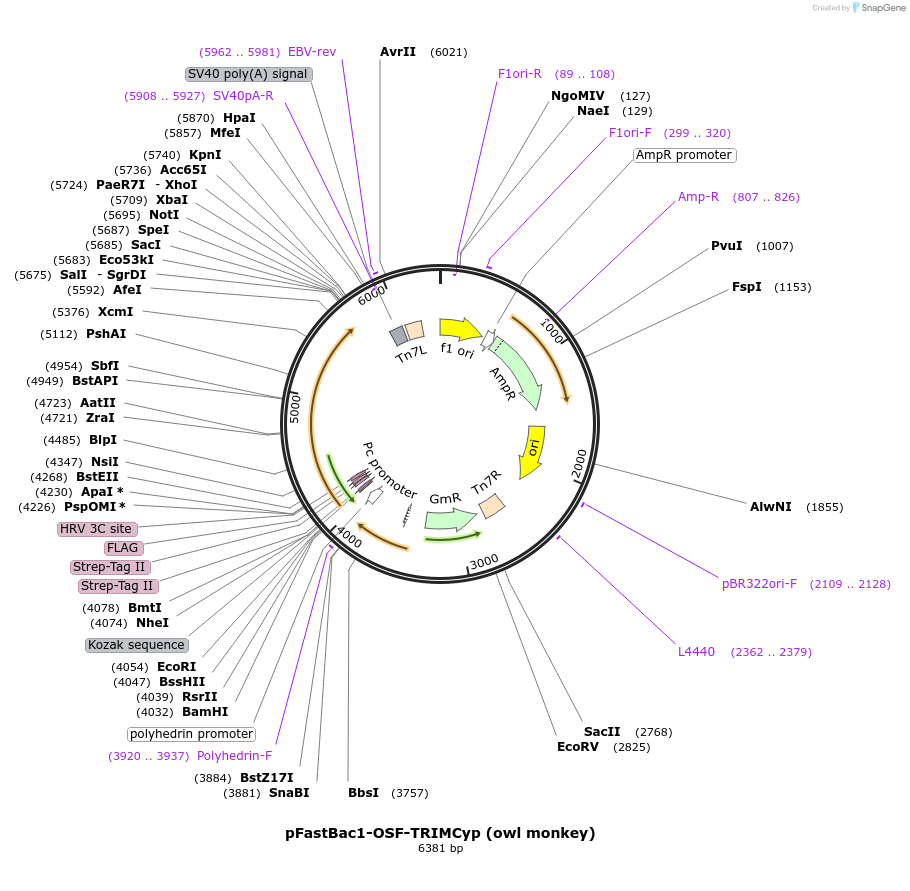
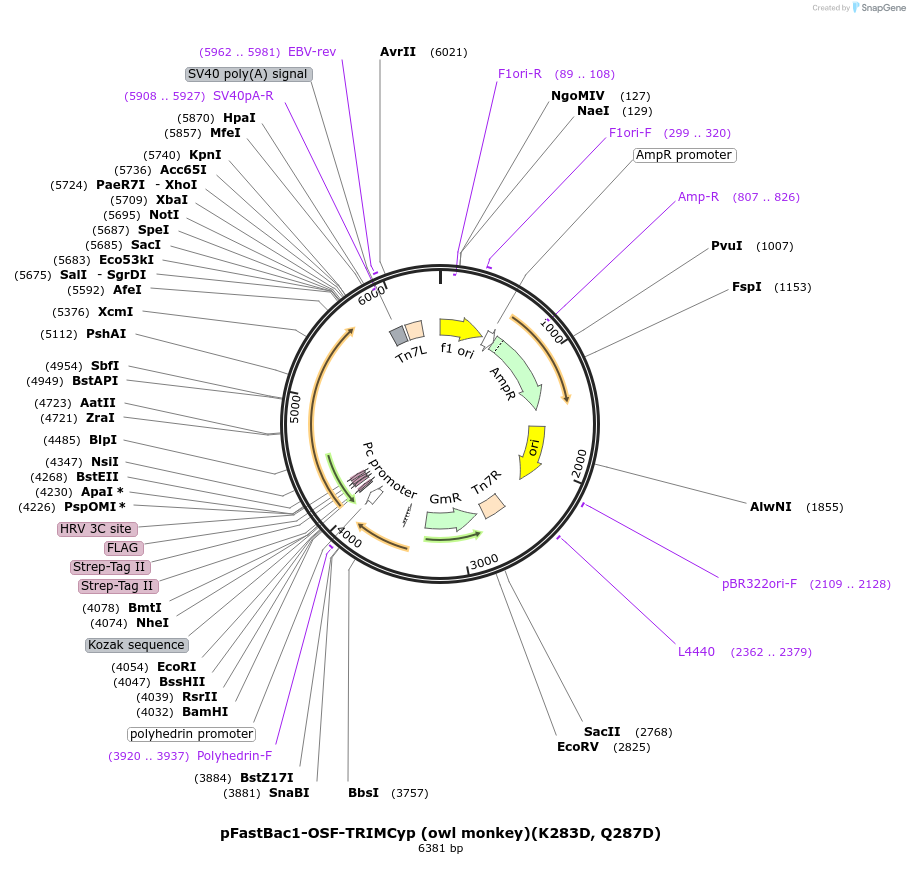
pFastBac1-OSF-TRIMCyp (owl monkey)
Plasmid#79035PurposeBaculoviral entry vector to produce Bac-to-bac baculovirusesDepositorInsertOSF-TRIM5Cyp (owl monkey)
TagsOneSTrEP-FLAGExpressionInsectPromoterpolyhedrinAvailable SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
Flag-TRIM24-C840W/F869D/F979A/N980A
Plasmid#28144DepositorAvailable SinceJune 6, 2011AvailabilityAcademic Institutions and Nonprofits only -
Flag-TRIM24-F869D/F979A/N980A shRescue
Plasmid#28145DepositorInsertTRIM24 (TRIM24 Human)
TagsFLAGX2ExpressionMammalianMutationF869D, F979A, N980A and aa 308 and 309 mutated fo…Available SinceJune 6, 2011AvailabilityAcademic Institutions and Nonprofits only -
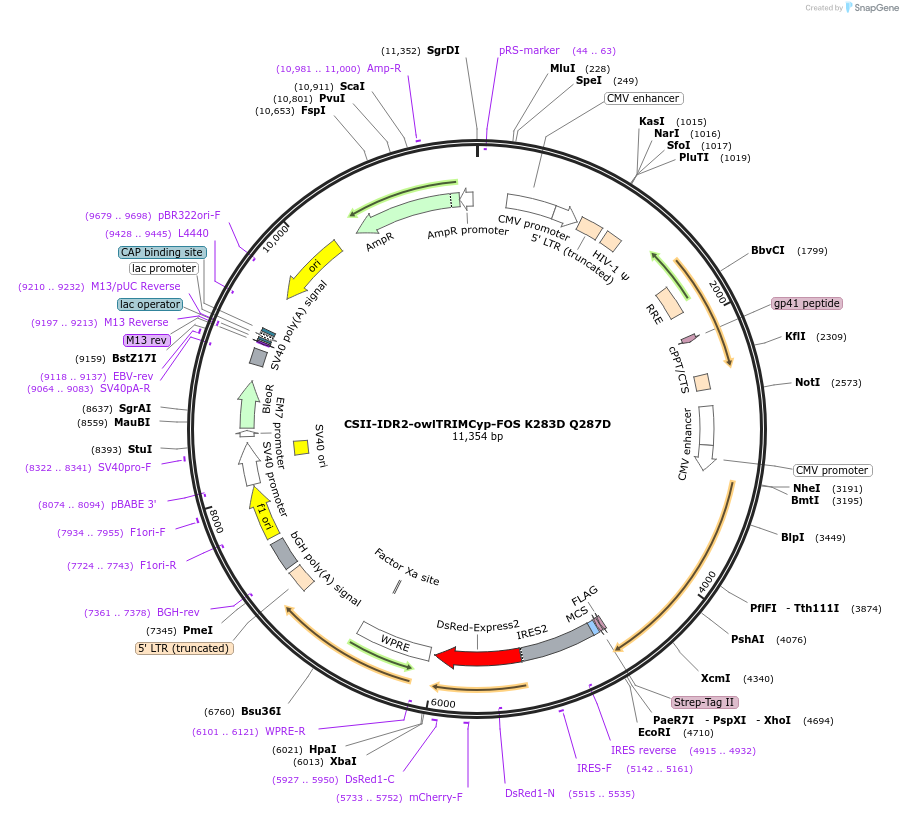
CSII-IDR2-owlTRIMCyp-FOS K283D Q287D
Plasmid#79068PurposeExpress owl monkey TRIMCyp K283D, Q287D in mammalan cellsDepositorInsertTRIM5Cyp (owl monkey) (K283D, Q287D)-FOS
TagsOneSTrEP-FLAGExpressionMammalianMutationChanged Lys 283 to Asp and Gln 287 to AspPromoterCMVAvailable SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
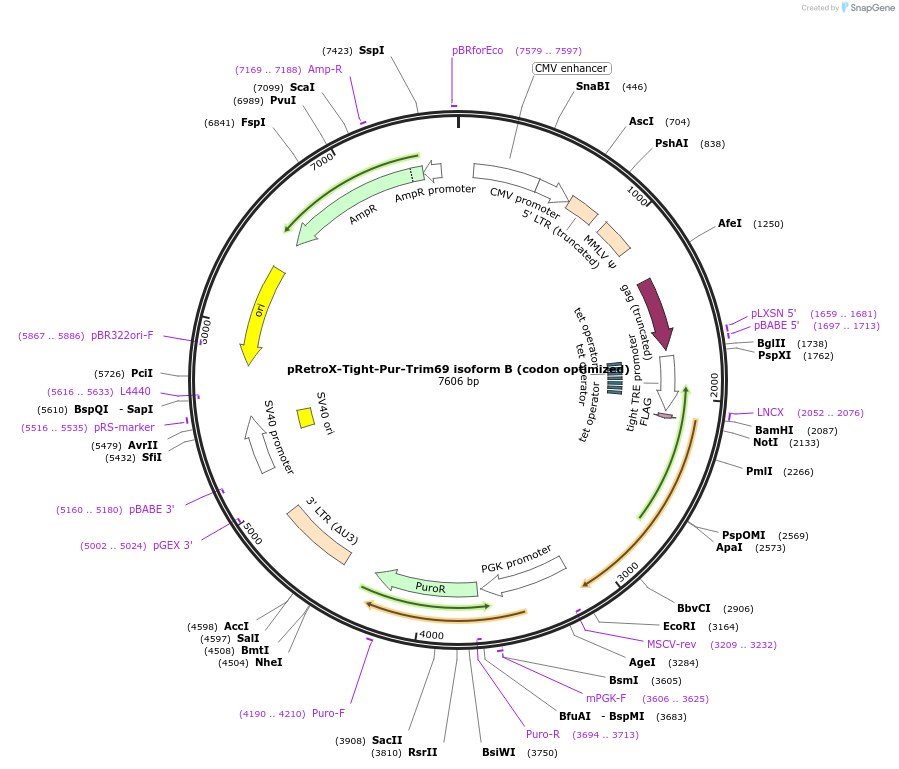
pRetroX-Tight-Pur-Trim69 isoform B (codon optimized)
Plasmid#199567PurposeStable and dox. inducible expression of Trim69 isoform B (codon optimized) after retroviral-mediated gene transferDepositorInsertTrim69 isoform B (codon optimized) (TRIM69 Human)
UseRetroviral; Allows for puromycin selectionTagsFlagMutationcodon optimizedAvailable SinceMay 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
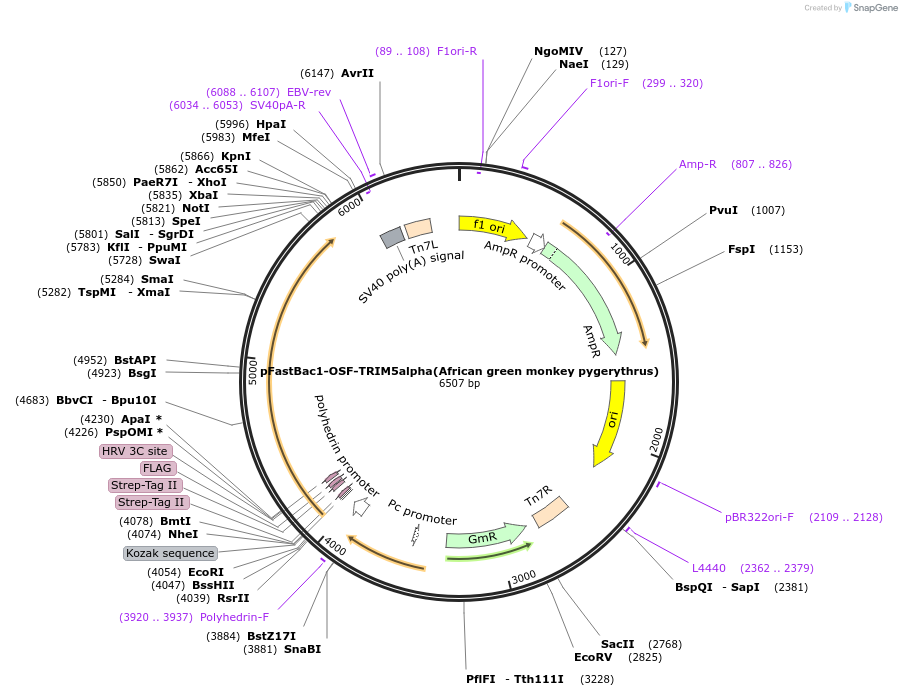
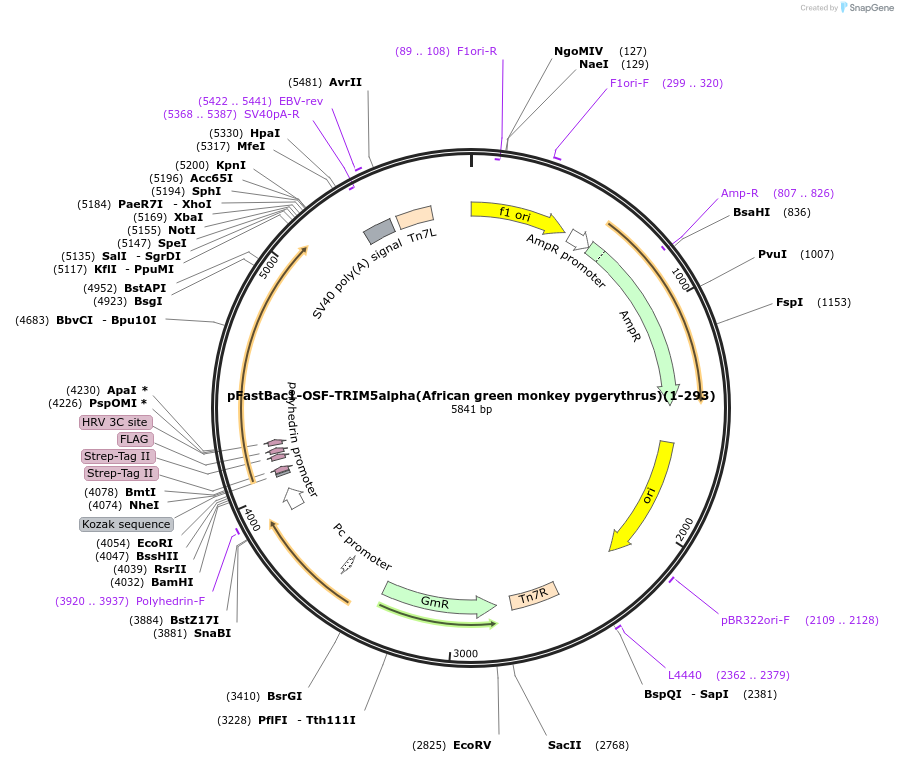
pFastBac1-OSF-TRIM5alpha(African green monkey pygerythrus)
Plasmid#79031PurposeBaculoviral entry vector to produce Bac-to-bac baculovirusesDepositorInsertOSF-TRIM5alpha(African green monkey pygerythrus)
TagsOneSTrEP-FLAGExpressionInsectPromoterpolyhedrinAvailable SinceMarch 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pFastBac1-OSF-TRIMCyp (owl monkey)(K283D, Q287D)
Plasmid#79036PurposeBaculoviral entry vector to produce Bac-to-bac baculovirusesDepositorInsertOSF-TRIM5Cyp (owl monkey) (K283D, Q287D)
TagsOneSTrEP-FLAGExpressionInsectMutationChanged Lys 283 to Asp and Gln 287 to AspPromoterpolyhedrinAvailable SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
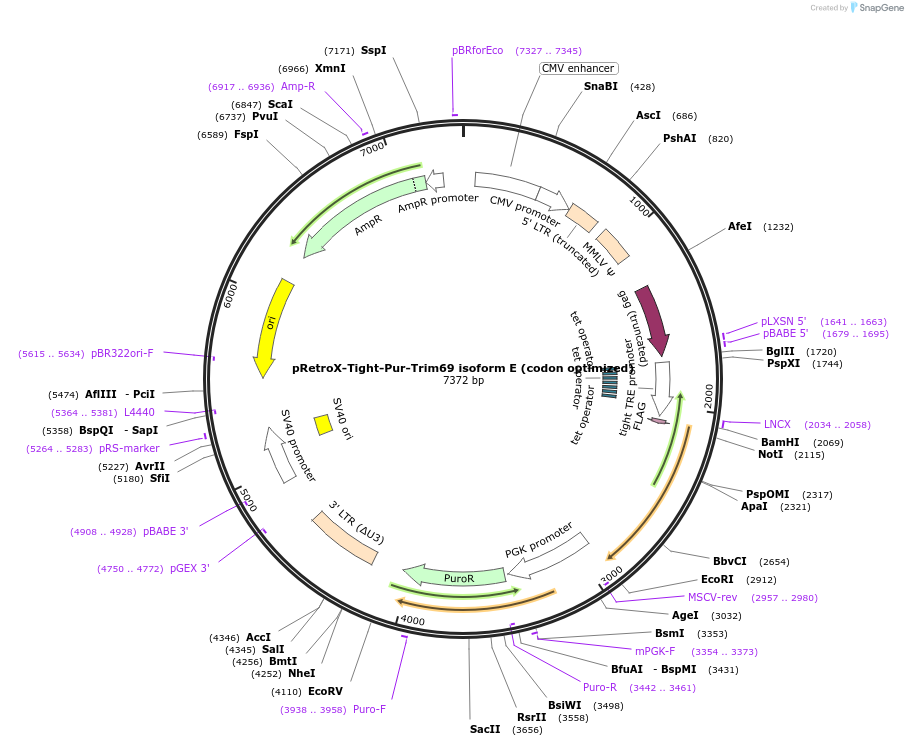
pRetroX-Tight-Pur-Trim69 isoform E (codon optimized)
Plasmid#199570PurposeStable and dox. inducible expression of Trim69 isoform E (codon optimized) after retroviral-mediated gene transferDepositorInsertTrim69 isoform E (codon optimized) (TRIM69 Human)
UseRetroviral; Allows for puromycin selectionTagsFlagMutationcodon optimizedAvailable SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
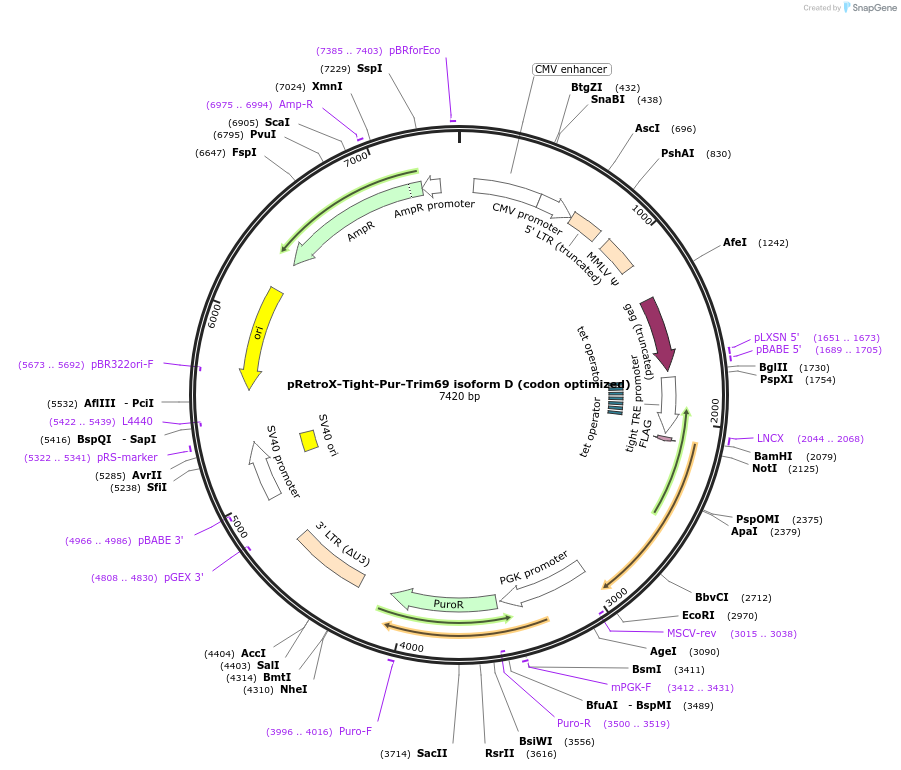
pRetroX-Tight-Pur-Trim69 isoform D (codon optimized)
Plasmid#199569PurposeStable and dox. inducible expression of Trim69 isoform D (codon optimized) after retroviral-mediated gene transferDepositorInsertTrim69 isoform D (codon optimized) (TRIM69 Human)
UseRetroviral; Allows for puromycin selectionTagsFlagMutationcodon optimizedAvailable SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
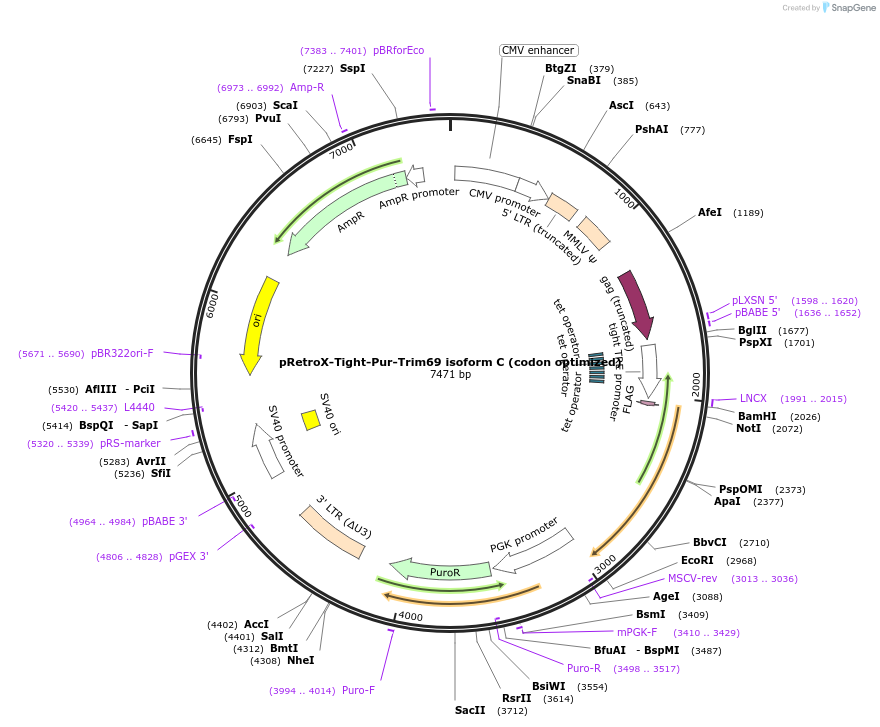
pRetroX-Tight-Pur-Trim69 isoform C (codon optimized)
Plasmid#199568PurposeStable and dox. inducible expression of Trim69 isoform C (codon optimized) after retroviral-mediated gene transferDepositorInsertTrim69 isoform C (codon optimized) (TRIM69 Human)
UseRetroviral; Allows for puromycin selectionTagsFlagMutationcodon optimizedAvailable SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
pFastBac1-OSF-TRIM5alpha(African green monkey pygerythrus)(1-293)
Plasmid#79030PurposeBaculoviral entry vector to produce Bac-to-bac baculovirusesDepositorInsertOSF-TRIM5alpha(African green monkey pygerythrus)(1-293)
TagsOneSTrEP-FLAGExpressionInsectMutationdeleted amino acid 294-515 from frican green monk…PromoterpolyhedrinAvailable SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
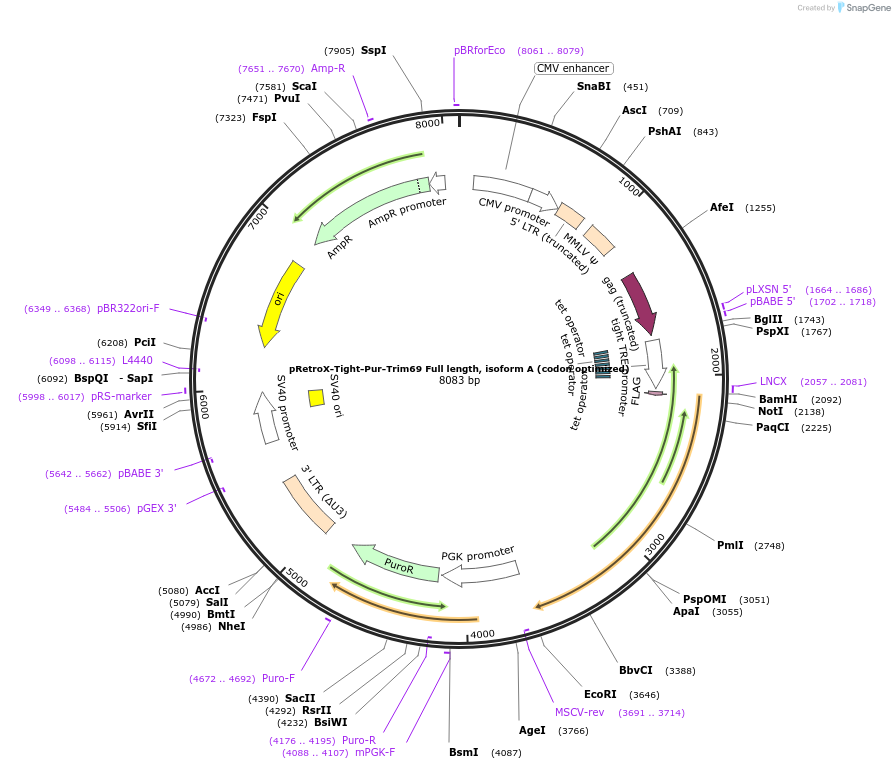
pRetroX-Tight-Pur-Trim69 Full length, isoform A (codon optimized)
Plasmid#199566PurposeStable and dox. inducible expression of Trim69 Full length, isoform A (codon optimized) after retroviral-mediated gene transferDepositorInsertTrim69 Full length, isoform A (codon optimized) (TRIM69 Human)
UseRetroviral; Allows for puromycin selectionTagsFlagMutationcodon optimizedAvailable SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
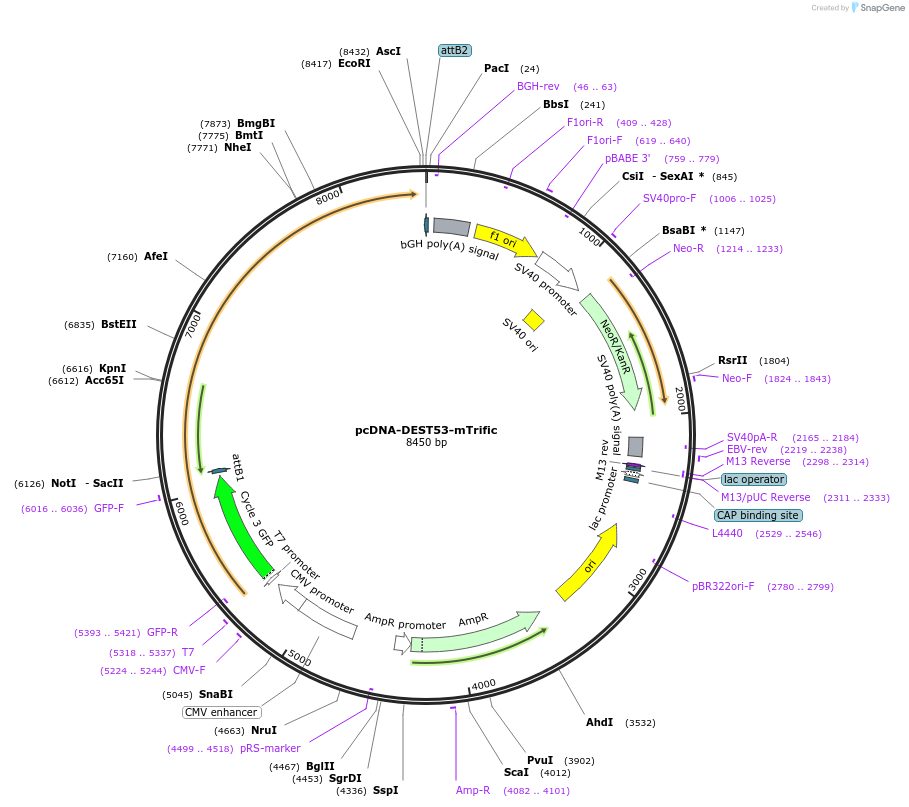
pcDNA-DEST53-mTrific
Plasmid#51031Purposemammalian expression of mouse Trific fused to GFPDepositorAvailable SinceFeb. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
pAS-1 (perf)
Plasmid#40836PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–perf
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-2 (mm21)
Plasmid#40837PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-13
Plasmid#40848PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–Bartel
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-EGFP-4xlet-7 (pAW-AS27)
Plasmid#40834PurposemiRNA sensor used in Drosophila cell linesDepositorInsertFour target sites perfectly complementary to let-7
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-GFP-2xmiR-34 (pAW-AS-25)
Plasmid#40833PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-34
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAW-EGFP-2xmiR-1 (pAW-AS-21)
Plasmid#40832PurposemiRNA sensor used in Drosophila cell linesDepositorInsertTwo target sites perfectly complementary to miR-1
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-8 (seed1-8)
Plasmid#40843PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–seed1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-7 (mm12-21)
Plasmid#40842PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm12-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-6 (mm17-21)
Plasmid#40841PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm17-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-5 (mm18-21)
Plasmid#40840PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm18-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-4 (mm19-21)
Plasmid#40839PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm19-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-3 (mm20-21)
Plasmid#40838PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm20-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-16 (mm14-21)
Plasmid#40851PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm14-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-15 (mm15-21)
Plasmid#40850PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm15-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS14 (mm16-21)
Plasmid#40849PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm16-21
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-12 (mm1-8)
Plasmid#40847PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm1-8
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-11 (bulge 9-15)
Plasmid#40846PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge9-15
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-10 (bulge 9-13)
Plasmid#40845PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-13
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-9 (bulge 9-11)
Plasmid#40844PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-11
UseMirna reporterTagsEGFPExpressionInsectPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
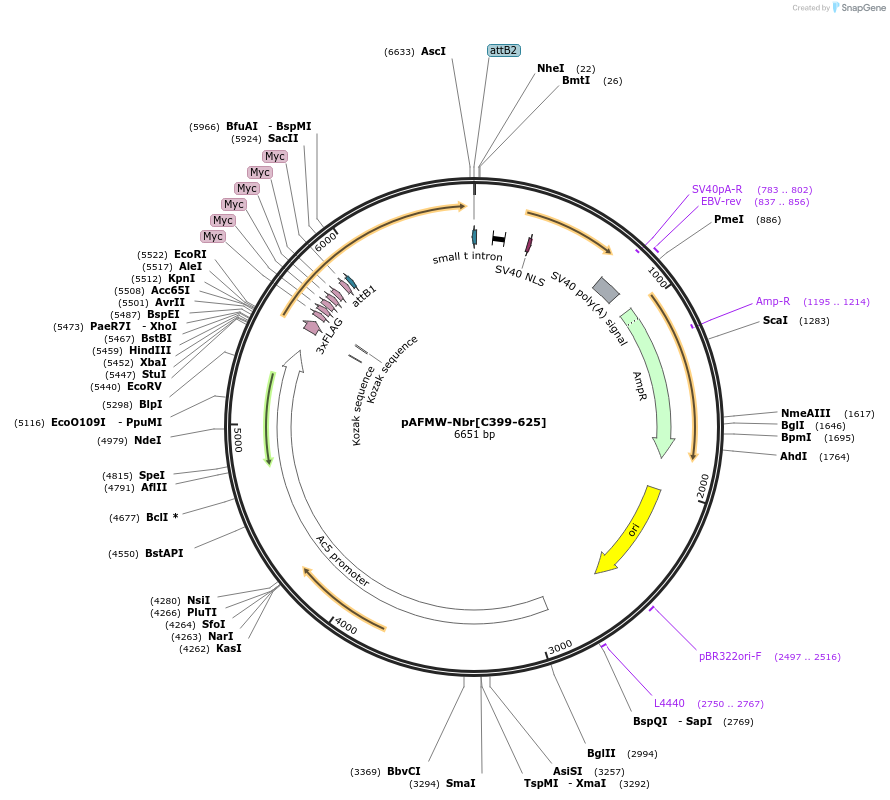
pAFMW-Nbr[C399-625]
Plasmid#188587PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorInsertNibbler C-terminal domain
TagsFLAG-MycExpressionInsectAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
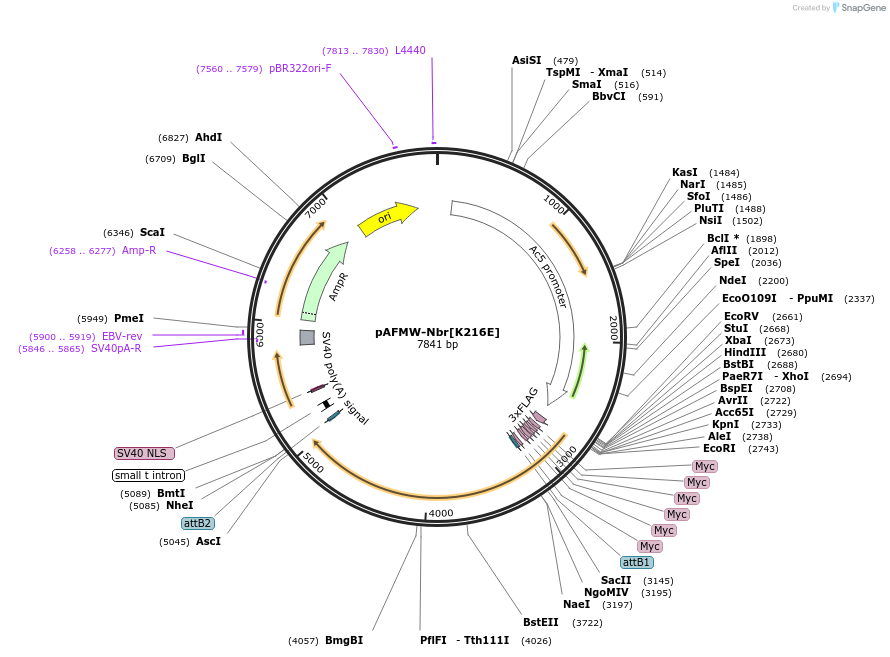
pAFMW-Nbr[K216E]
Plasmid#188600PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
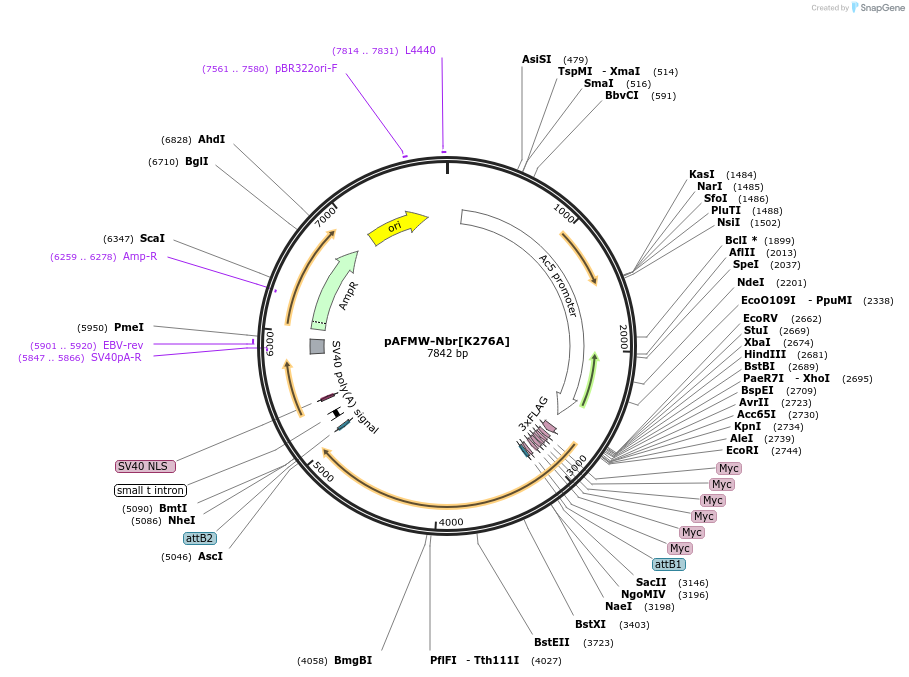
pAFMW-Nbr[K276A]
Plasmid#188597PurposeFlag-tagged Nibbler variants expression in Drosophila S2 cellsDepositorAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only