We narrowed to 4,622 results for: TIL;
-
Plasmid#17518DepositorInsertB. subtilis DEAD-box helicase YxiN
Tagsintein-chitin binding domainExpressionBacterialMutationE152Q mutationAvailable SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
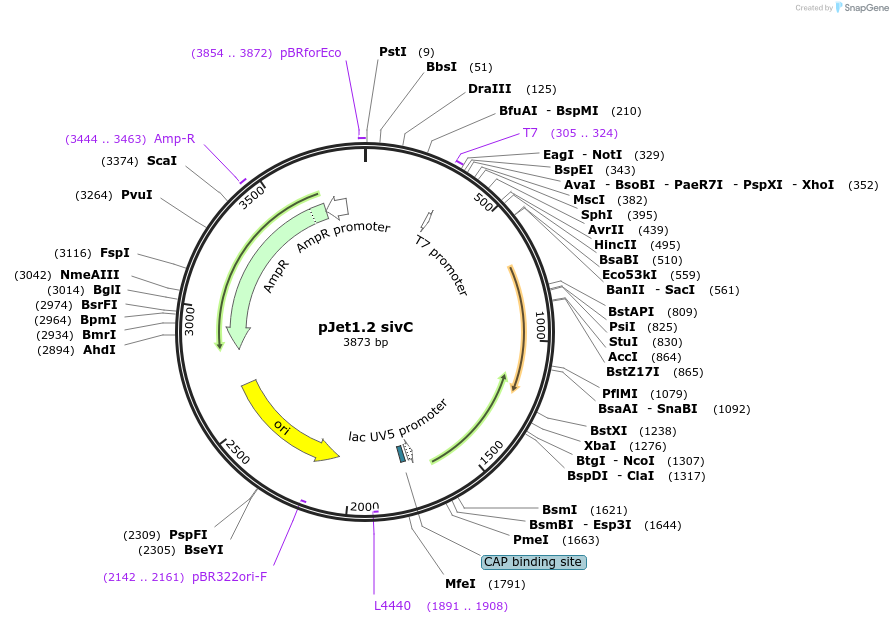
pJet1.2 sivC
Plasmid#117131Purposesmc downstream homology regionDepositorInsertsivC
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
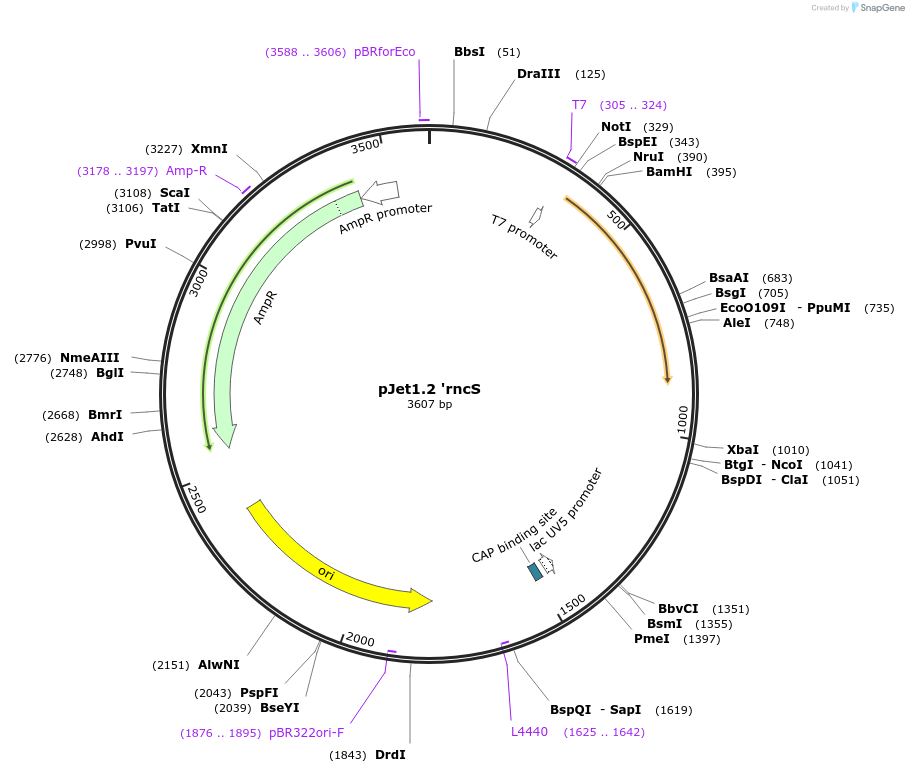
pJet1.2 'rncS
Plasmid#117129Purposesmc gene upstream homology regionDepositorInsert'rncS
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a207-479.wt
Plasmid#17516DepositorInsertB. subtilis DEAD-box helicase YxiN second domain
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 1-206Available SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
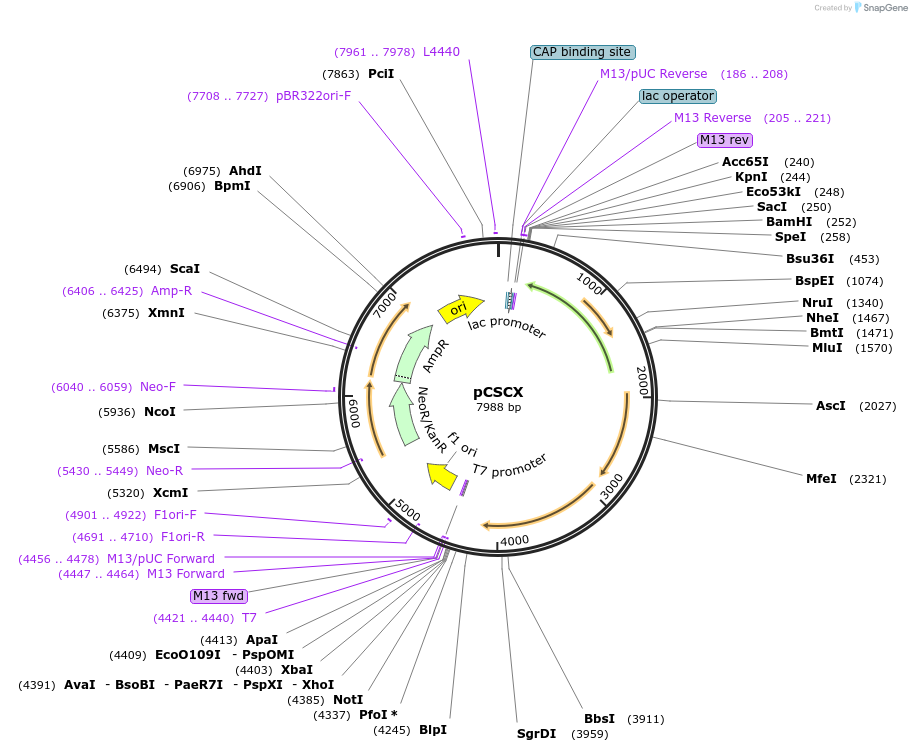
pCSCX
Plasmid#63918PurposeExpression of the sucrose utilization phenotypeDepositorInsertcscAKB
UseBacterial cloning vectorAvailable SinceMay 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
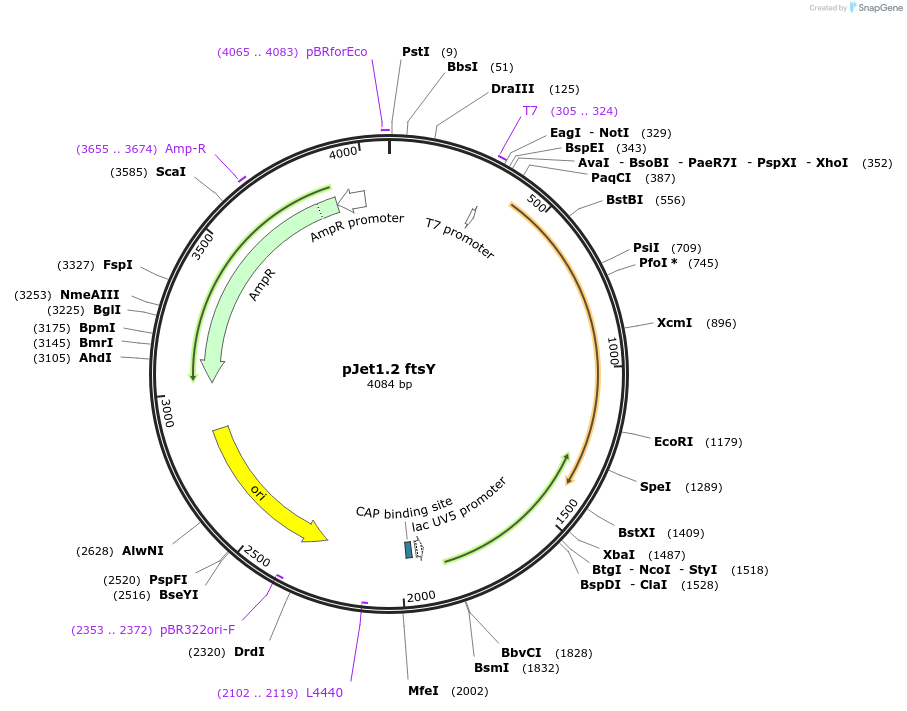
pJet1.2 ftsY
Plasmid#117130PurposeFtsY gene cassetteDepositorInsertftsY
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a1-368.E152Q
Plasmid#17517DepositorInsertB. subtilis DEAD-box helicase YxiN first two domains
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 369-479; E152Q mutat…Available SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
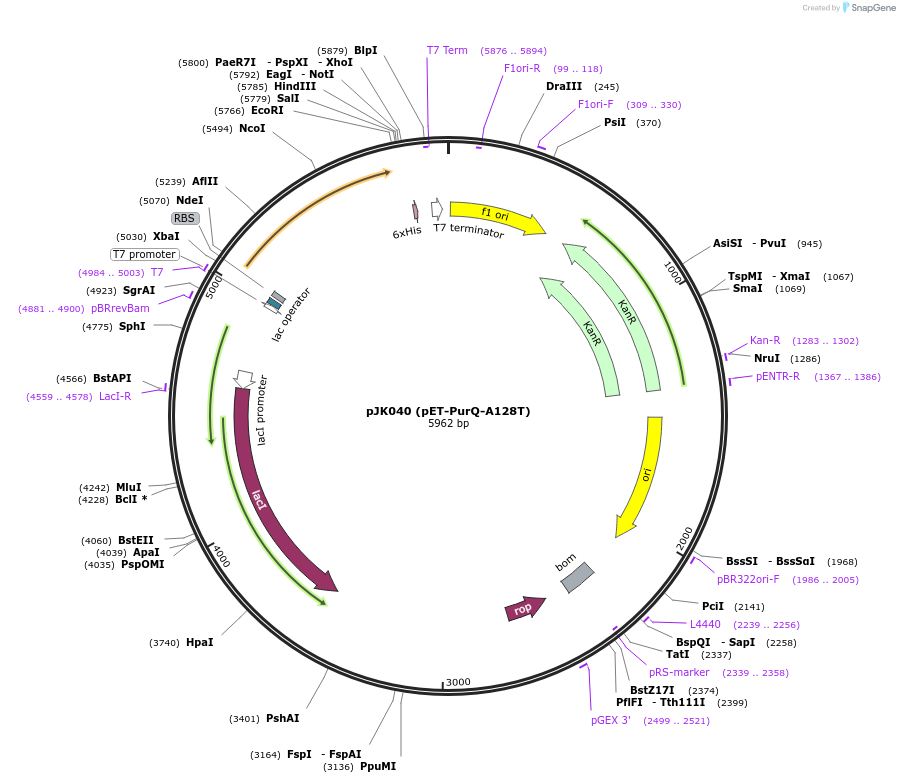
pJK040 (pET-PurQ-A128T)
Plasmid#73840PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, A128T mutant (PurQ-A128T)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialMutationA128T mutantPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
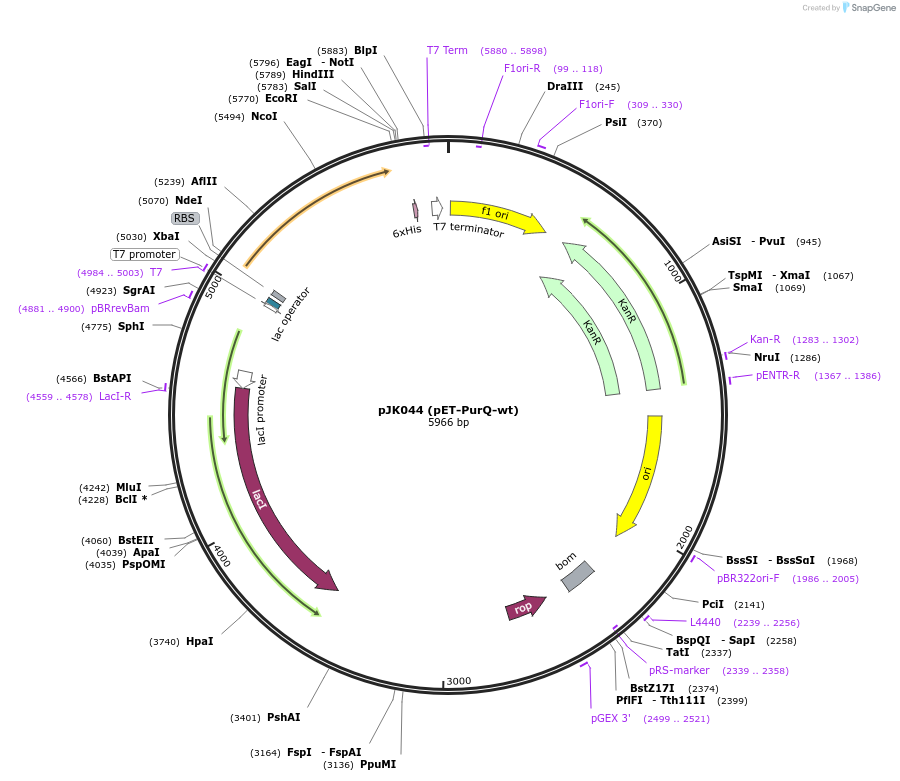
pJK044 (pET-PurQ-wt)
Plasmid#73839PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, (PurQ)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
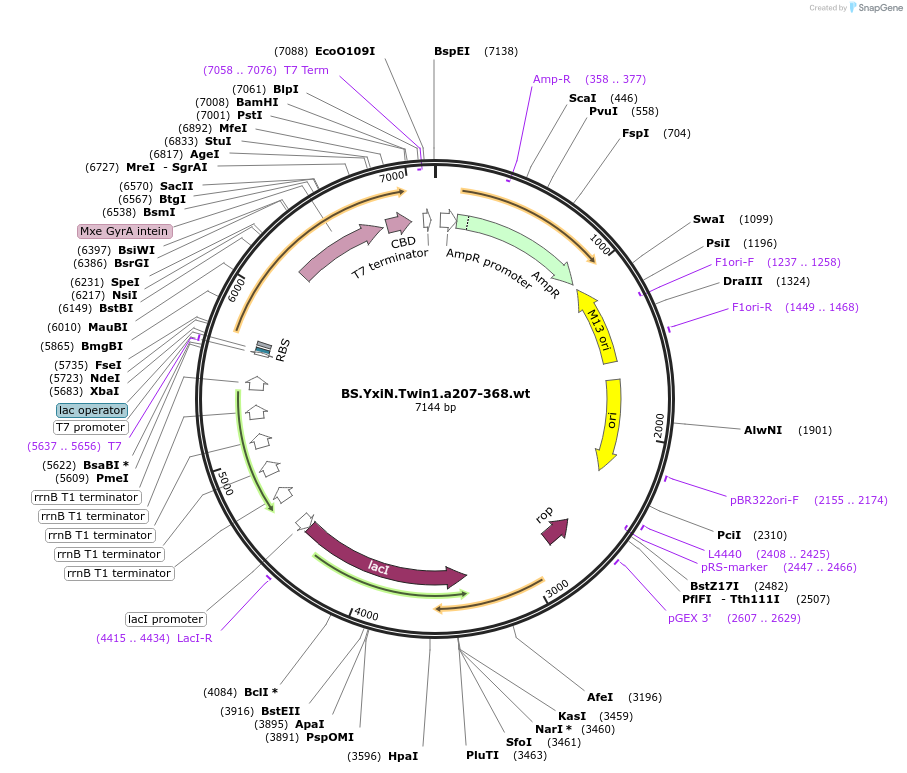
BS.YxiN.Twin1.a207-368.wt
Plasmid#13531DepositorInsertB. subtilis DEAD-box helicase YxiN second domain
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 1-206 and 369-479Available SinceJan. 4, 2007AvailabilityAcademic Institutions and Nonprofits only -
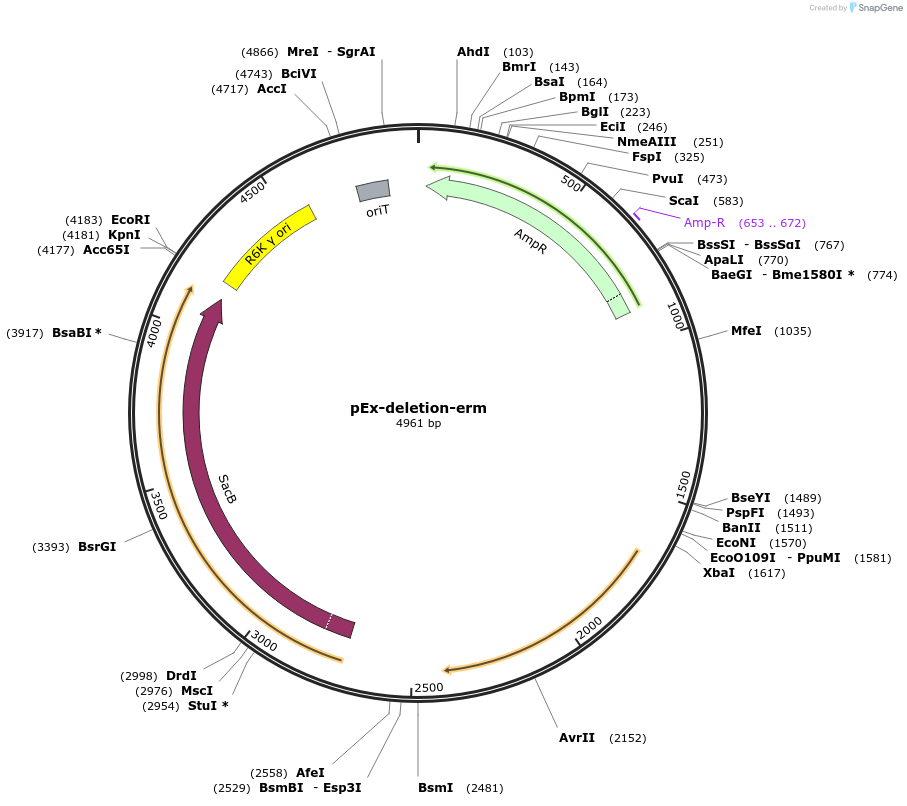
pEx-deletion-erm
Plasmid#172213PurposeFor allelic exchange in erythromycin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseAllelic exchange in prevotella copriAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
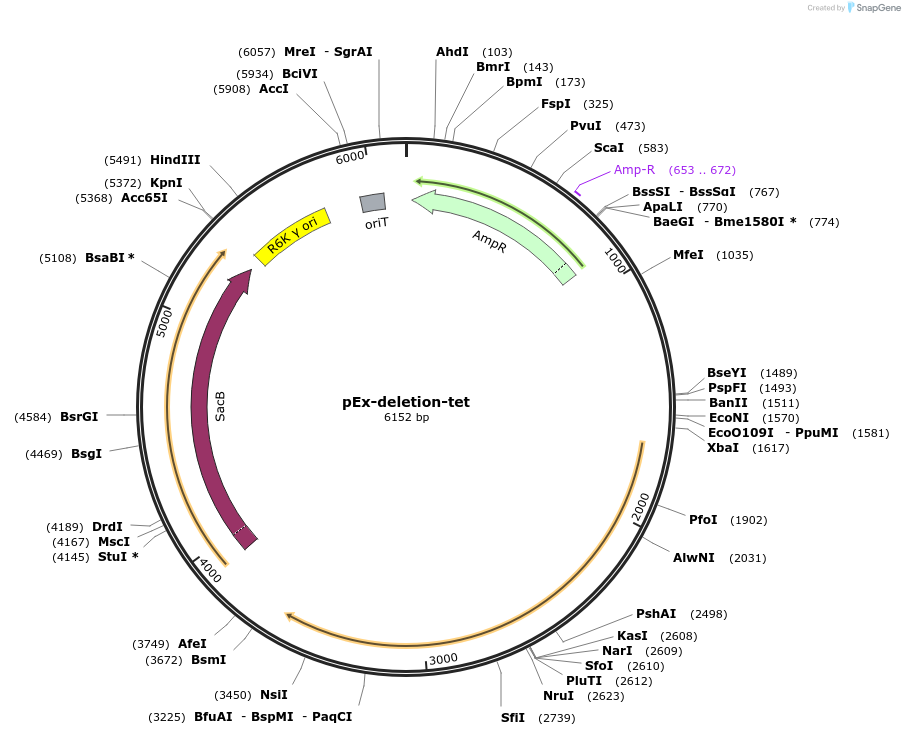
pEx-deletion-tet
Plasmid#172214PurposeFor allelic exchange in tetracyclin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseAllelic exchange in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
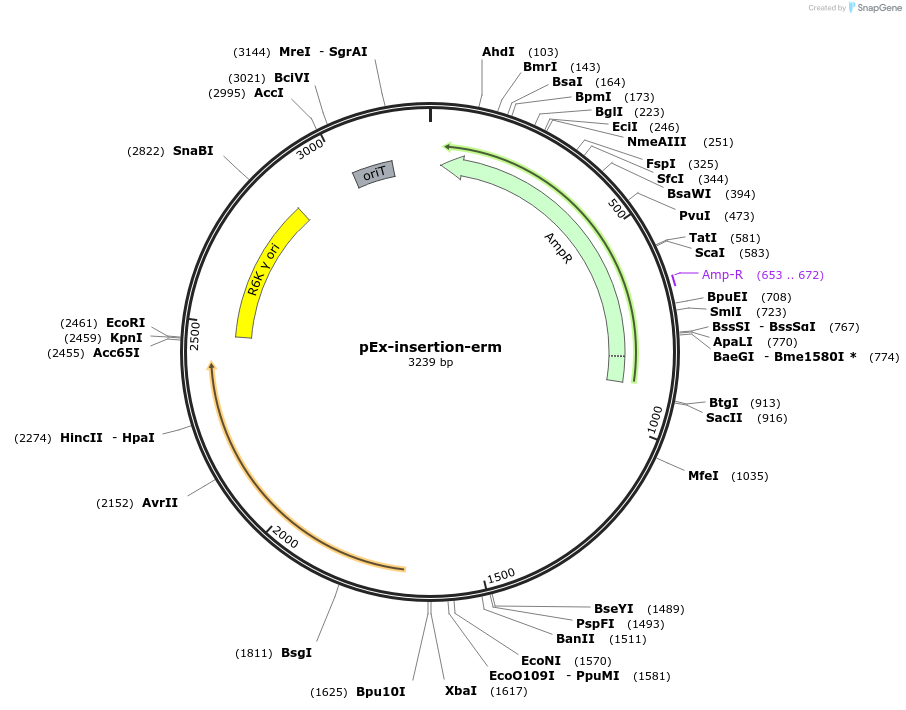
pEx-insertion-erm
Plasmid#172210PurposeFor genetic insertion in erythromycin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
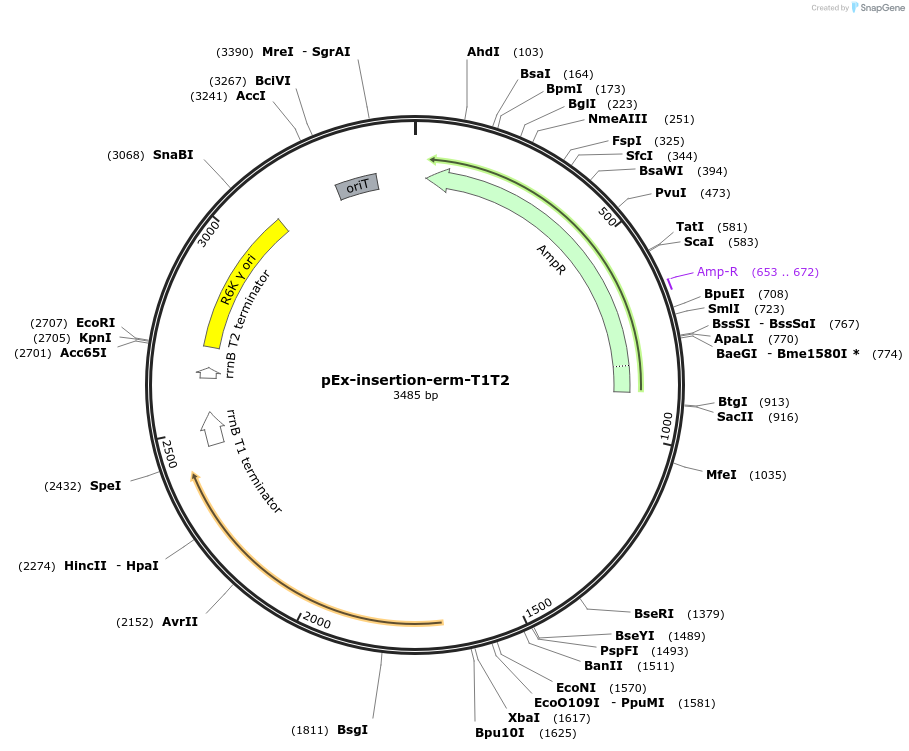
pEx-insertion-erm-T1T2
Plasmid#172212PurposeFor genetic insertion in erythromycin-sensitive Prevotella copri strains, with T1-T2 terminatorDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
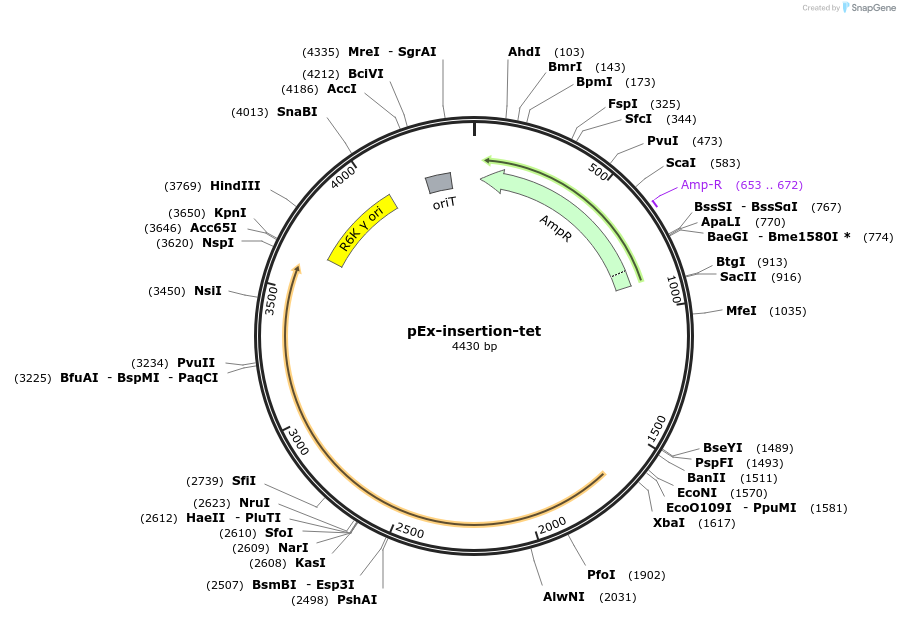
pEx-insertion-tet
Plasmid#172211PurposeFor genetic insertion in tetracyclin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
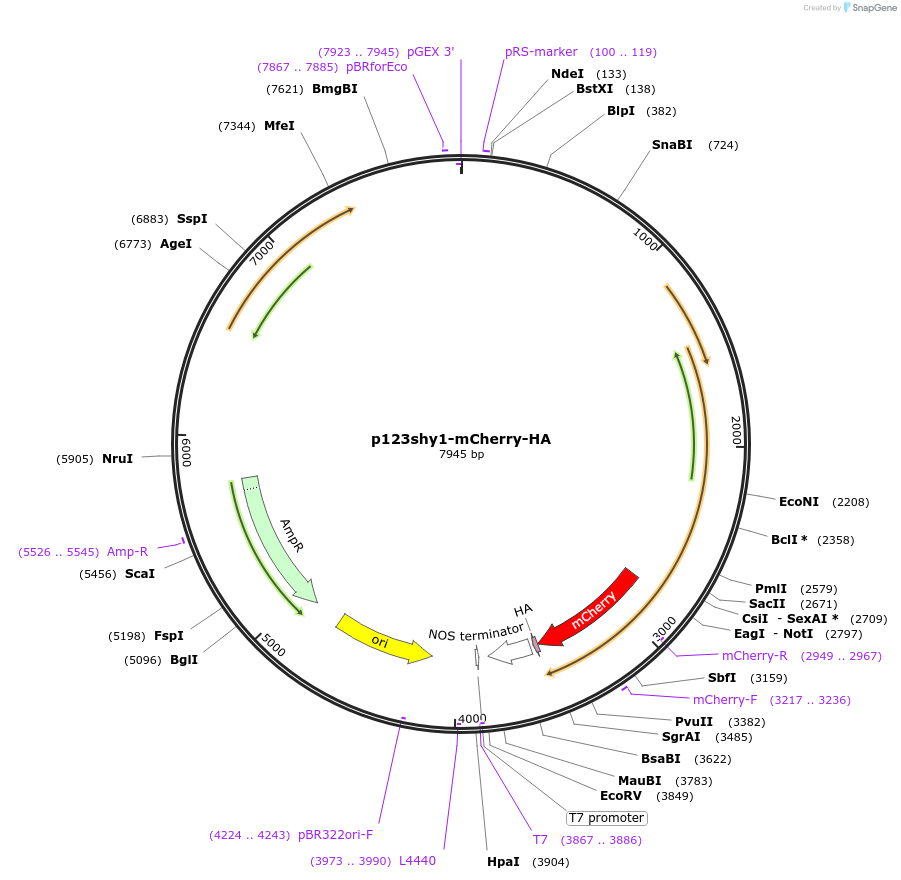
p123shy1-mCherry-HA
Plasmid#67875Purposeencodes shy1 with a C-terminal mCherry-HA tag under control of its endogenous promoter.It's suitable for localization studies in Ustilago maydis.DepositorInsertshy1
UseExpression in ustilago maydisTagsHA and mCherryPromotershy1Available SinceAug. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
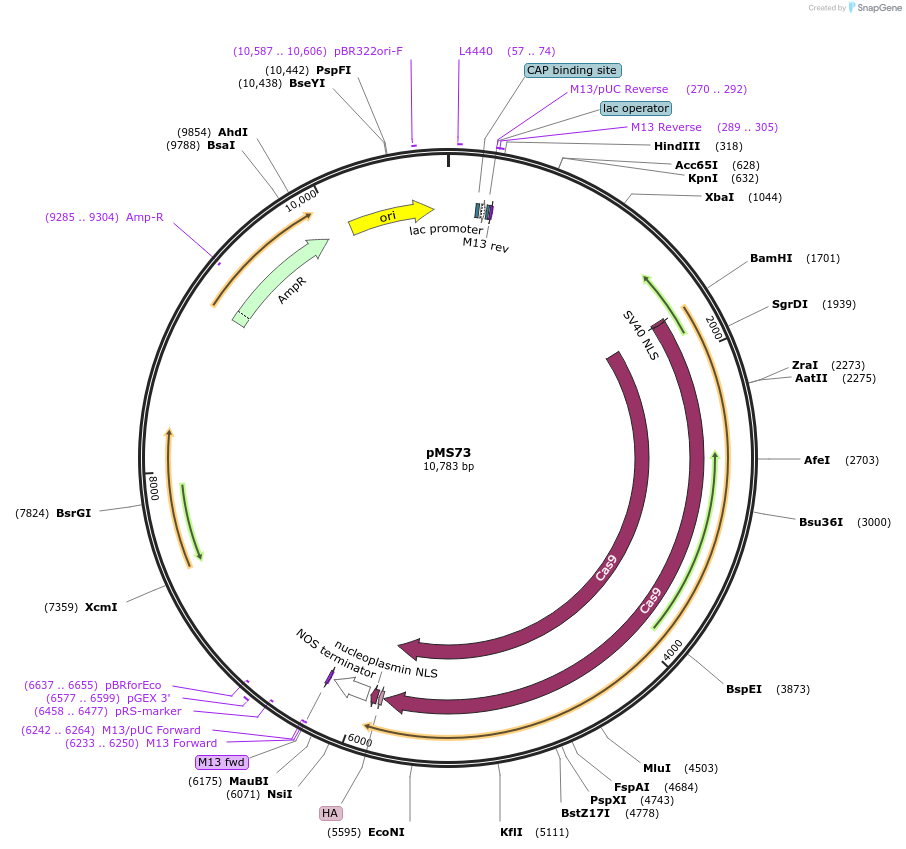
pMS73
Plasmid#110629PurposeCRISPR/Cas9 vector for mutliplexed genome editing in Ustilago maydis. Expresses U. maydis codon-optimized Cas9 under the hsp70 promoter, contains a short version of U6 promoter, is self-replicatingDepositorInsertsshort U6 promoter
cas9
tnos terminator
UseCRISPR; Self-replicating in ustilago maydis, conf…TagsN-terminal NLS, C-terminal HA-tag +NLSExpressionBacterialMutationStreptococcus pyogenes gene codon-optimized for U…PromoterU. maydis hsp70 promoter (Kronstad and Leong, 198…Available SinceMay 16, 2018AvailabilityAcademic Institutions and Nonprofits only -
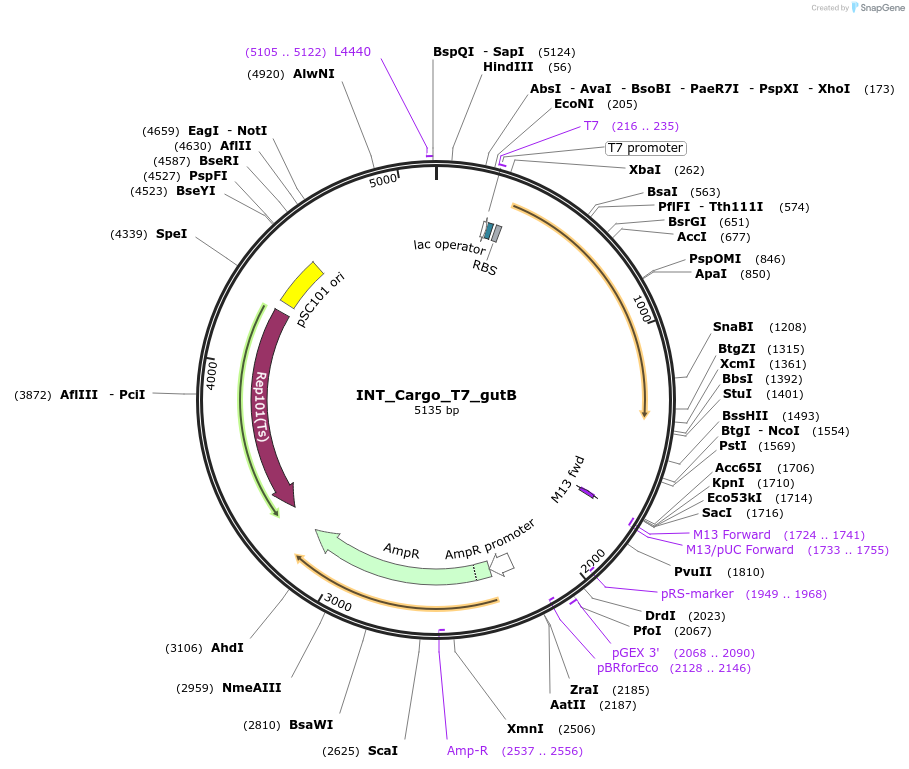
INT_Cargo_T7_gutB
Plasmid#212139PurposeCargo plasmid for integrating PT7 expression cassette of xylitol dehydrogenase in in genome.Plasmid can be removed by incubating cells at 37 C.DepositorInsertxylitol dehydrogenase
ExpressionBacterialPromoterPT7Available SinceMarch 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
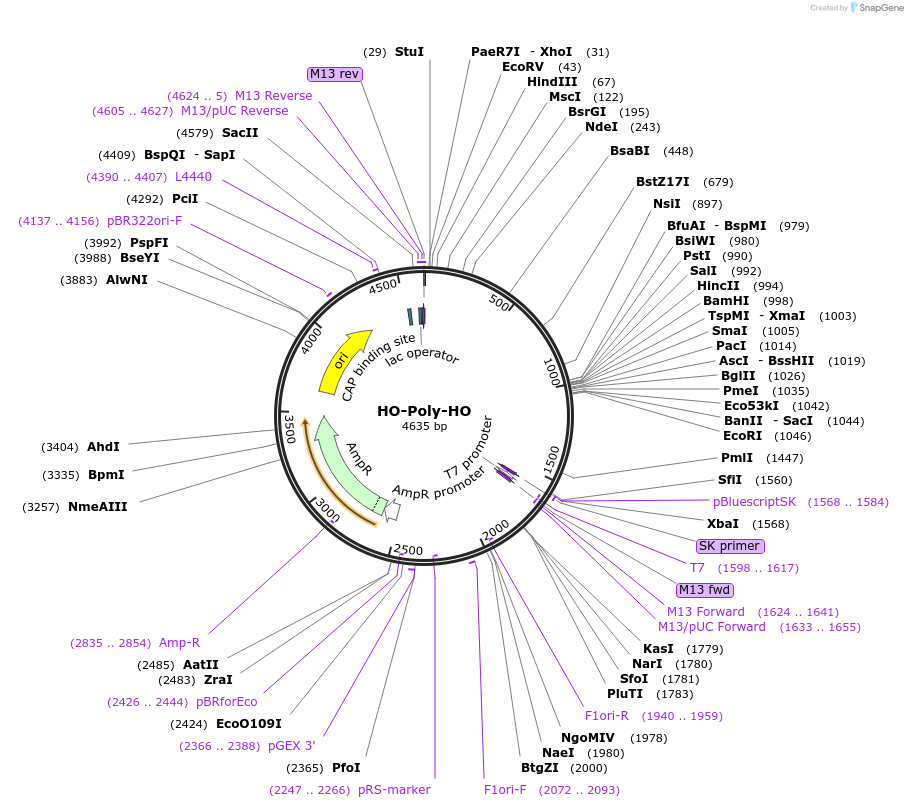
HO-Poly-HO
Plasmid#51661Purposeyeast plasmid for integration of target sequence at HO locusDepositorTypeEmpty backboneUseYeast integrationExpressionYeastAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
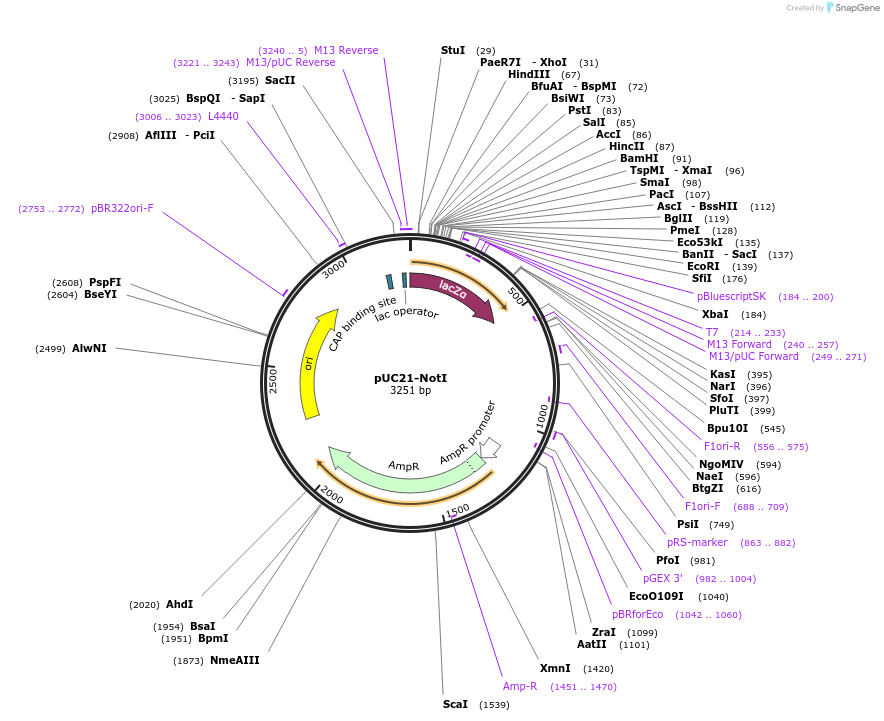
pUC21-NotI
Plasmid#51659PurposeBacterial cloning vector based on pUC21 (Amp)DepositorTypeEmpty backboneUseCloning vectorAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only