We narrowed to 1,880 results for: FAST
-
Plasmid#170906PurposeGoldenBraid Double Transcriptional Unit - pAtUBQ10_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpAtUBQ10Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGP-CMV-NES-fRGECO1
Plasmid#125247PurposeExpresses red fluorescent calcium sensor in mammalian cellsDepositorInsertfRGECO2
TagsNES signalExpressionMammalianAvailable SinceNov. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pJ414-PCY1
Plasmid#98150PurposeExpresses PCY1 from Saponaria vaccaria possessing a N-terminal His6-tag (TEV cleavable) in E. coli.DepositorInsertPeptide cyclase 1
TagsHis6-TEVExpressionBacterialMutationWTPromoterT7Available SinceMarch 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
pZA1
Plasmid#158479PurposeEntry clone for Golden Gate assembly. Golden Gate compatible wild-type A. thaliana ZAR1 with STOP codonDepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
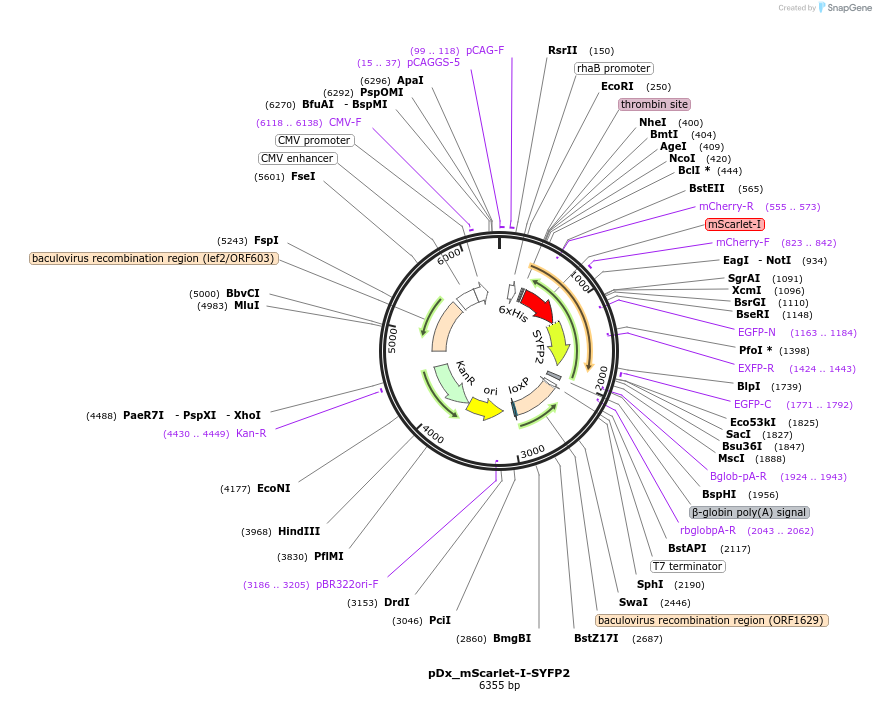
pDx_mScarlet-I-SYFP2
Plasmid#189761PurposeDual expression vector for bacteria and mammalian cells producing mScarlet-I-SYFP2 fusion protein for FRET analysisDepositorInsertmScarlet-I-SYFP2
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
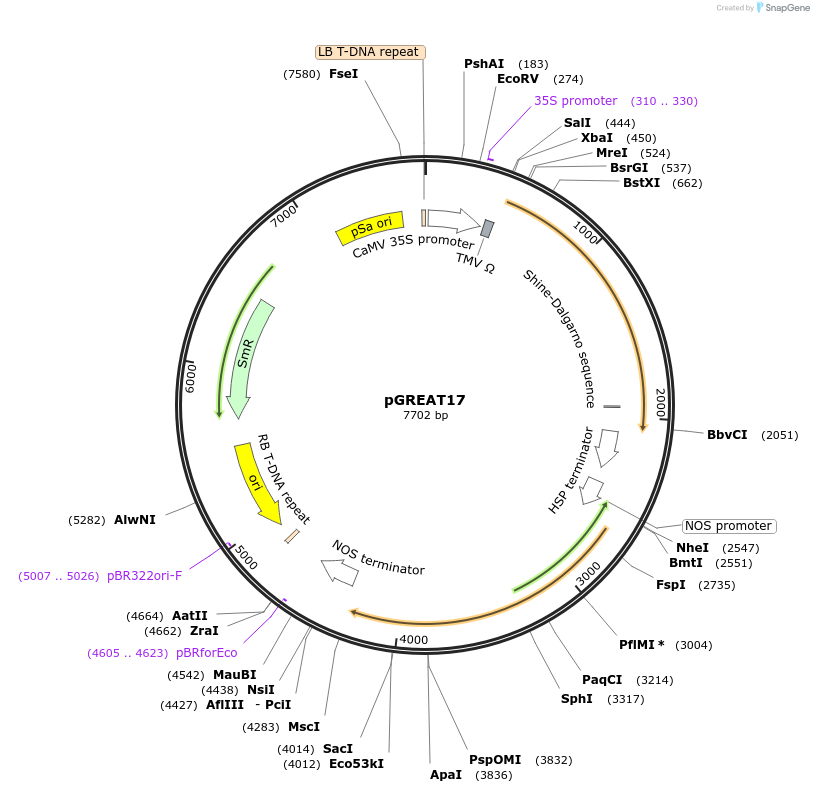
pGREAT17
Plasmid#170905PurposeGoldenBraid Double Transcriptional Unit - p35SoTMV_E-Luc_AtHSP18.2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…Promoterp35SoTMVAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
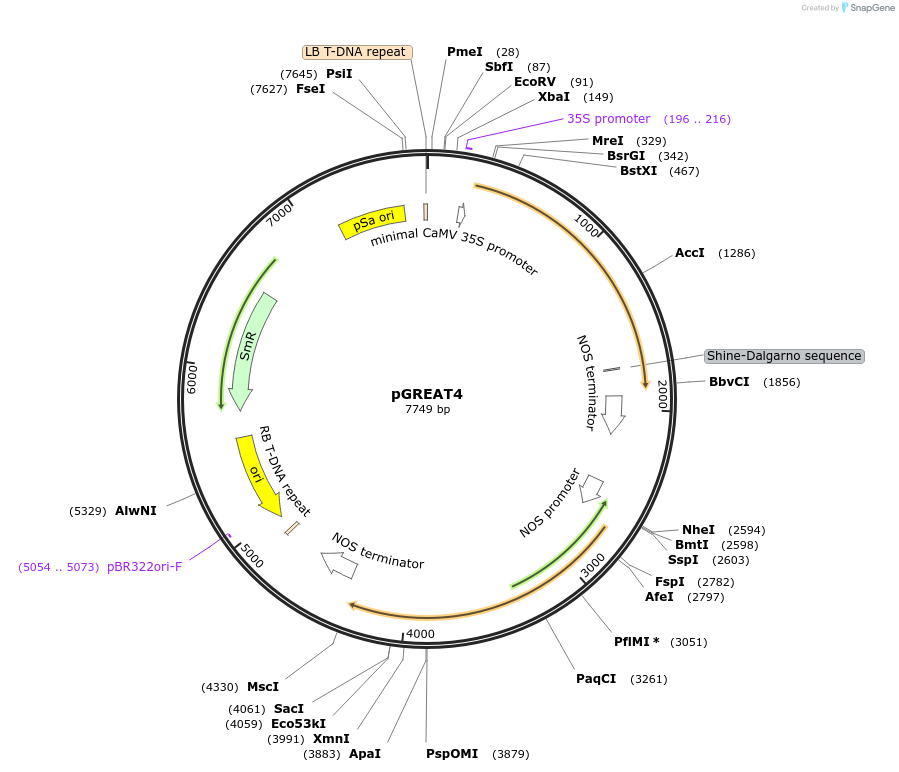
pGREAT4
Plasmid#170892PurposeGoldenBraid Double Transcriptional Unit - pG10-90_E-Luc_NOSt+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpG10-90Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
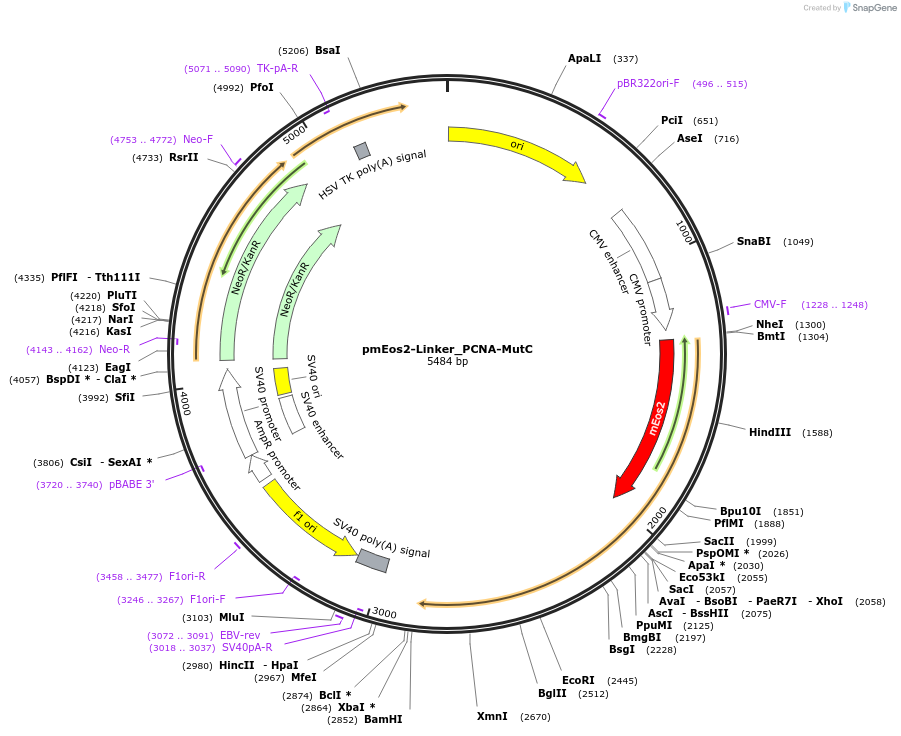
pmEos2-Linker_PCNA-MutC
Plasmid#98272Purposefor low level expression of mEos2-PCNA in mammalian cellsDepositorInsertPCNA (PCNA Human)
TagsmEos2ExpressionMammalianMutationSilent mutated Sequence: GACGCCGTAGTTATATCTTGC (O…PromoterCMVAvailable SinceJuly 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pNB785
Plasmid#60164PurposeFirefly luciferase (Promega) yeast codon-optimized yellow fluorescent protein (yEVenus) fusion with PEST degron driven by SIC1 promoter.DepositorInsertFLuc-yEVenus-PEST
TagsPEST degronExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterSIC1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
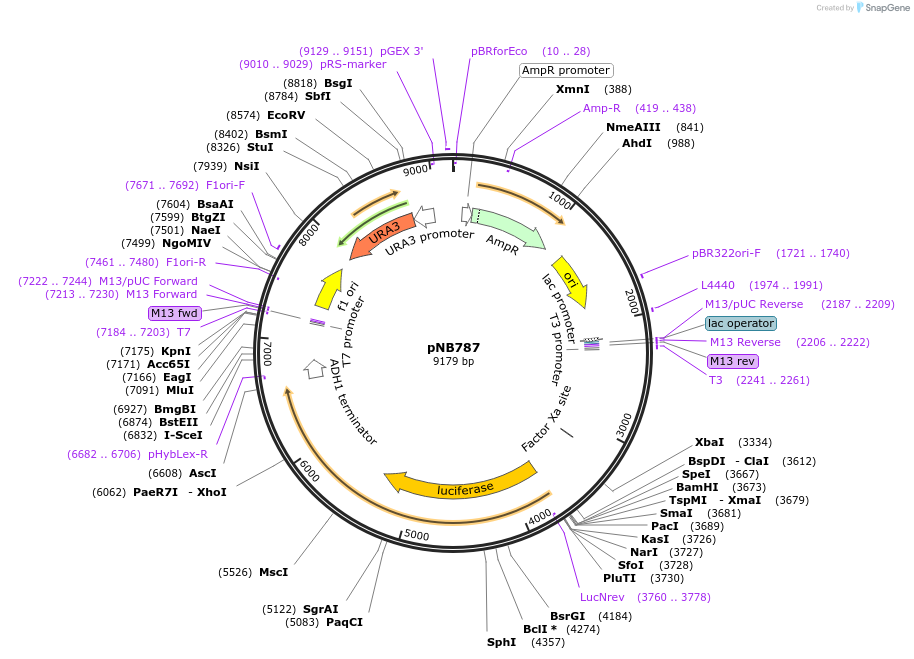
pNB787
Plasmid#60152PurposeFirefly luciferase (Promega) and yeast codon-optimized yellow fluorescent protein (yEVenus) fusion with PEST degron driven by LEU1 promoter.DepositorInsertFLuc-yEVenus-PEST
TagsPEST degronExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterLEU1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
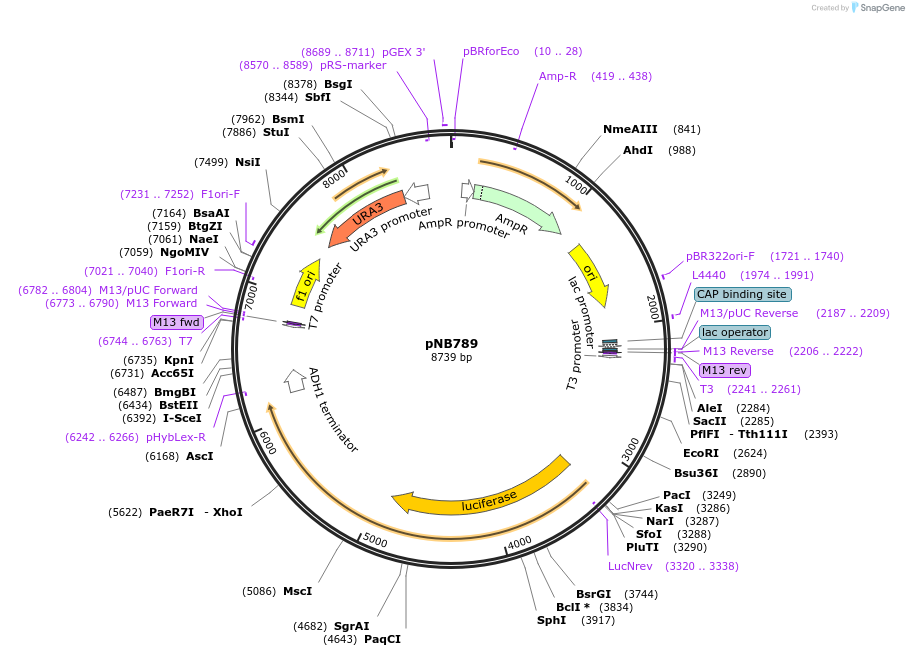
pNB789
Plasmid#60166PurposeFirefly luciferase (Promega) yeast codon-optimized yellow fluorescent protein (yEVenus) fusion with PEST degron driven by RNR1 promoter.DepositorInsertFLuc-yEVenus-PEST
TagsPEST degronExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterRNR1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
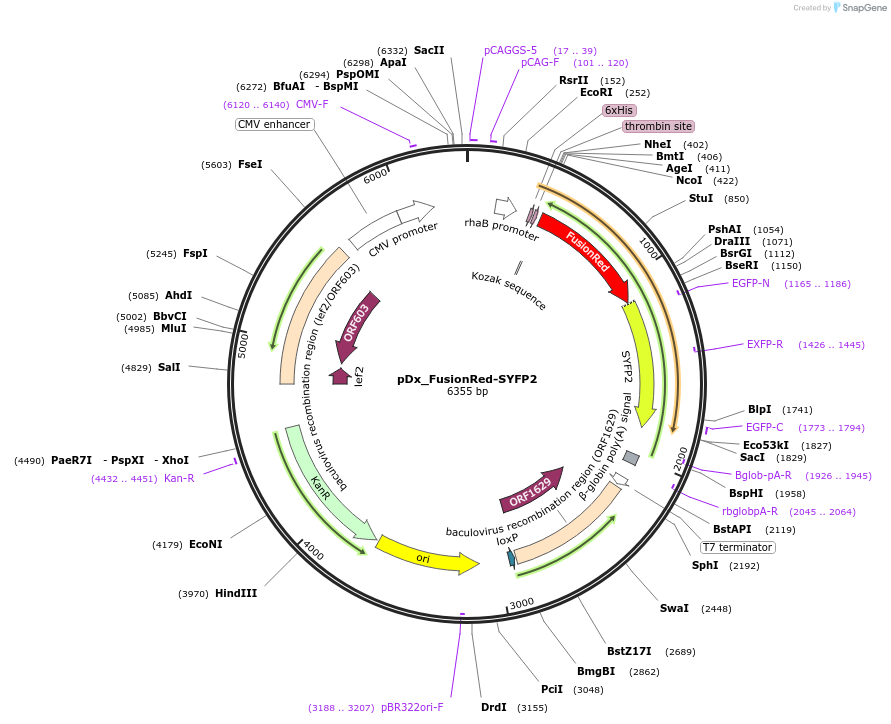
pDx_FusionRed-SYFP2
Plasmid#189760PurposeDual expression vector for bacteria and mammalian cells producing FusionRed-SYFP2 fusion protein for FRET analysisDepositorInsertFusionRed-SYFP2
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
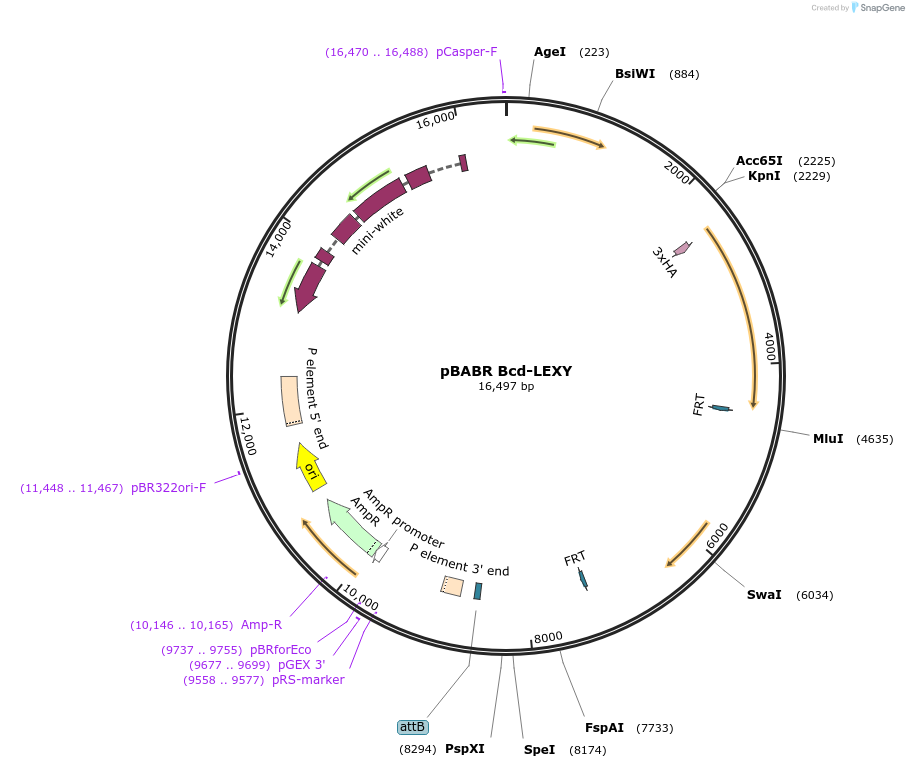
pBABR Bcd-LEXY
Plasmid#182594PurposeDrosophila integration plasmid expressing Bcd-LEXY fusion protein under the maternal tubulin promoter.DepositorAvailable SinceAug. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
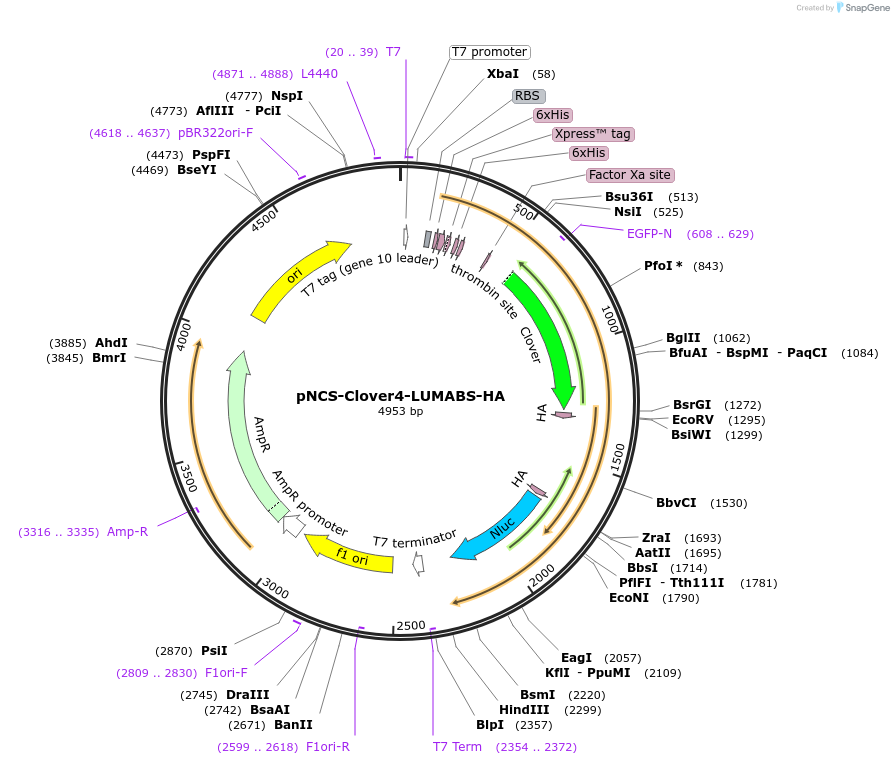
pNCS-Clover4-LUMABS-HA
Plasmid#183486PurposeTo express the Clover4-LUMABS-HA sensor for Avian Influenza Virus antibody detection.DepositorInsertClover4-LUMABS-HA
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
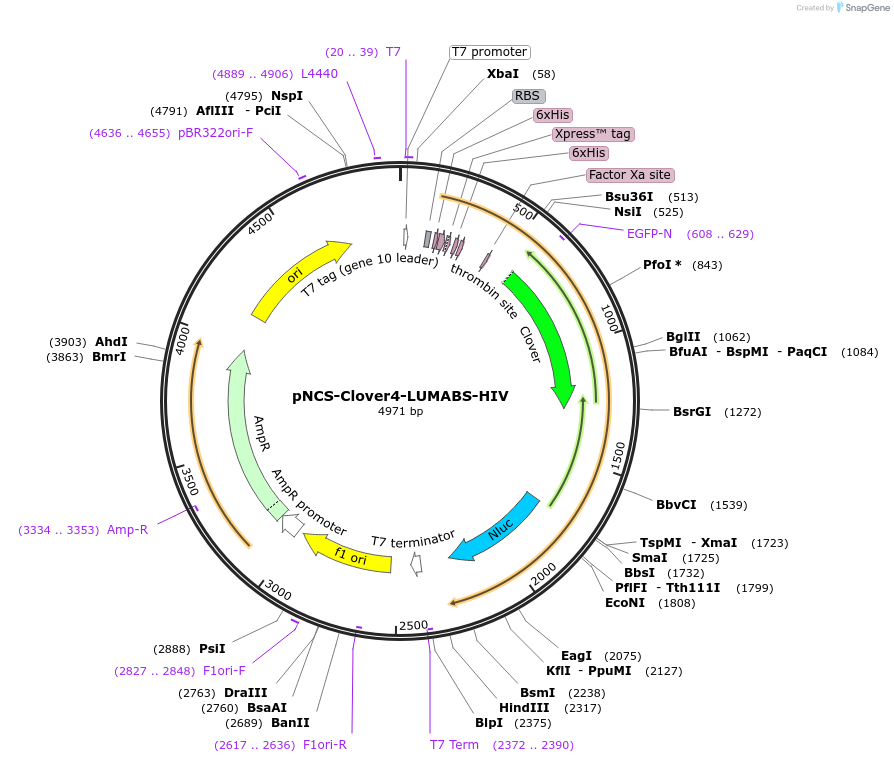
pNCS-Clover4-LUMABS-HIV
Plasmid#183485PurposeTo express the Clover4-LUMABS-HIV sensor for Acquired Immunodeficiency Virus antibody dection.DepositorInsertClover4-LUMABS-HIV
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
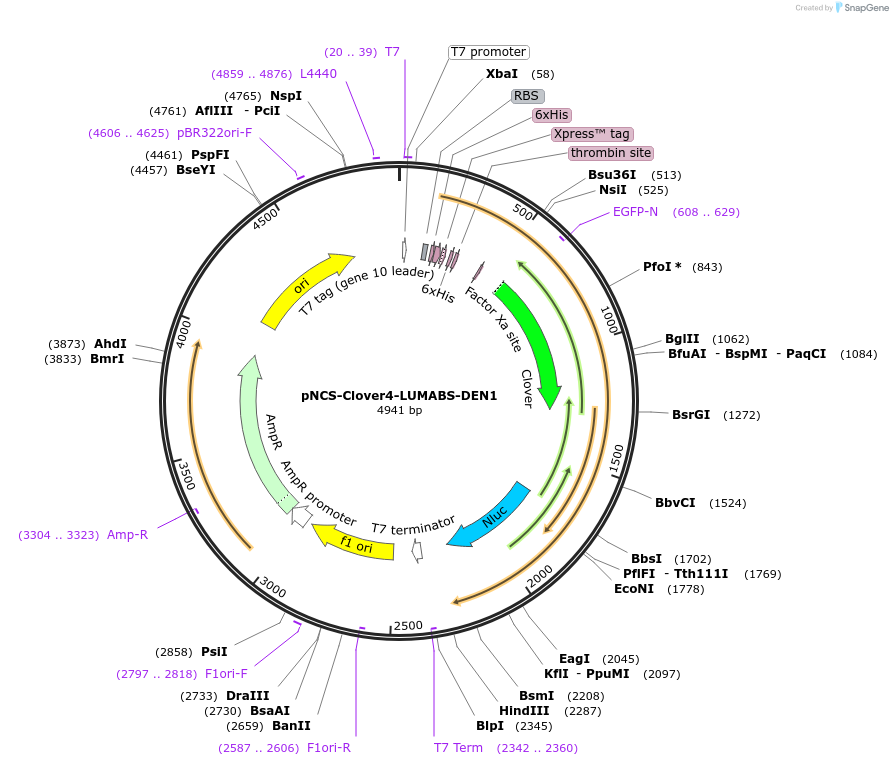
pNCS-Clover4-LUMABS-DEN1
Plasmid#183487PurposeTo express the Clover4-LUMABS-DEN1 sensor for Dengue Virus antibody dection.DepositorInsertClover4-LUMABS-DEN1
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
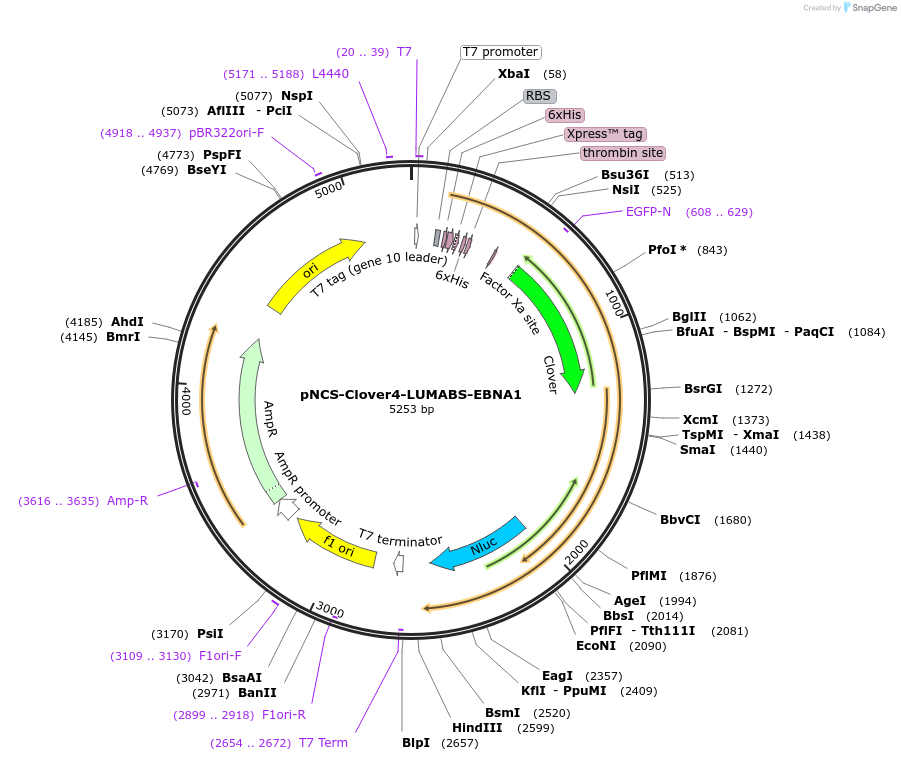
pNCS-Clover4-LUMABS-EBNA1
Plasmid#183488PurposeTo express the Clover4-LUMABS-EBNA1 sensor for Epstein-barr Virus antibody dection.DepositorInsertClover4-LUMABS-EBNA1
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
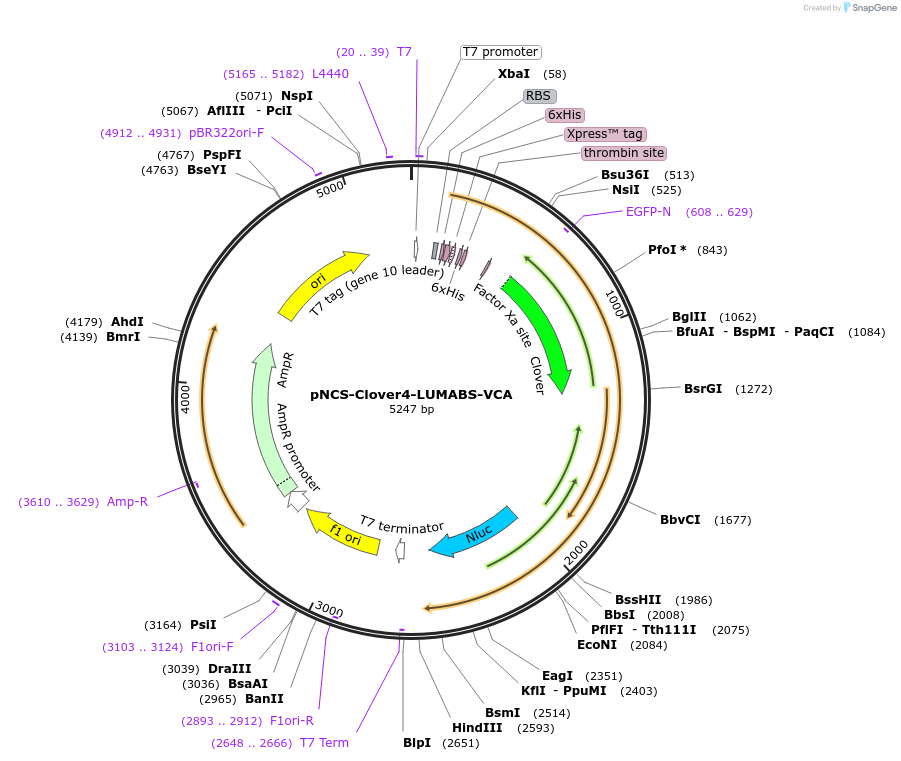
pNCS-Clover4-LUMABS-VCA
Plasmid#183489PurposeTo express the Clover4-LUMABS-VCA sensor for Epstein-barr Virus antibody dection.DepositorInsertClover4-LUMABS-VCA
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
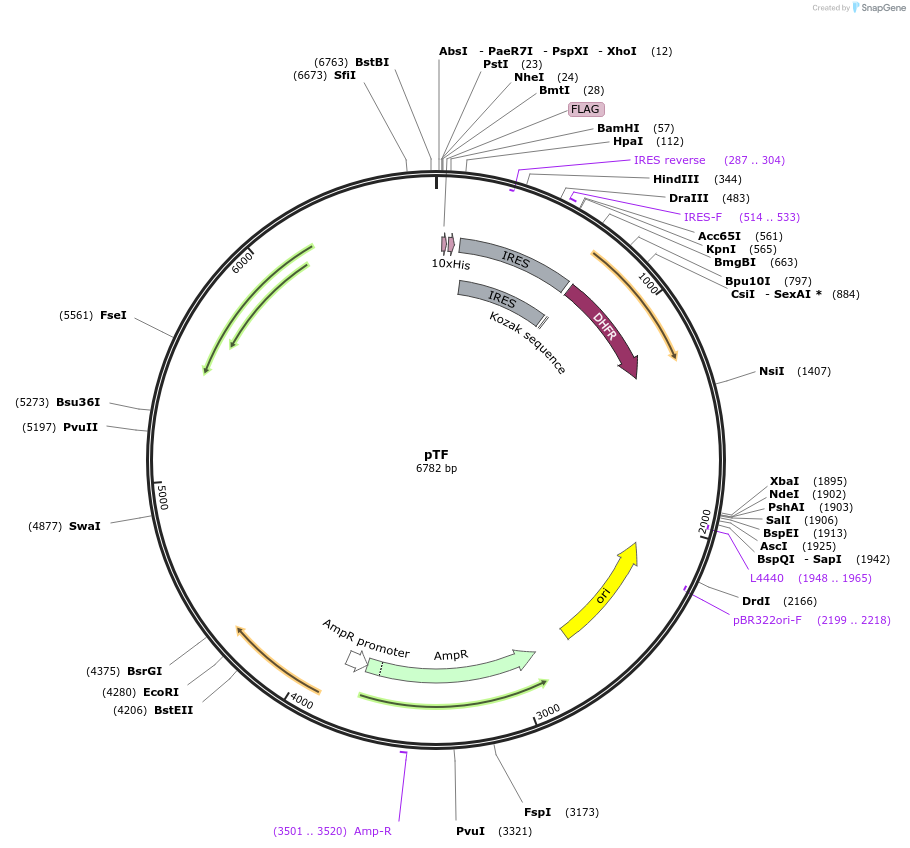
pTF
Plasmid#162784PurposeProtein expression in CHO cells - C-terminal FLAG and 10xhis tags are codedDepositorTypeEmpty backboneTags10xHis and FLAGExpressionMammalianPromoterCHO Elongation factor 1AAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
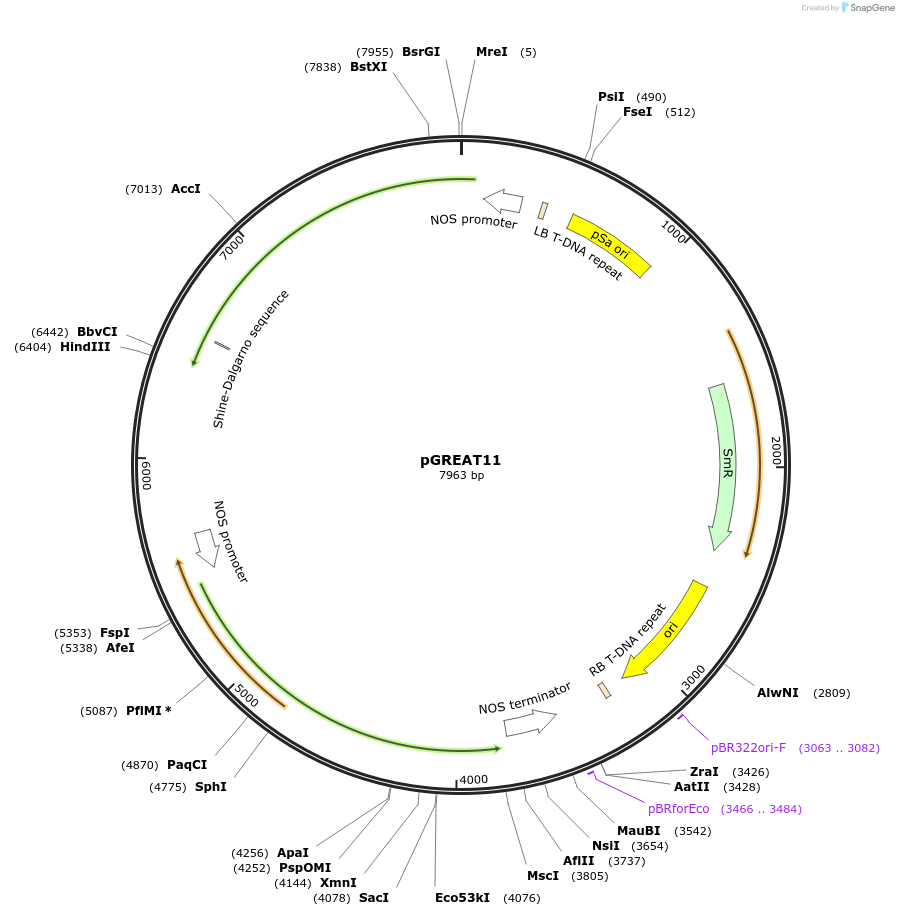
pGREAT11
Plasmid#170899PurposeGoldenBraid Double Transcriptional Unit - pNOS_E-Luc_AtACT2t+pNOS_Red-F_NOStDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyExpressionPlantMutationc.375C>T silent mutation to remove BbsI site (…PromoterpNOSAvailable SinceAug. 25, 2021AvailabilityAcademic Institutions and Nonprofits only