We narrowed to 13,272 results for: LIC
-
Plasmid#20644DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only
-
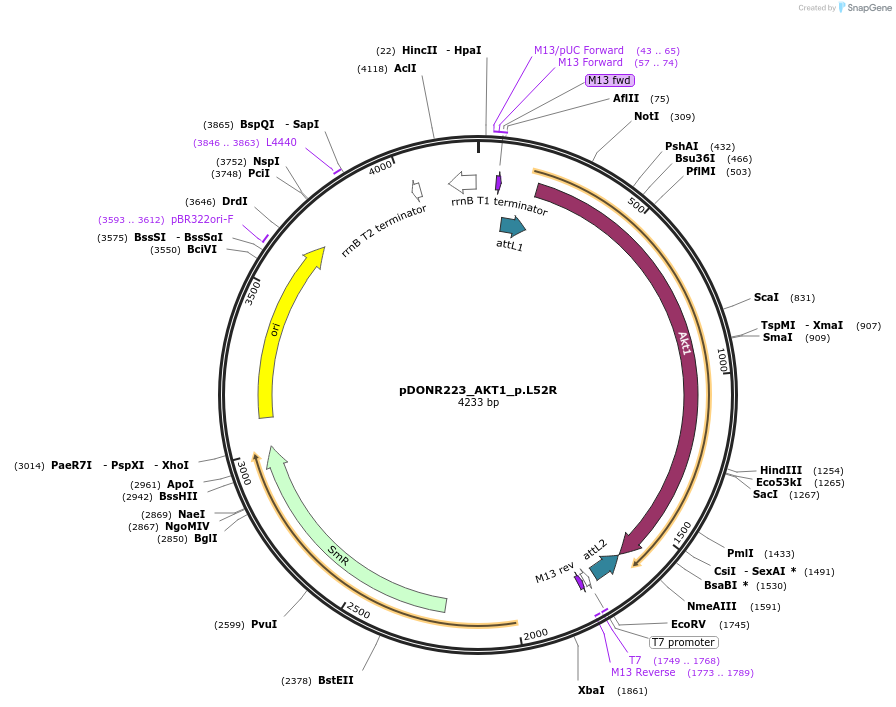
pDONR223_AKT1_p.L52R
Plasmid#81604PurposeGateway Donor vector containing AKT1 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
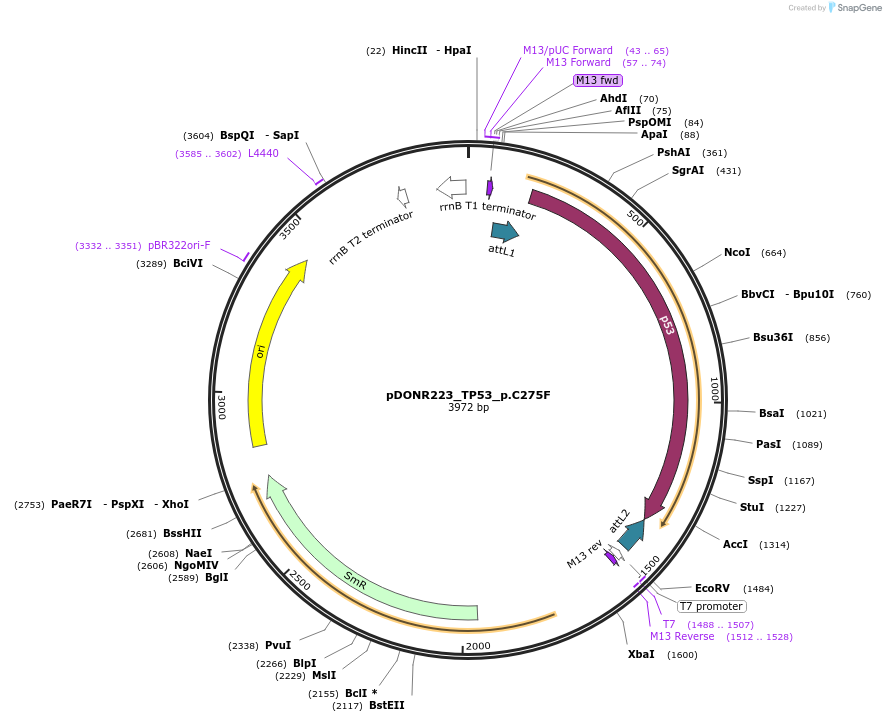
pDONR223_TP53_p.C275F
Plasmid#81399PurposeGateway Donor vector containing TP53 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag PMVK
Plasmid#20593DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
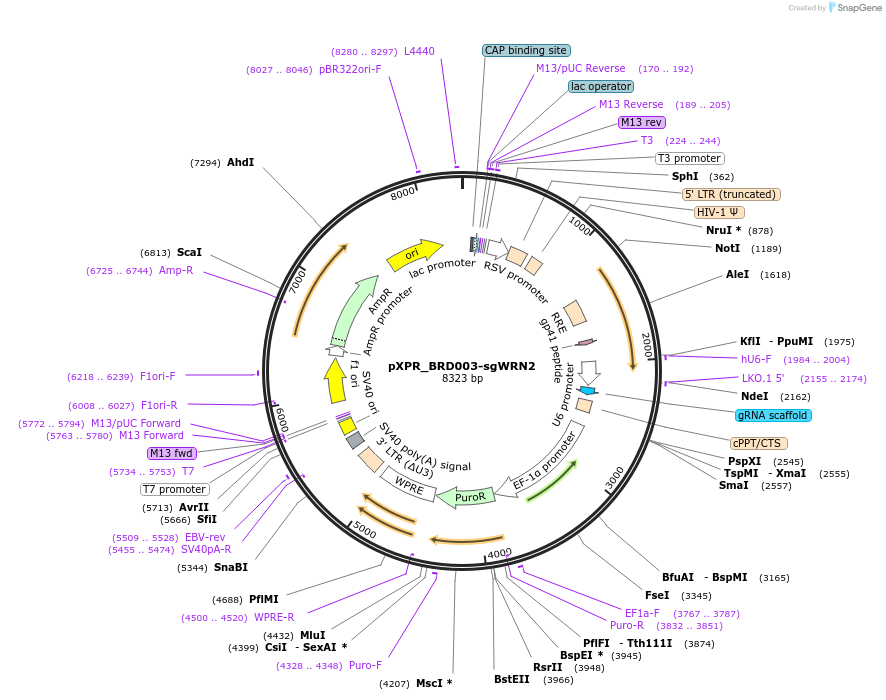
pXPR_BRD003-sgWRN2
Plasmid#125772Purposeconstitutive expression of a guide RNA targeting human WRNDepositorInsertsgWRN2 (WRN Human)
UseCRISPRAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
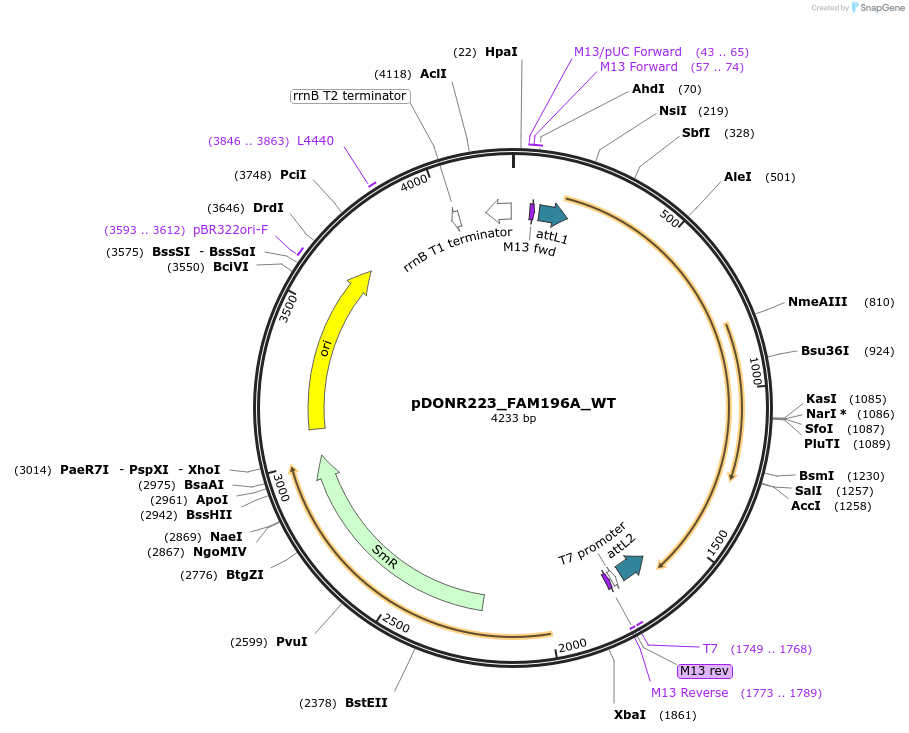
pDONR223_FAM196A_WT
Plasmid#81882PurposeGateway Donor vector containing FAM196A , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
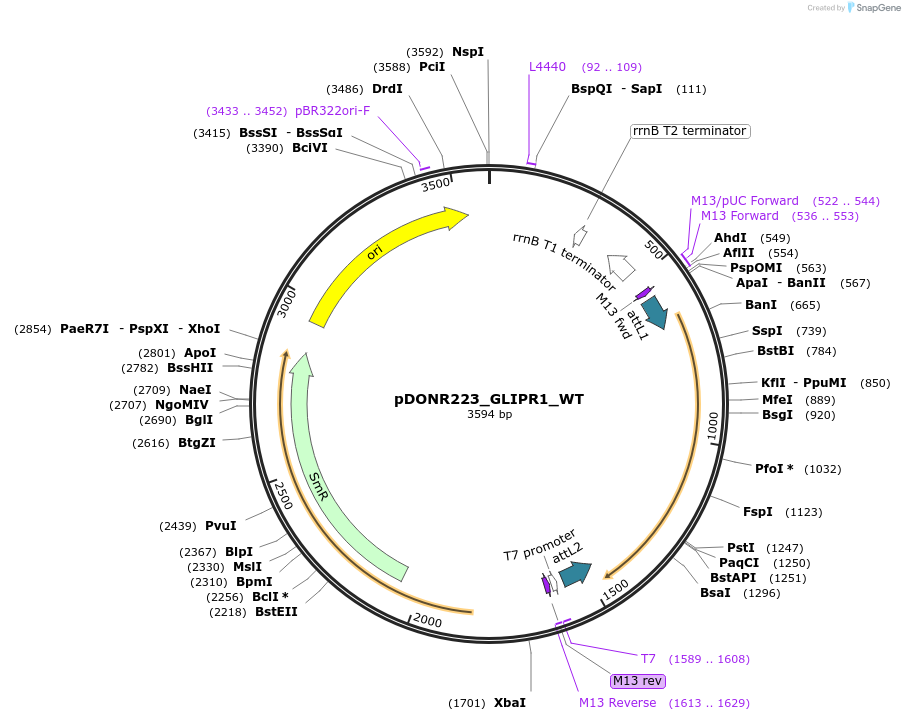
pDONR223_GLIPR1_WT
Plasmid#81780PurposeGateway Donor vector containing GLIPR1 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
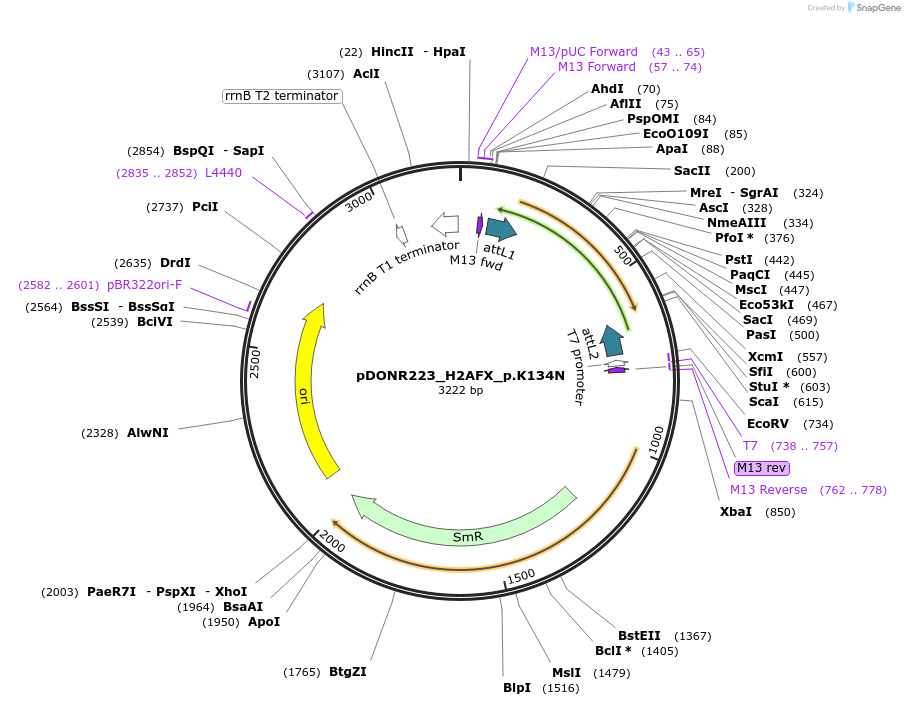
pDONR223_H2AFX_p.K134N
Plasmid#81275PurposeGateway Donor vector containing H2AFX , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
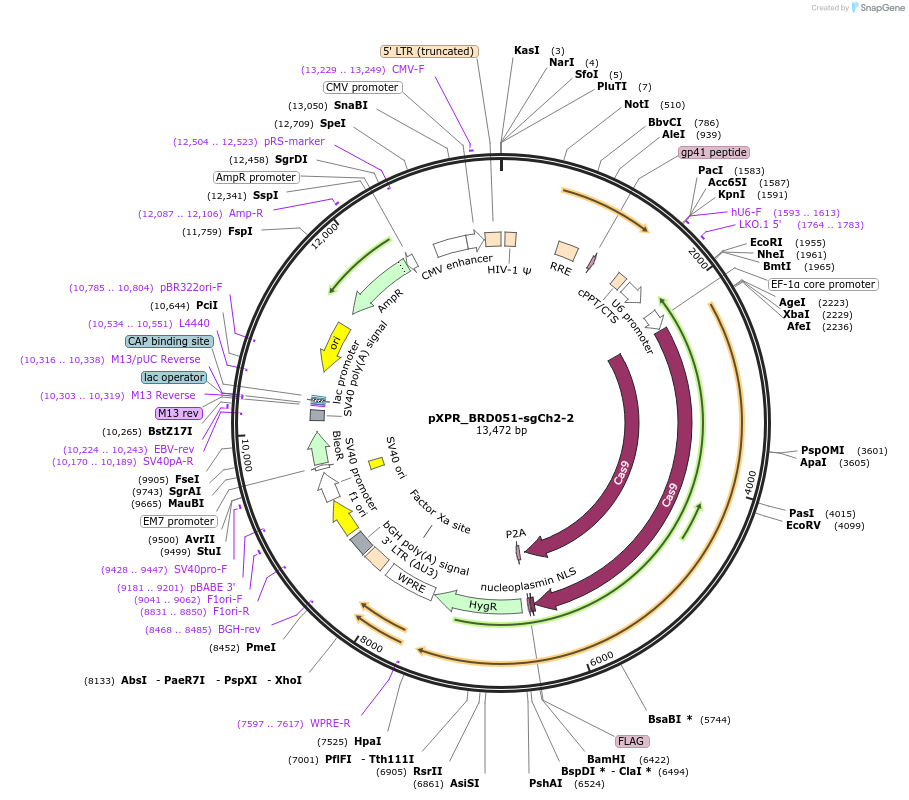
pXPR_BRD051-sgCh2-2
Plasmid#125775Purposeconstitutive expression of a guide RNA targeting an intergenic region of human chromosome 2 (CRISPR cutting control) and S. pyogenes Cas9DepositorInsertsgCh2-2
UseCRISPRAvailable SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
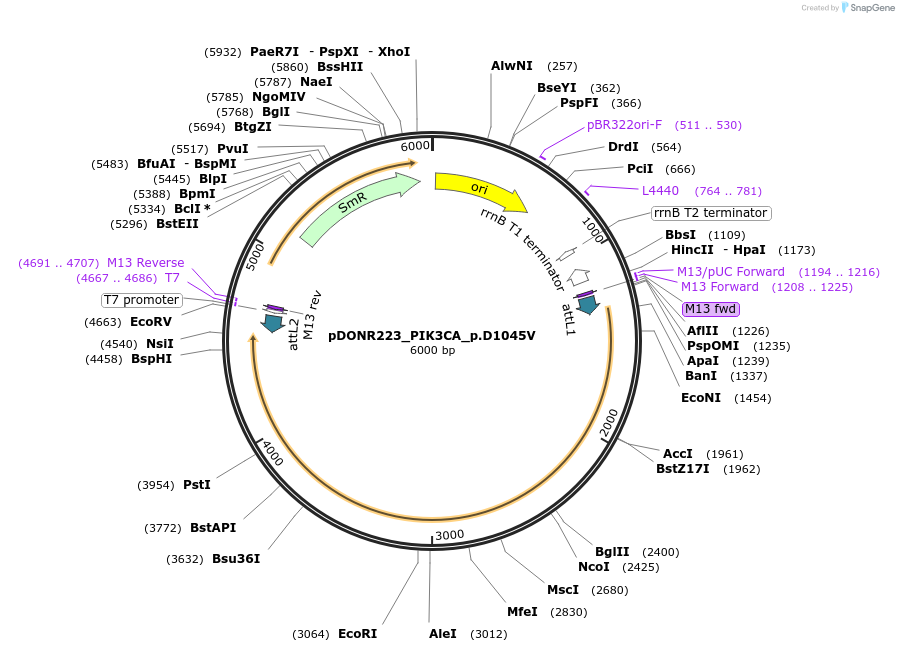
pDONR223_PIK3CA_p.D1045V
Plasmid#81702PurposeGateway Donor vector containing PIK3CA, part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
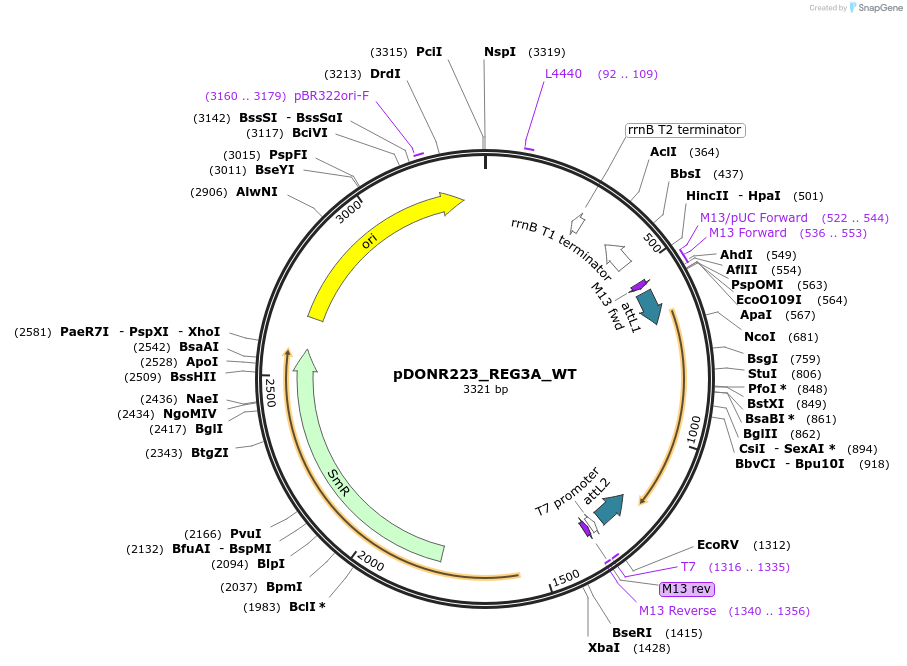
pDONR223_REG3A_WT
Plasmid#81826PurposeGateway Donor vector containing REG3A , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
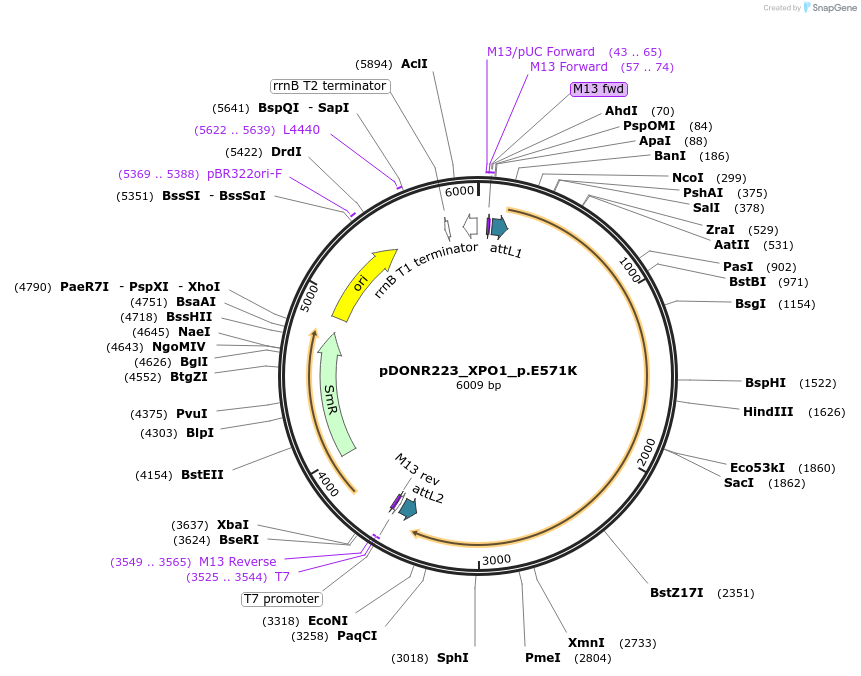
pDONR223_XPO1_p.E571K
Plasmid#81554PurposeGateway Donor vector containing XPO1 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag TIE1
Plasmid#20651DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
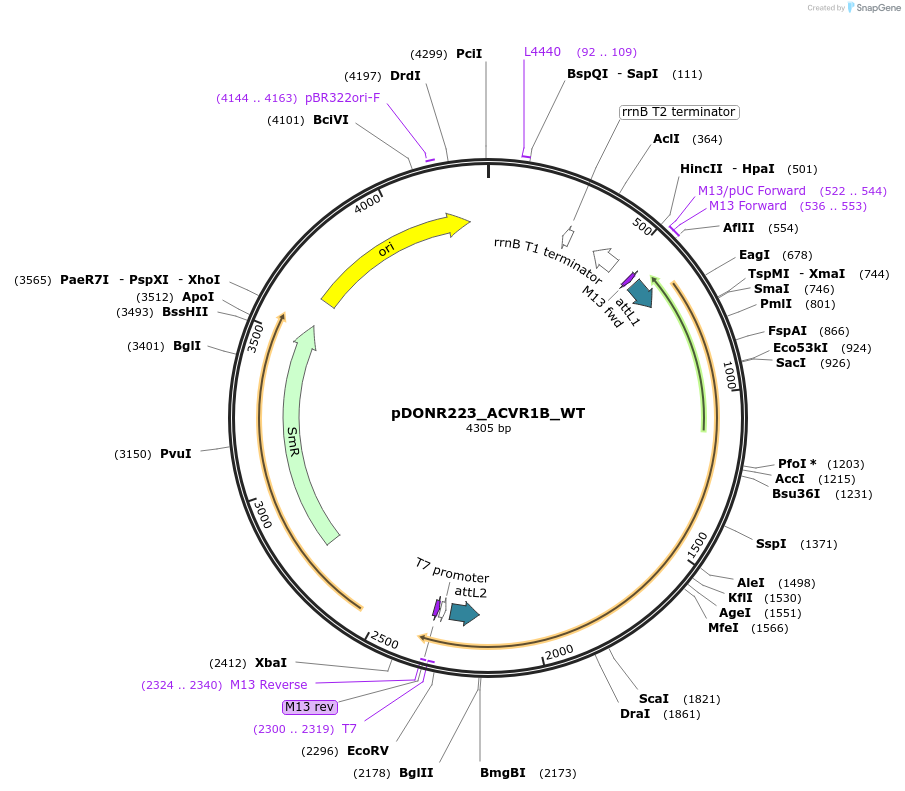
pDONR223_ACVR1B_WT
Plasmid#81738PurposeGateway Donor vector containing ACVR1B , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
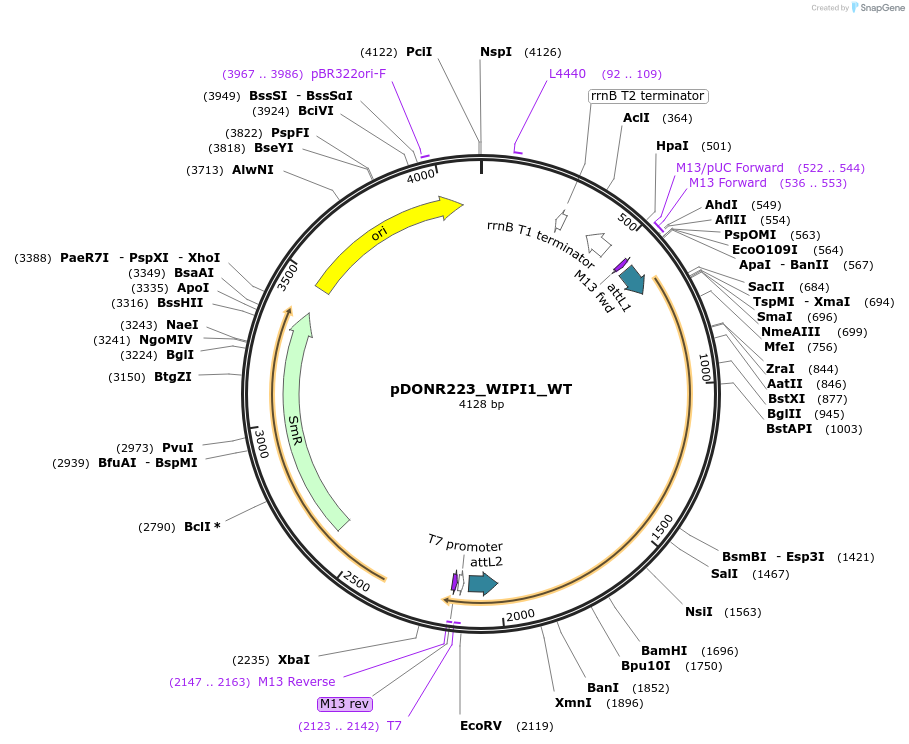
pDONR223_WIPI1_WT
Plasmid#82055PurposeGateway Donor vector containing WIPI1 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag VRK2
Plasmid#20663DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag STK32C
Plasmid#20517DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag OXSR1
Plasmid#20553DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
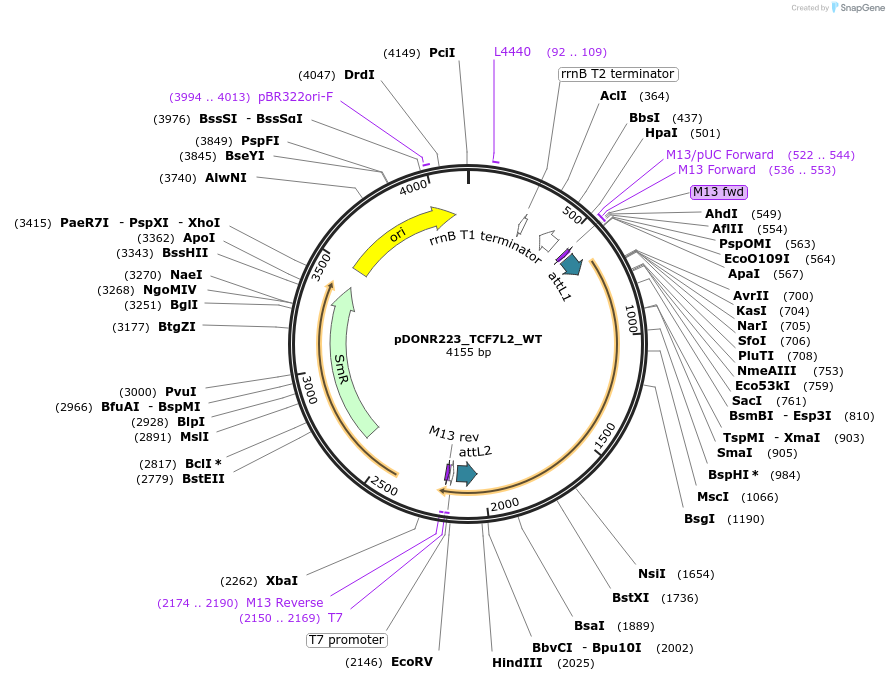
pDONR223_TCF7L2_WT
Plasmid#81745PurposeGateway Donor vector containing TCF7L2 , part of the Target Accelerator Plasmid Collection.DepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag ULK4
Plasmid#20662DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only