We narrowed to 9,373 results for: Pol;
-
Plasmid#31795DepositorInsertTAZ
UseRNAi and RetroviralPromoterRNA Pol-IIIAvailable SinceAug. 29, 2011AvailabilityAcademic Institutions and Nonprofits only -
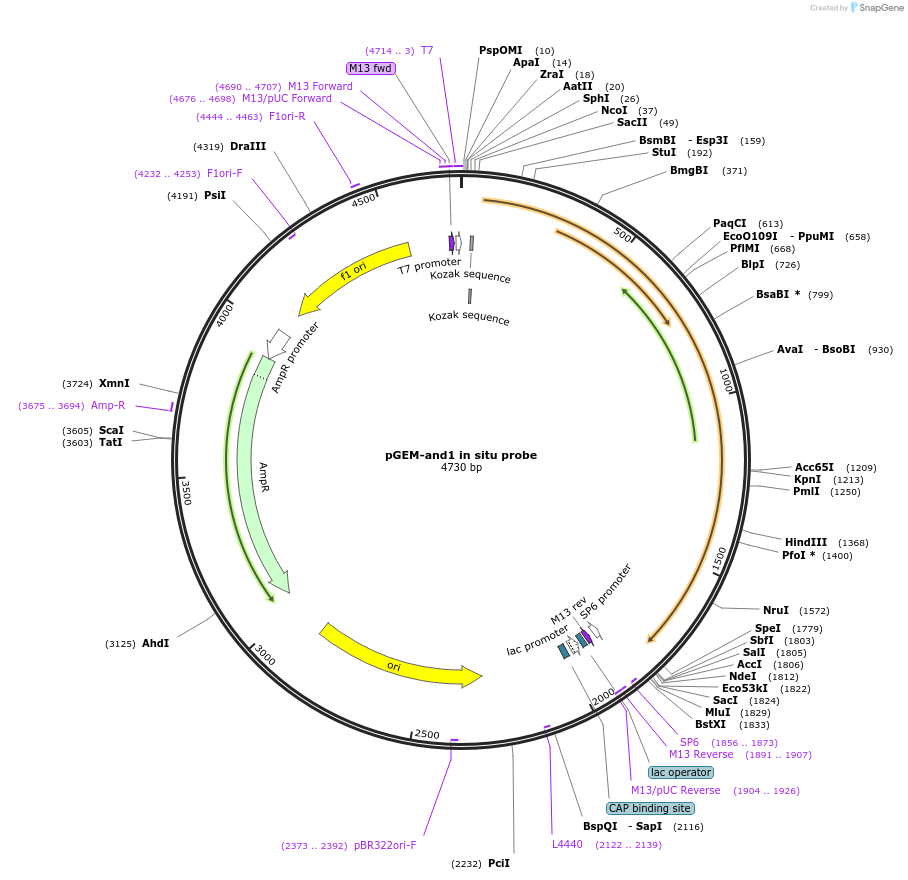
pGEM-and1 in situ probe
Plasmid#52102Purposeactinodin 1 and1 antisense in situ probe linearize w/ ApaI SP6 polDepositorInsertactinodin 1 in situ probe (and1 Zebrafish)
ExpressionBacterialAvailable SinceApril 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
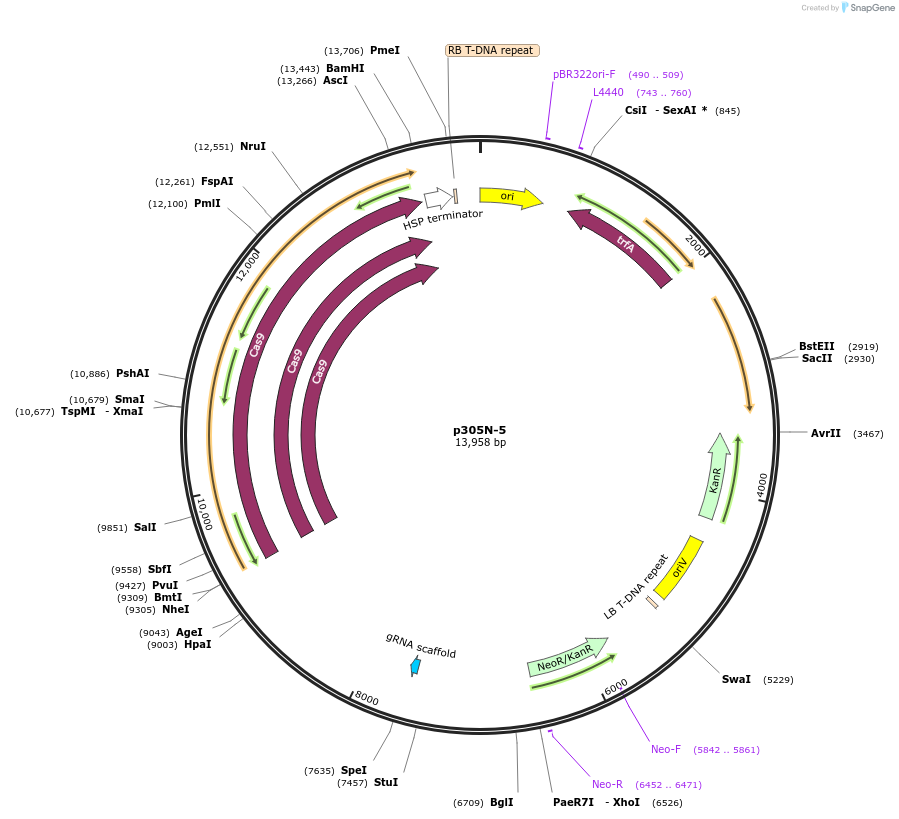
p305N-5
Plasmid#246288PurposeEvaluation of AtU6.1m2 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1m2 promoter
UseCRISPRExpressionPlantMutationA-to-C (-63 from TSS) and G-to-T (-57) within USEPromoterAtU6.1m2 promoterAvailable SinceDec. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
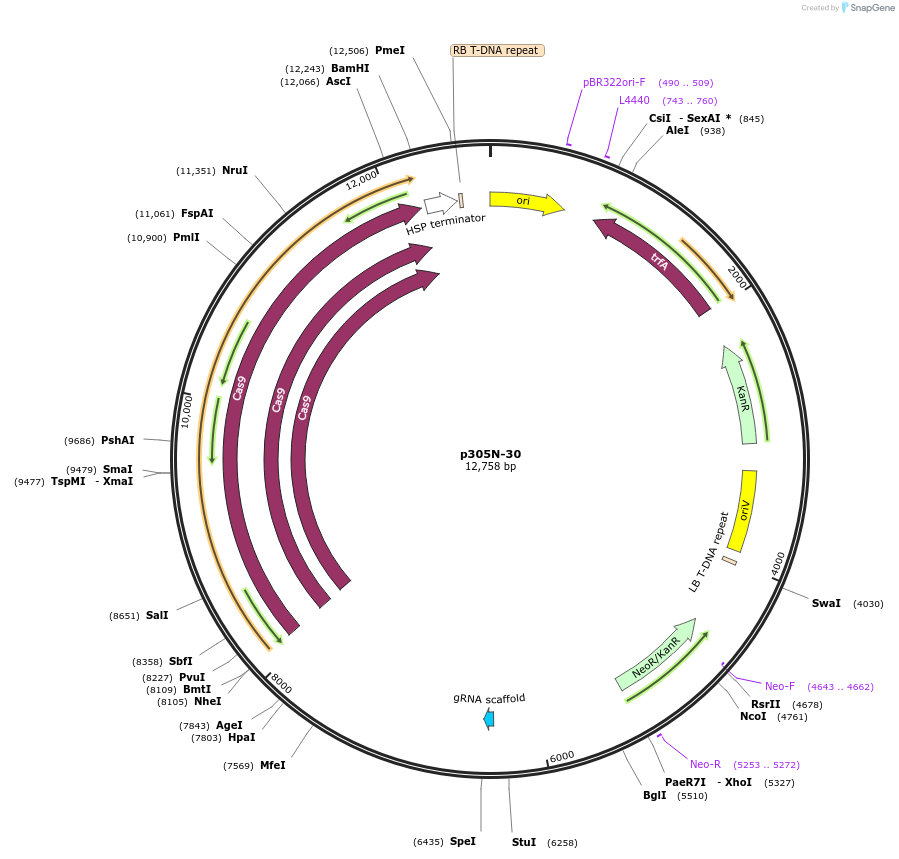
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
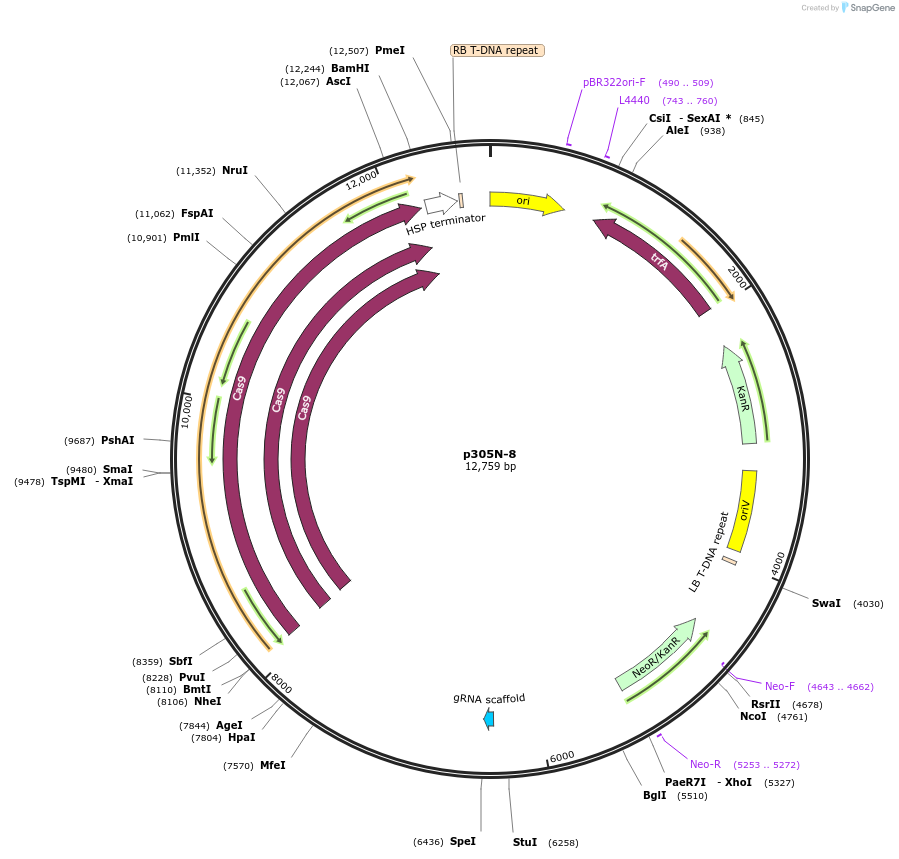
p305N-8
Plasmid#246291PurposeEvaluation of AtU6.29c14 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c14 promoter
UseCRISPRExpressionPlantMutationT-to-C (-1 from TSS)PromoterAtU6.29c14 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
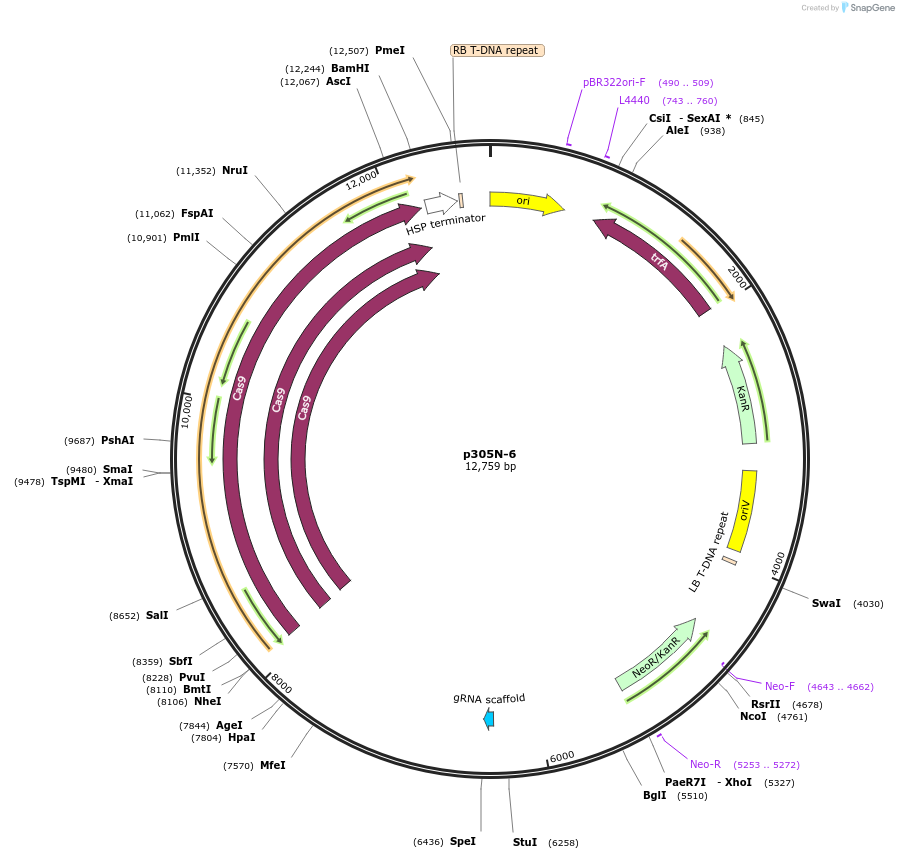
p305N-6
Plasmid#246289PurposeEvaluation of AtU6.1m3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1m3 promoter
UseCRISPRExpressionPlantMutationG-to-T (-57 from TSS) within USEPromoterAtU6.1m3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
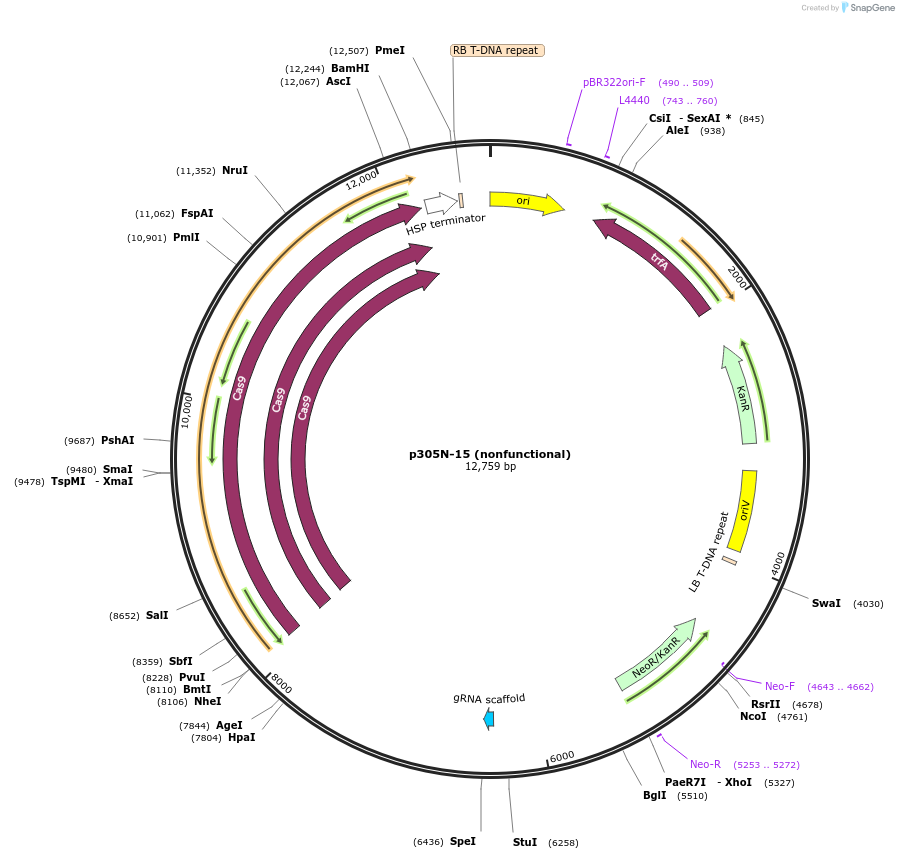
p305N-15 (nonfunctional)
Plasmid#246298PurposeEvaluation of HbU6.2 promoter (nonfunctional) (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2 promoter (nonfunctional)
UseCRISPRExpressionPlantPromoterHbU6.2 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
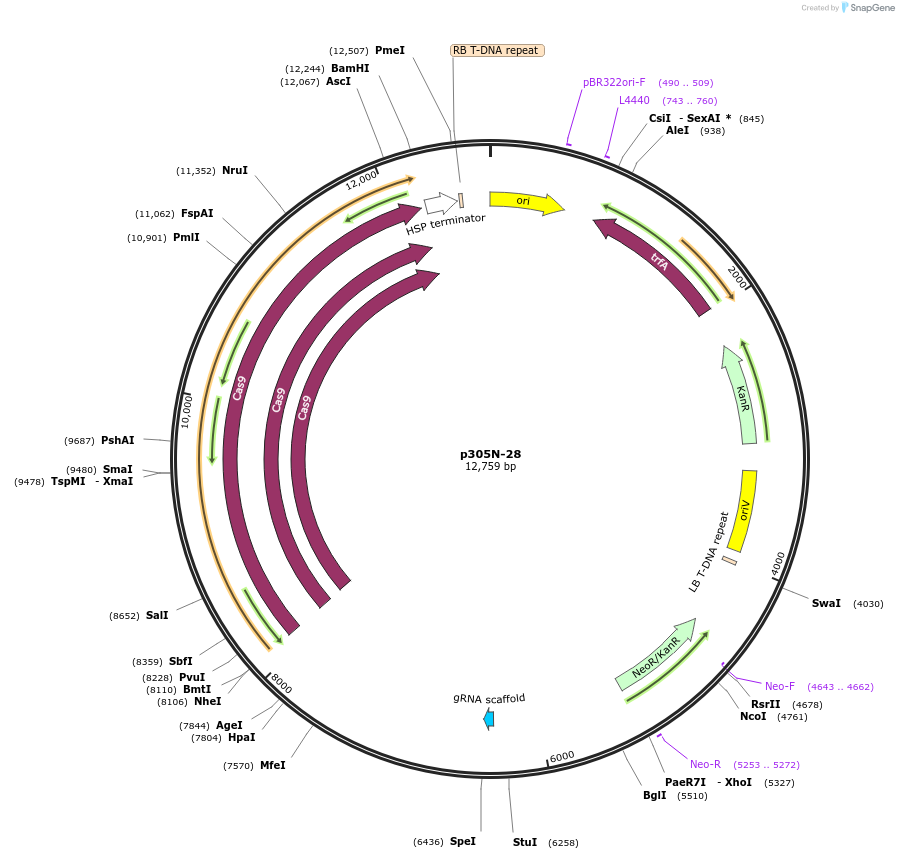
p305N-28
Plasmid#246311PurposeEvaluation of PtU6.1c1 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.1c1 promoter
UseCRISPRExpressionPlantMutationC-to-A (-42 from TSS), A-to-C (-41)PromoterPtU6.1c1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
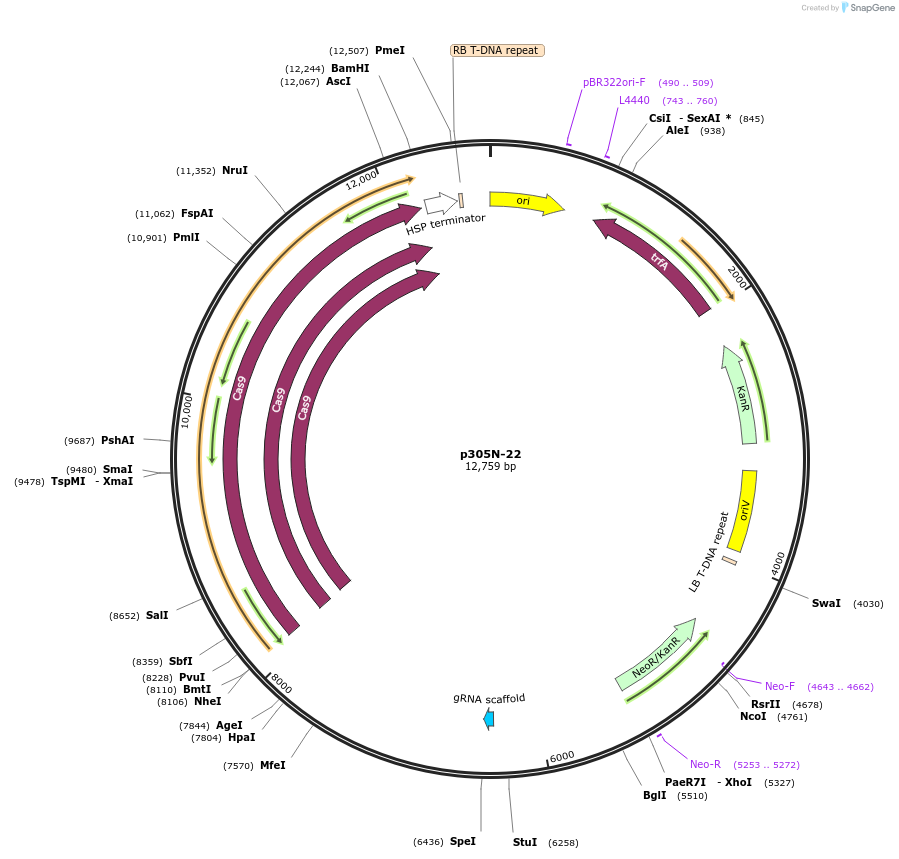
p305N-22
Plasmid#246305PurposeEvaluation of MtU6.6-70 promoter (Pol III promoter) deletion (70 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-70 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-70 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
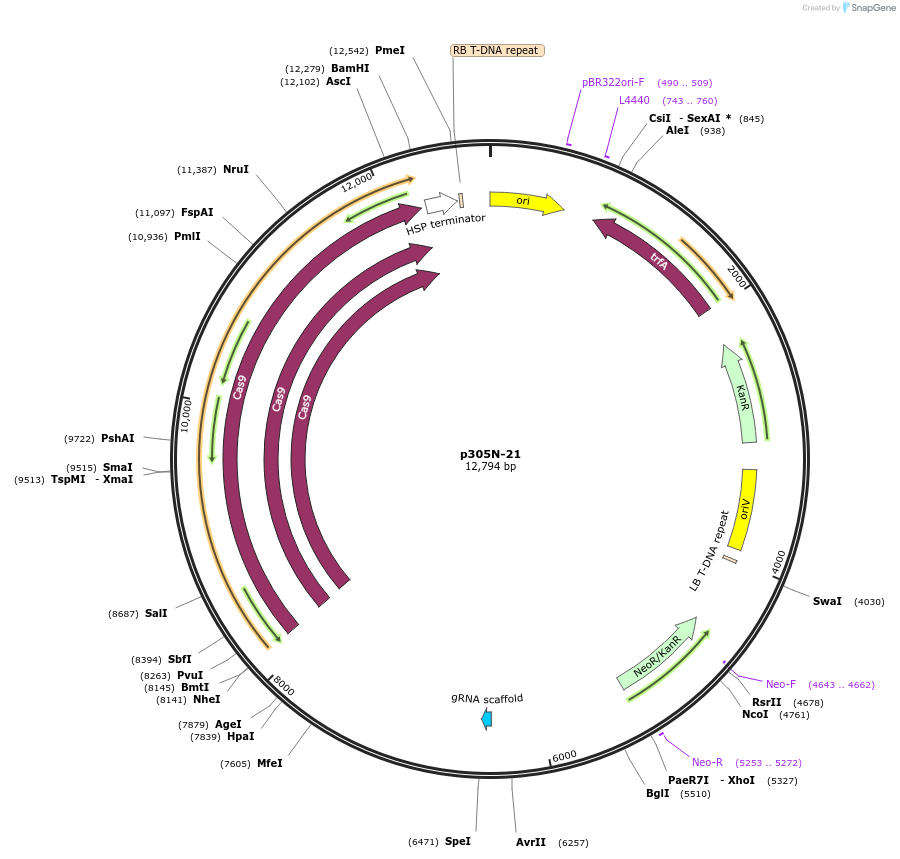
p305N-21
Plasmid#246304PurposeEvaluation of MtU6.6-104 promoter (Pol III promoter) deletion (104 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-104 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-104 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
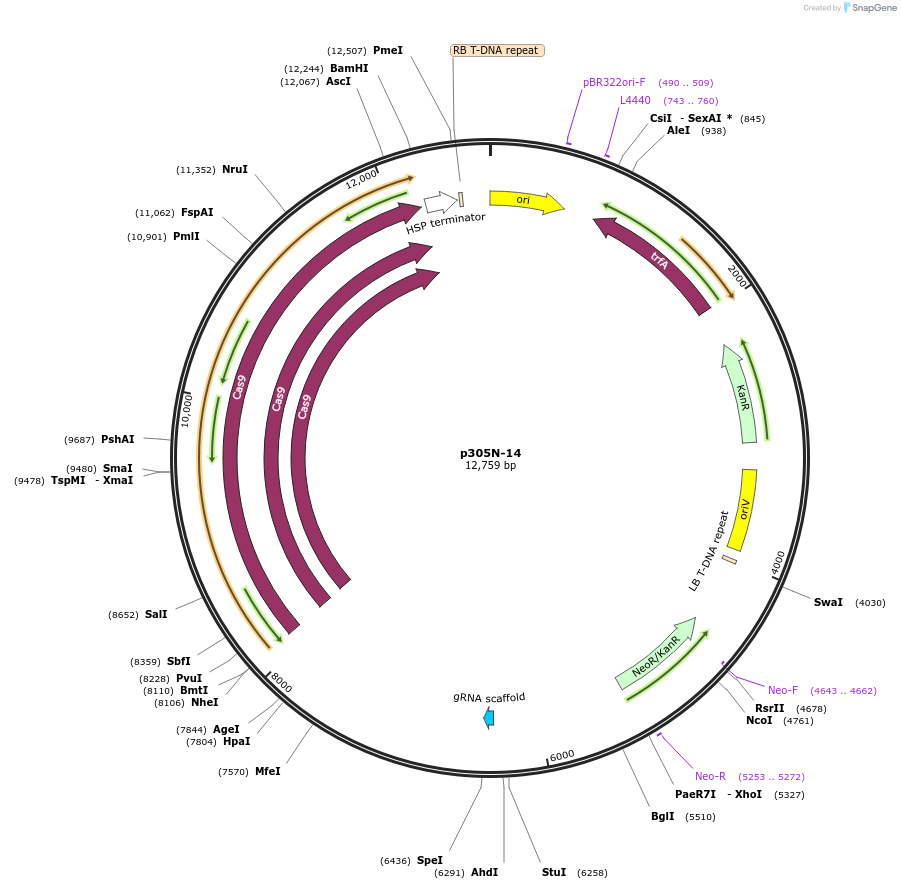
p305N-14
Plasmid#246297PurposeEvaluation of CiU6.6c16 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.6c16 promoter
UseCRISPRExpressionPlantMutationT-to-C (-2 from TSS)PromoterCiU6.6c16 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
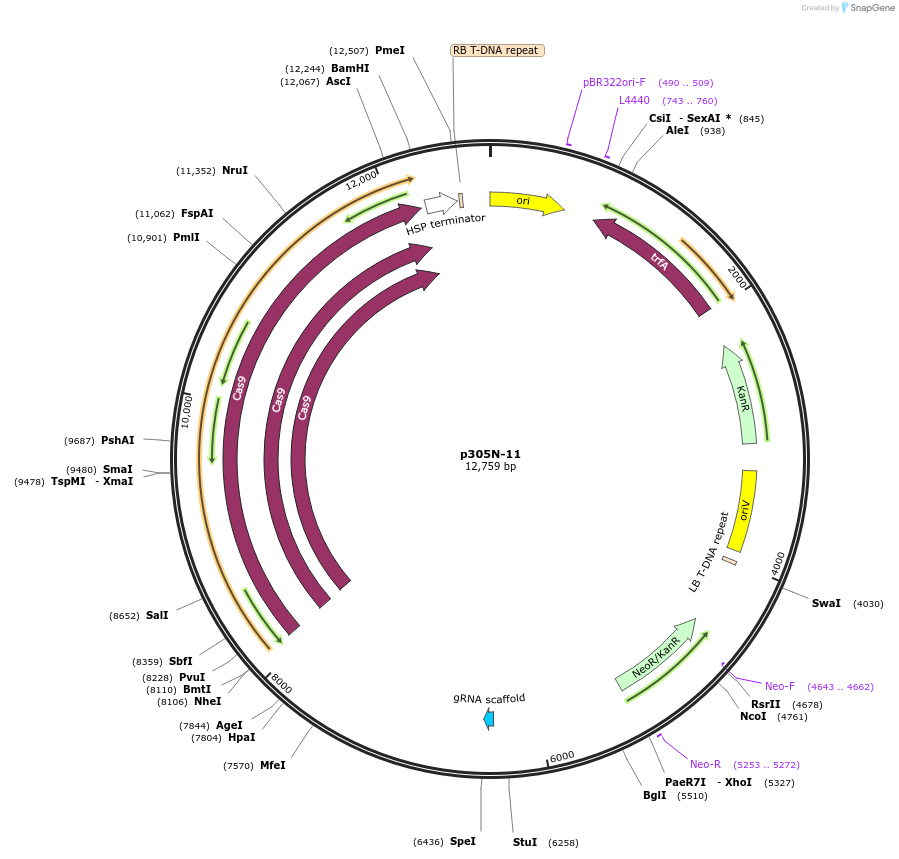
p305N-11
Plasmid#246294PurposeEvaluation of CiU6.3c8 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.3c8 promoter
UseCRISPRExpressionPlantMutationA-to-C (-23 from TSS)PromoterCiU6.3c8 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
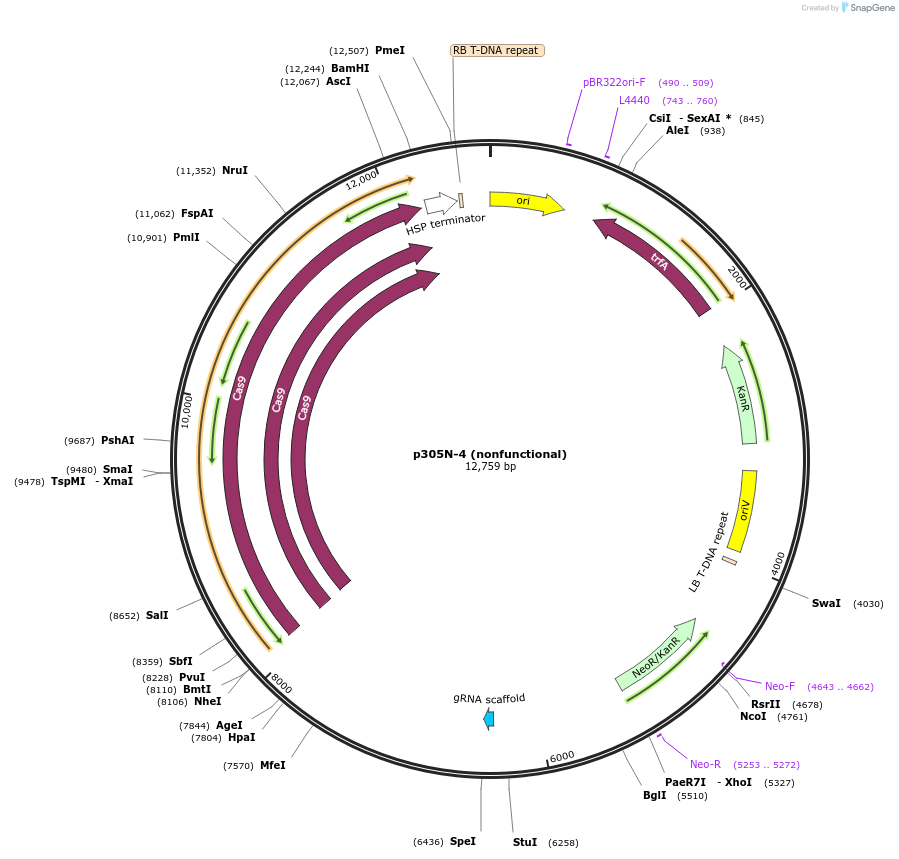
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
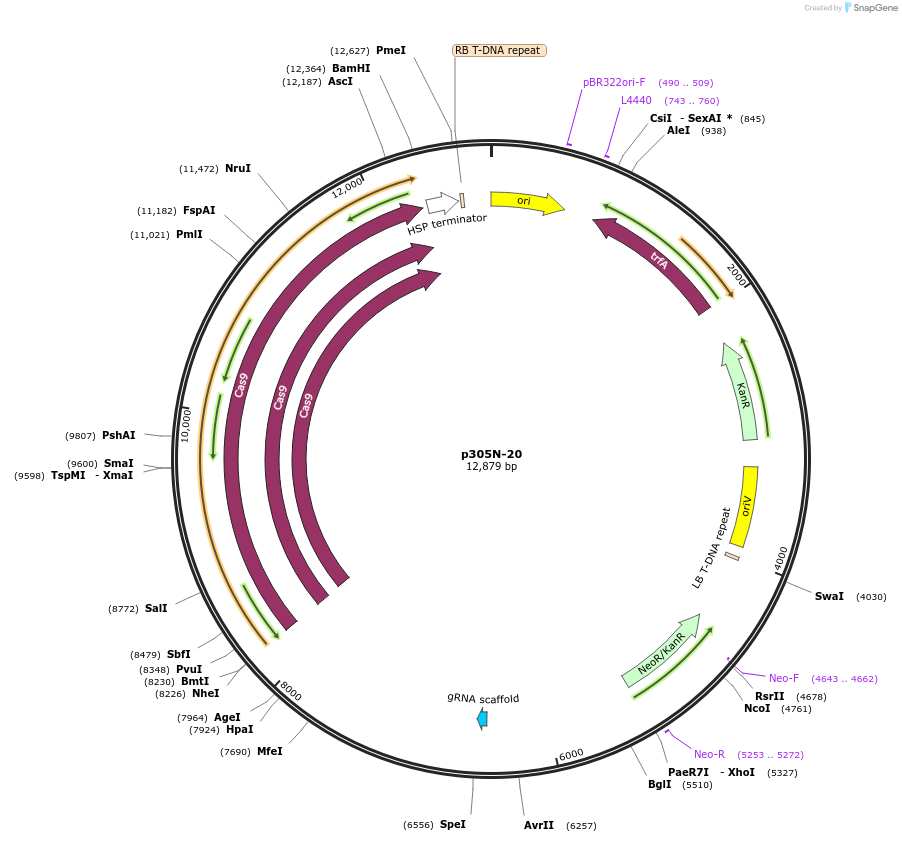
p305N-20
Plasmid#246303PurposeEvaluation of MtU6.6-189 promoter (Pol III promoter) deletion (189 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-189 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-189 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
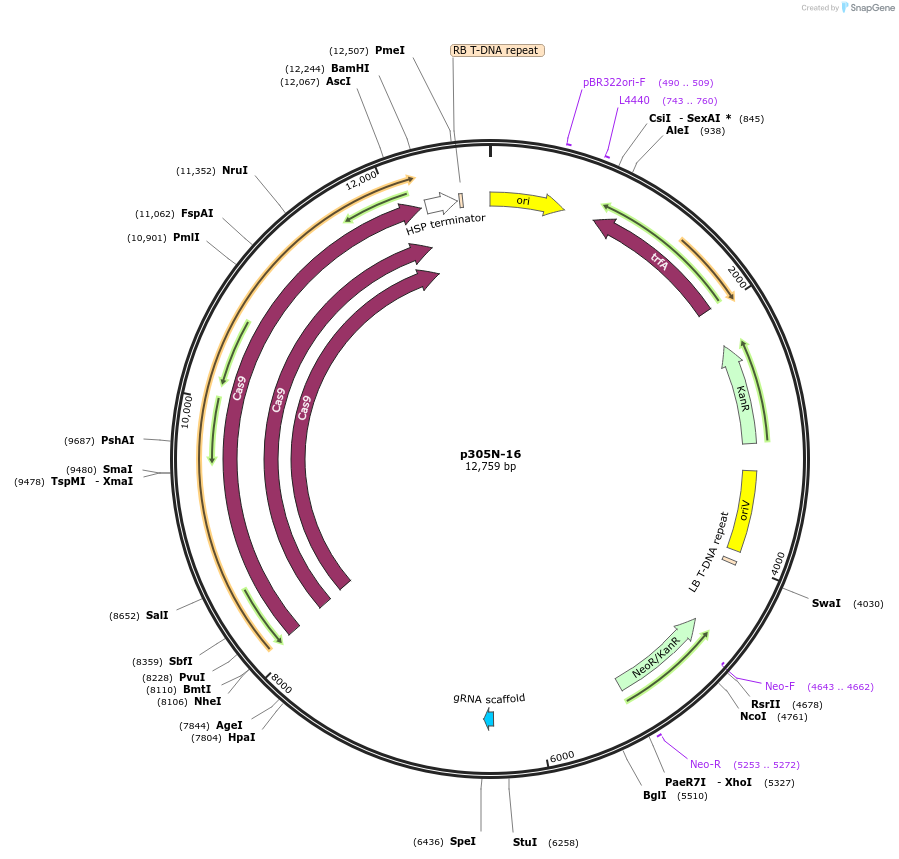
p305N-16
Plasmid#246299PurposeEvaluation of HbU6.2m1 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m1 promoter
UseCRISPRExpressionPlantMutationA-to-G (-56 from TSS), C-to-T (-29), G-to-A (-24)PromoterHbU6.2m1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
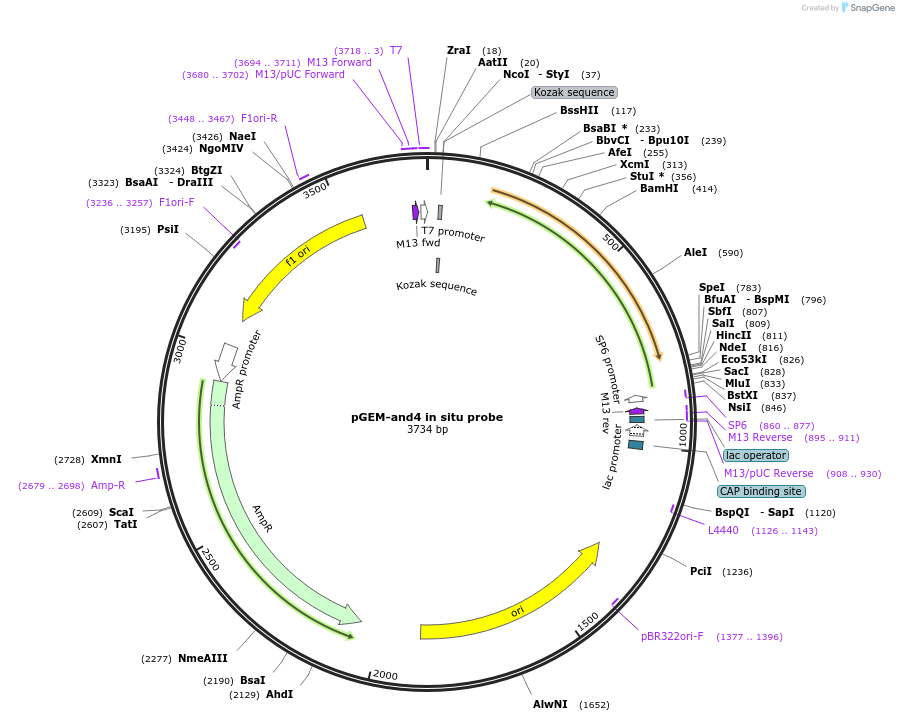
pGEM-and4 in situ probe
Plasmid#52105Purposeactinodin 4 and4 antisense in situ probe linearize w/ AatII SP6 polDepositorInsertactinodin 4 in situ probe (and4 Zebrafish)
ExpressionBacterialAvailable SinceApril 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
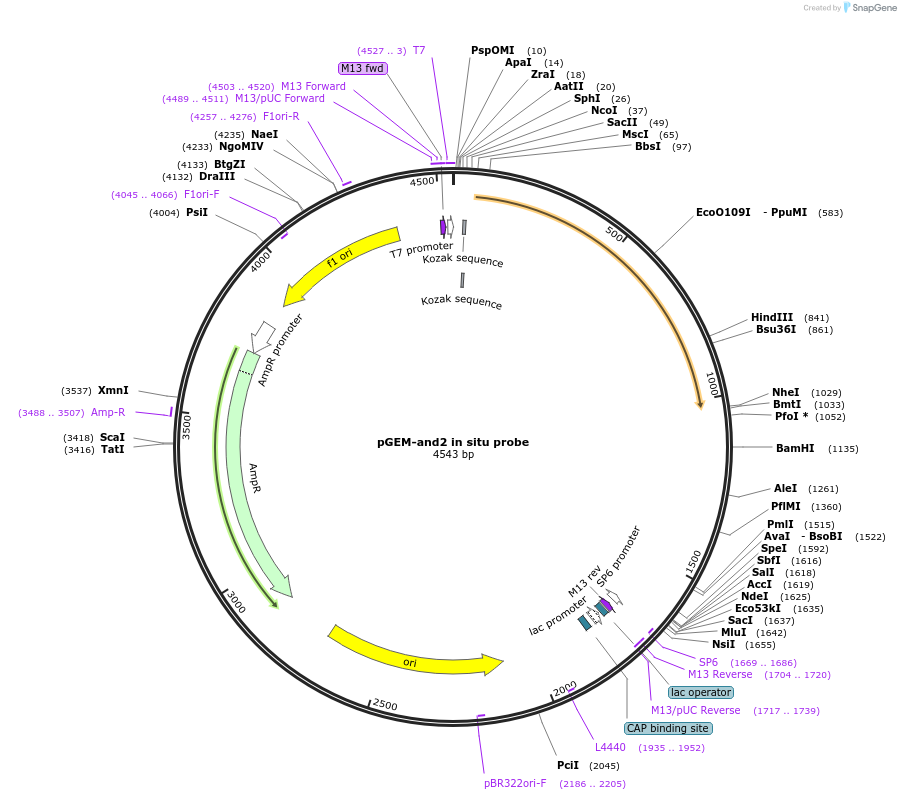
pGEM-and2 in situ probe
Plasmid#52103Purposeactinodin 2 and2 antisense in situ probe linearize w/ ApaI SP6 polDepositorInsertactinodin 2 in situ probe (and2 Zebrafish)
ExpressionBacterialAvailable SinceApril 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
Watermelon
Pooled Library#155257PurposeLineage tracing libraryDepositorExpressionMammalianUseLentiviralAvailable SinceOct. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
JS200 strain
Bacterial Strain#11794DepositorBacterial ResistanceNoneAvailable SinceJune 20, 2007AvailabilityAcademic Institutions and Nonprofits only -
Human CRISPR Poison Exon Knockout Library
Pooled Library#138084PurposeTargets 3' splice sites with paired gRNAs. Induces skipping of poison cassette exons and matching upstream constitutive exons. Compare loss of a constitutive coding exon with loss of a poison exon.DepositorSpeciesHomo sapiensUseCRISPR and LentiviralAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only