We narrowed to 11,850 results for: Vars
-
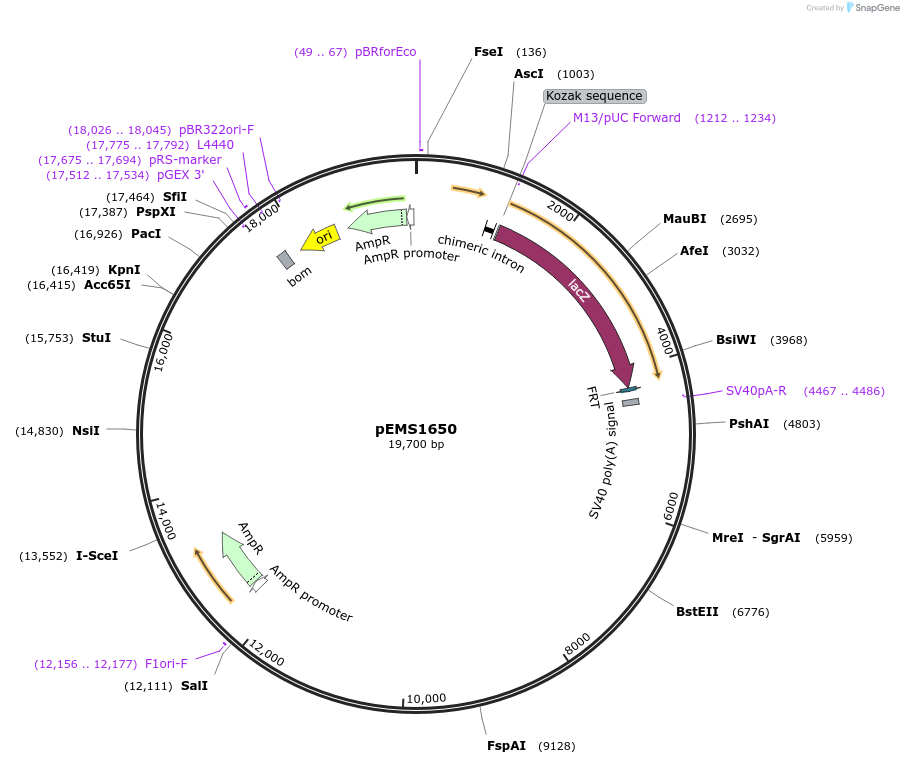
Plasmid#29285PurposeExpression of the reporter gene was not detected in the brain and eye using LacZ.DepositorInsertPle178 (RGS16 Human)
UsePleiades promoter project [sic, pleaides plieades]TagsIntron-LacZ-NLSAvailable SinceJune 10, 2011AvailabilityIndustry, Academic Institutions, and Nonprofits -
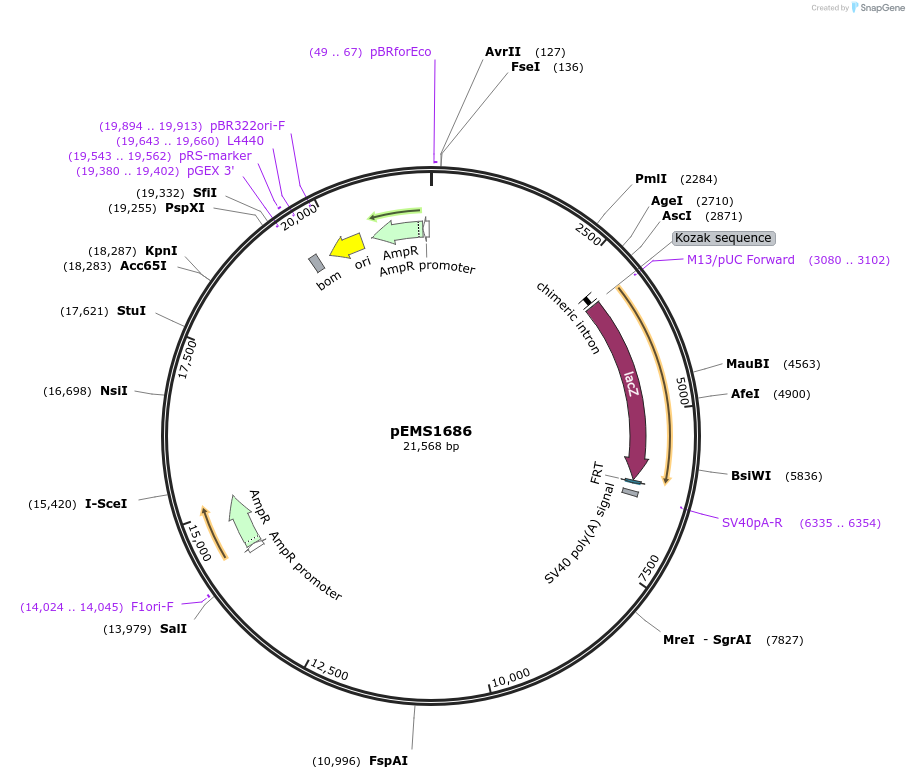
pEMS1686
Plasmid#29291PurposeExpression of the reporter gene was not detected in the brain and eye using LacZ.DepositorInsertPle214 (TAC1 Human)
UsePleiades promoter project [sic, pleaides plieades]TagsIntron-LacZ-NLSAvailable SinceJune 10, 2011AvailabilityIndustry, Academic Institutions, and Nonprofits -
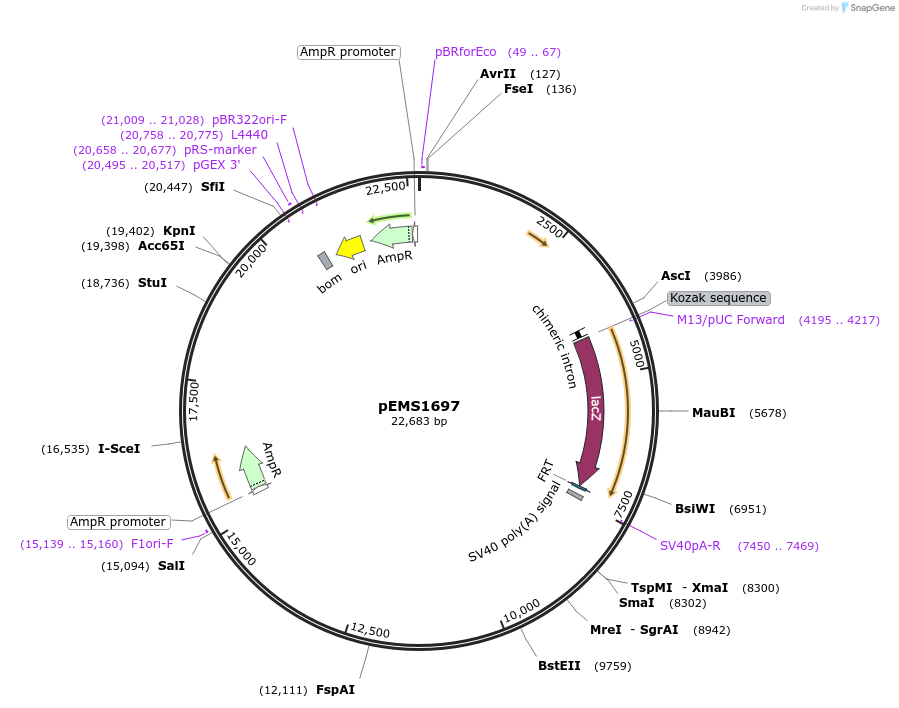
pEMS1697
Plasmid#29292PurposeExpression of the reporter gene under the control of the MiniPromoter in this plasmid has not been testedDepositorInsertPle225 (TBR1 Human)
UsePleiades promoter project [sic, pleaides plieades]TagsIntron-LacZ-NLSAvailable SinceJune 10, 2011AvailabilityIndustry, Academic Institutions, and Nonprofits -
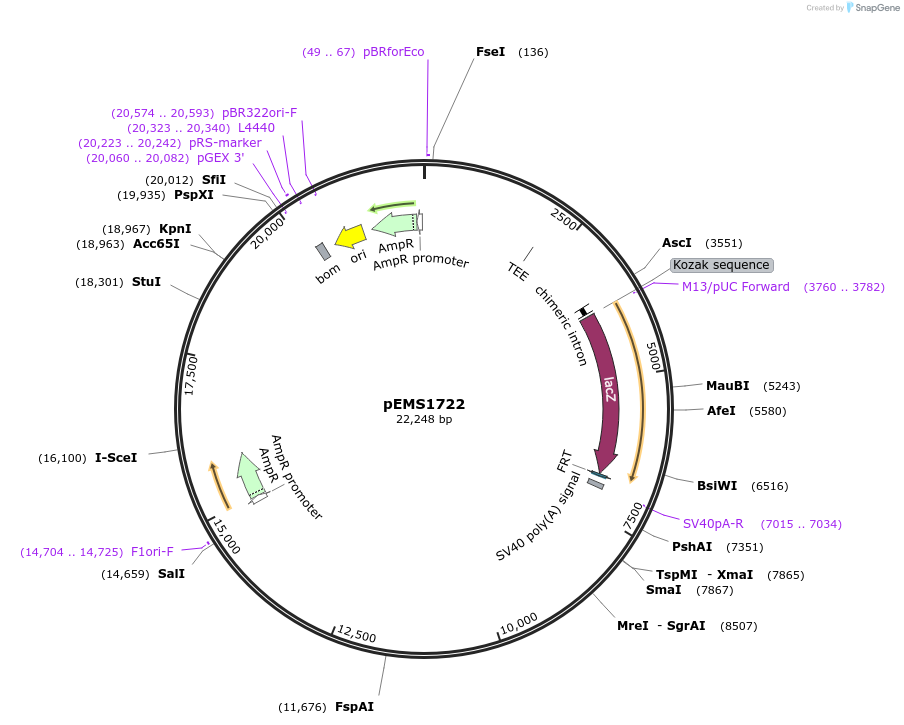
pEMS1722
Plasmid#29298PurposeExpression of the reporter gene was not detected in the brain and eye using LacZ.DepositorInsertPle250 (VIP Human)
UsePleiades promoter project [sic, pleaides plieades]TagsIntron-LacZ-NLSAvailable SinceJune 10, 2011AvailabilityIndustry, Academic Institutions, and Nonprofits -
C321.∆A.opt
Bacterial Strain#87359PurposeThis is C321.∆A with additional mutations engineered by Kuznetsov et al. to improve doubling time.DepositorBacterial ResistanceGentamicinSpeciesSyntheticAvailable SinceMay 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
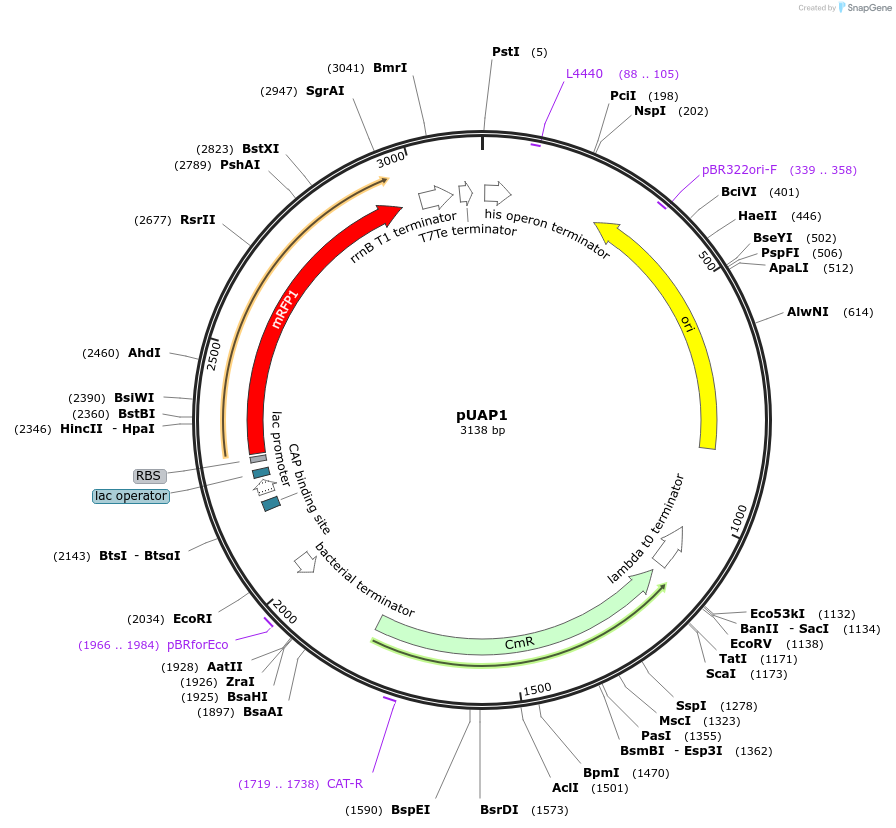
pUAP1
Plasmid#63674PurposeAccepts DNA sequences via BpiI cloning sites, resulting in Level 0 standard parts for Golden Gate cloning. Parts can be released with a user-defined 4-base-pair, 5 prime overhangs using BsaI.DepositorInsertRFP cloning selection cassette
UseSynthetic BiologyAvailable SinceApril 24, 2015AvailabilityAcademic Institutions and Nonprofits only