We narrowed to 4,663 results for: luciferase
-
Plasmid#63857PurposeThis pUC19 vector contains a tetramethylrosamine (TMR) binding riboswitch, controlling the translation initaition rate of luciferase gene.DepositorInsertTMR7 TMR riboswitch
UseSynthetic BiologyPromotertacAvailable SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
-
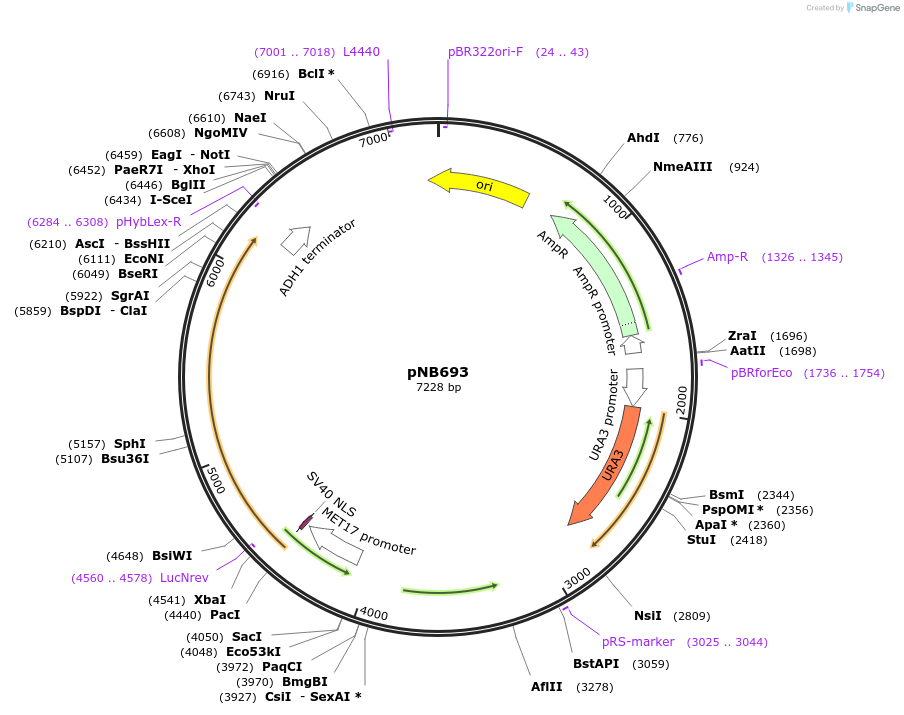
pNB693
Plasmid#60145PurposeDisintegrator plasmid encoding MET17 promoter driven yellow beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA).DepositorInsertYeLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterMET17prAvailable SinceDec. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
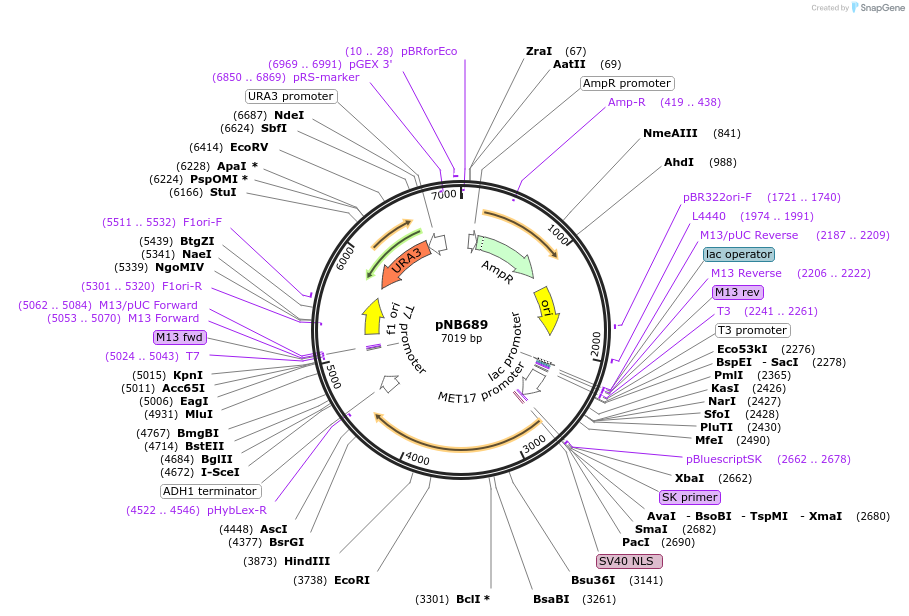
pNB689
Plasmid#60133PurposeGreen beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA), driven by MET17 promoter.DepositorInsertGrLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterMET17prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
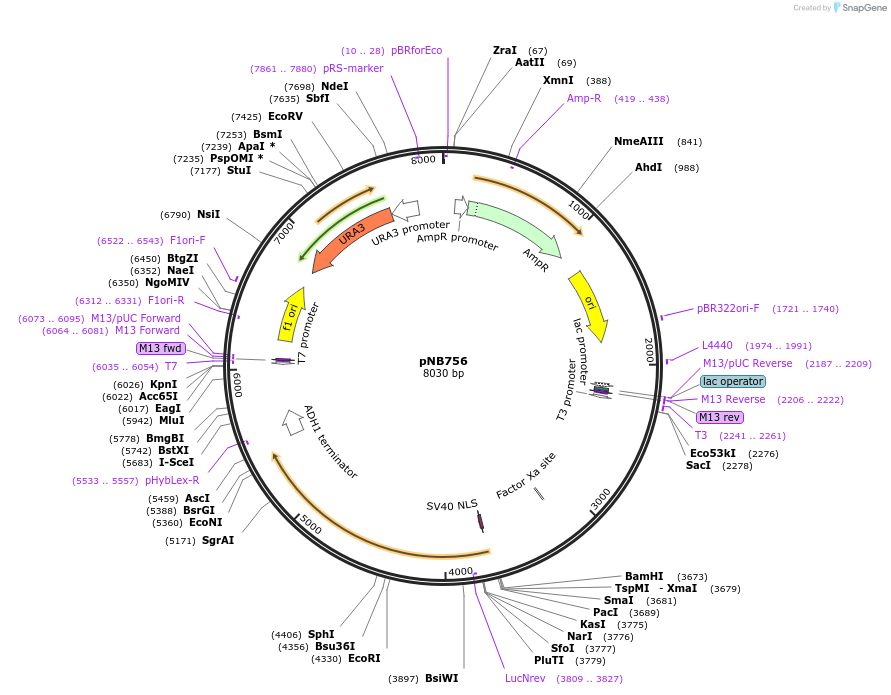
pNB756
Plasmid#60149PurposeYellow beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA), driven by LEU1 promoter.DepositorInsertYeLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterLEU1prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
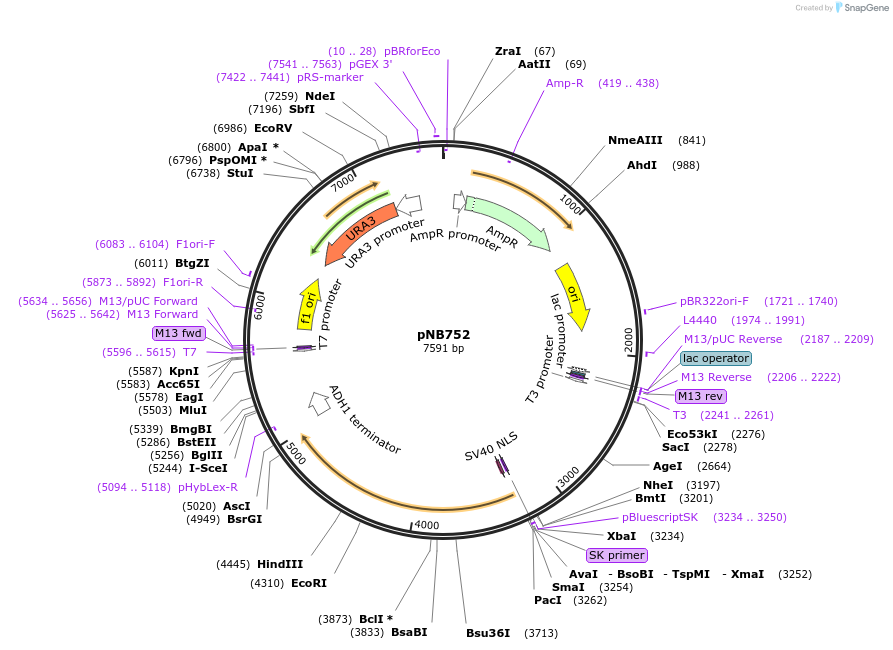
pNB752
Plasmid#60155PurposeGreen beetle luciferase with nuclear localization signal (NLS) and adjustable degron tag (ssrA), driven by ADE17 promoter.DepositorInsertGrLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterADE17prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
pGL2 enhancer- F6 Sp-1 mut
Plasmid#22439DepositorAvailable SinceFeb. 4, 2010AvailabilityAcademic Institutions and Nonprofits only -
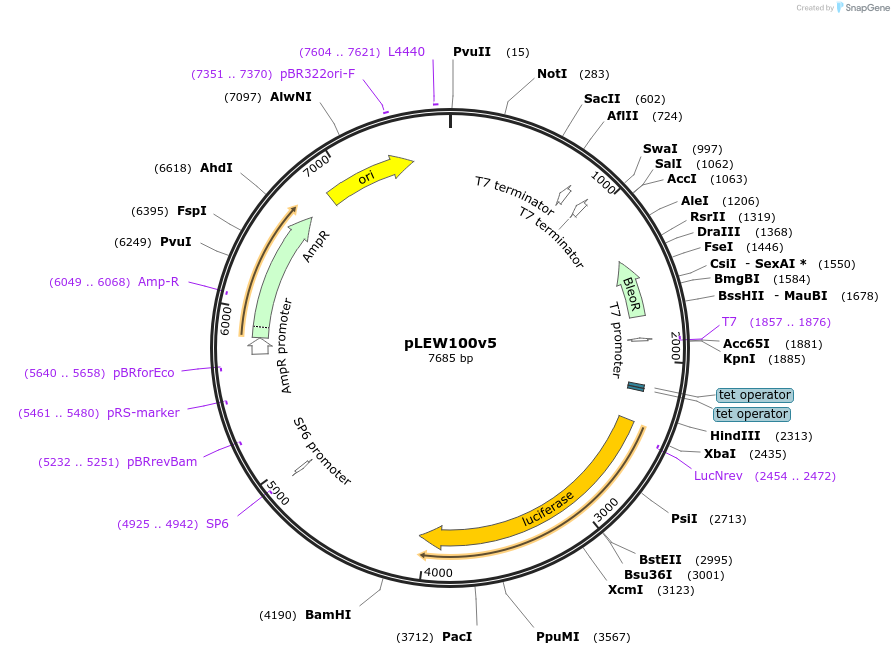
pLEW100v5
Plasmid#24011DepositorInsertLuciferase
UseTrypanosome expressionAvailable SinceFeb. 25, 2010AvailabilityAcademic Institutions and Nonprofits only -
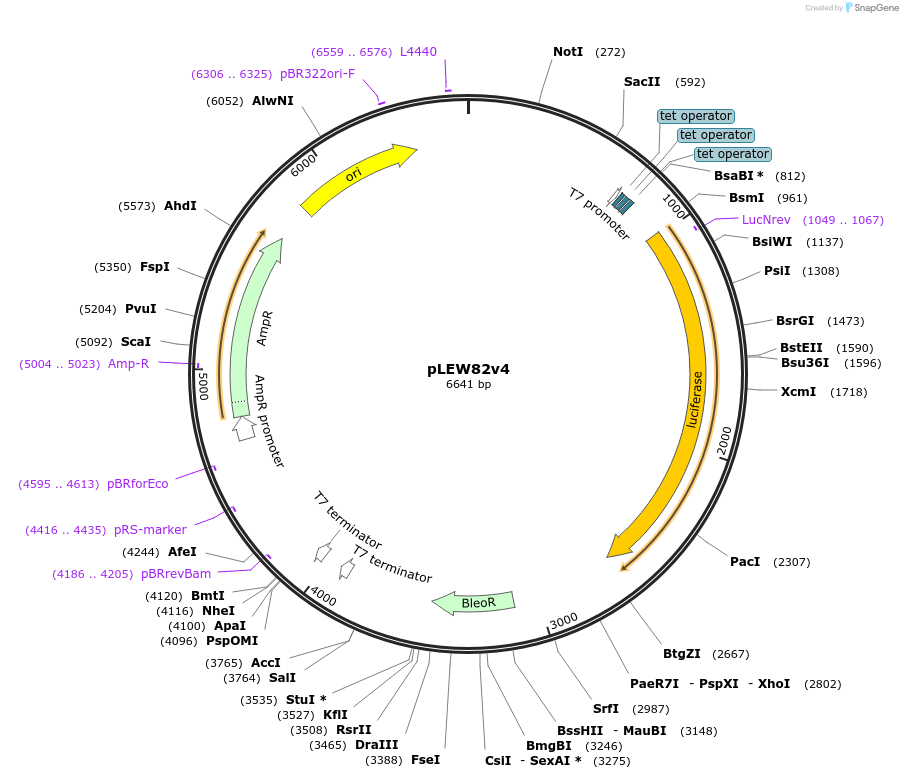
pLEW82v4
Plasmid#24009DepositorInsertLuciferase
UseTrypanosome expressionAvailable SinceApril 7, 2010AvailabilityAcademic Institutions and Nonprofits only -
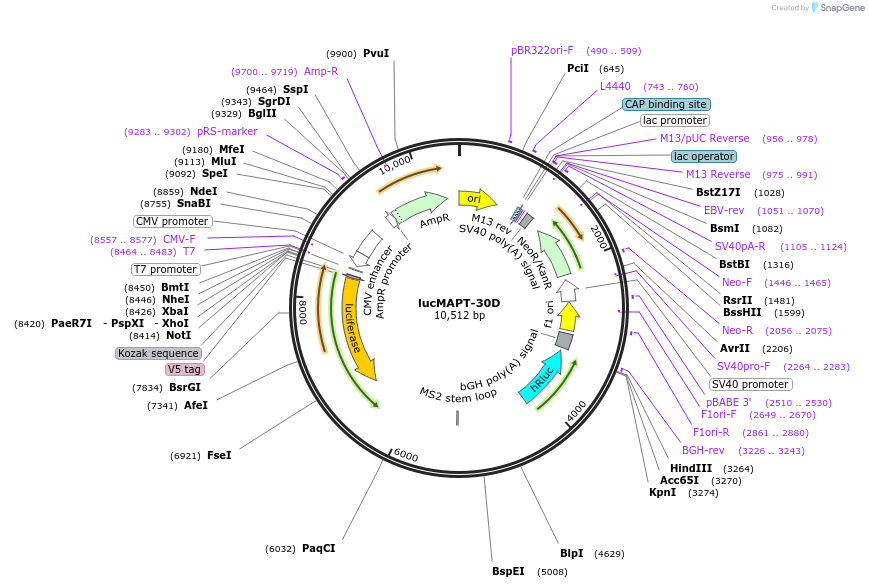
lucMAPT-30D
Plasmid#214672PurposeMAPT Exon 10 reporter containing the MS2 hairpin 30 base pairs downstream of the 5′ splice siteDepositorInsertMAPT (MAPT Human)
TagsFirefly Luciferase, Renilla Luciferase, and V5ExpressionMammalianMutationContains Exon 9, Exon 10 with a mutation to intro…PromoterCMVAvailable SinceMarch 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
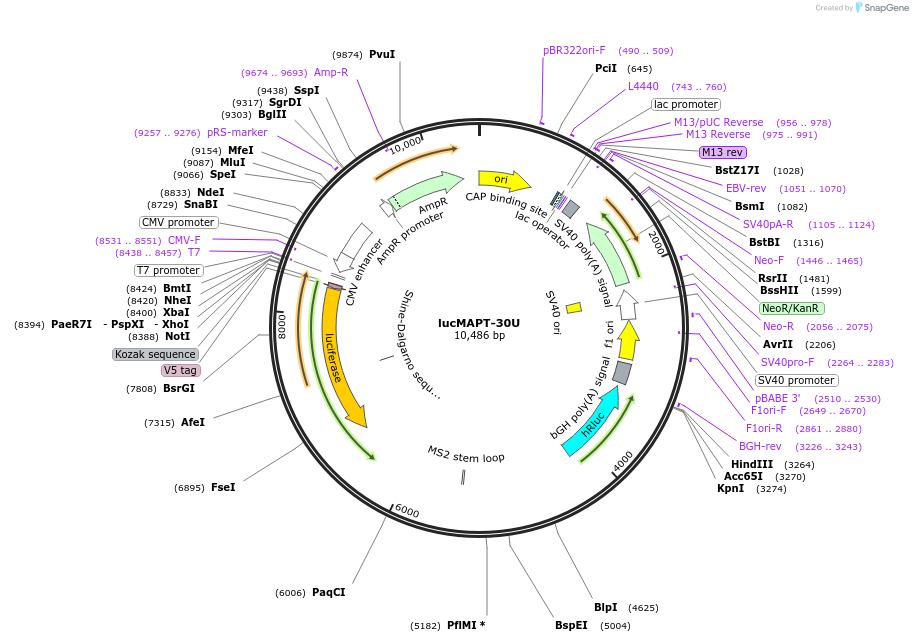
lucMAPT-30U
Plasmid#214673PurposeMAPT Exon 10 reporter containing the MS2 hairpin 30 base pairs upstream of the 3′ splice siteDepositorInsertMAPT (MAPT Human)
TagsFirefly Luciferase, Renilla Luciferase, and V5ExpressionMammalianMutationContains Exon 9, Exon 10 with a mutation to intro…PromoterCMVAvailable SinceMarch 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
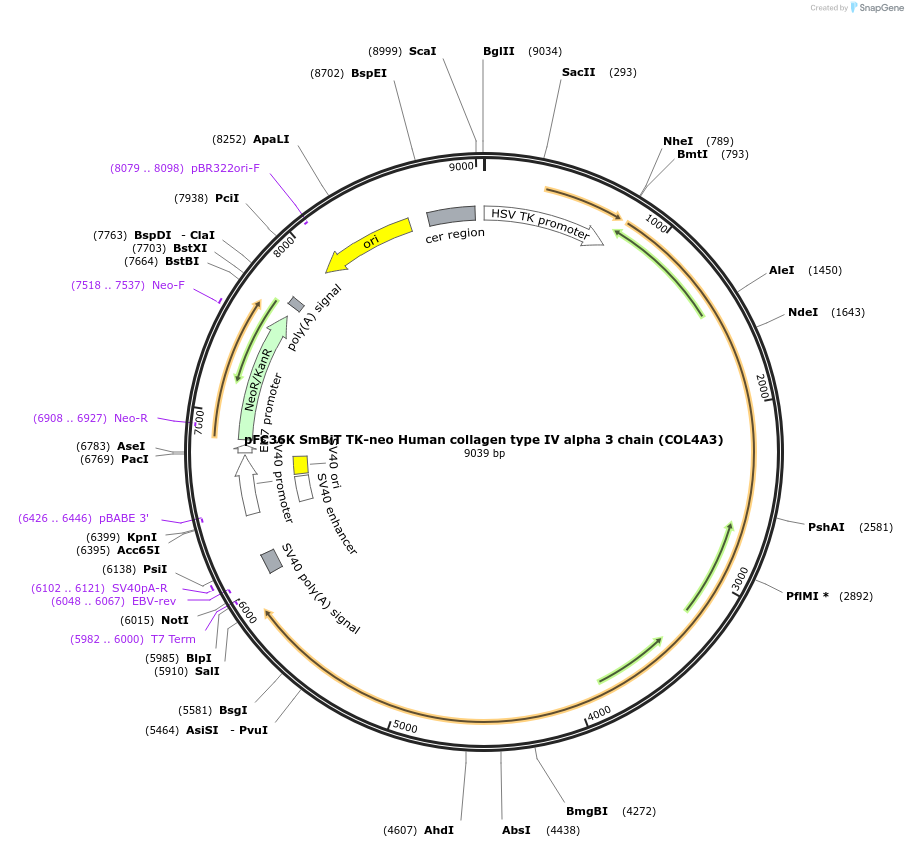
pFC36K SmBiT TK-neo Human collagen type IV alpha 3 chain (COL4A3)
Plasmid#229769PurposeHuman Col4A3 tagged at C-terminal with SmBiT to be used with C-tagged LgBiT hCol4A5 and Flag-tagged hCol4A4 in Nanobit assay systemDepositorAvailable SinceDec. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pF CAG luc-EGFP-cre puro
Plasmid#67503PurposeConstitutive expression of a firefly luciferase-enhanced green fluorescent protein-cre recombinase fusion protein in mammalian cellsDepositorInsertluciferase-EGFP-cre
UseCre/Lox, Lentiviral, and Luciferase ; Fluorescent…ExpressionMammalianPromoterCAGAvailable SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
pGL2 HNF4A P2 mut
Plasmid#60325PurposeMutated HNF4A P2 promoter region cloned into the pGL2-basic backbone. The -181 G to A mutation is in a HNF1alpha binding site and cosegregates with MODY.DepositorInsertHNF4A P2 mut (-181 G to A) (HNF4A Human)
UseLuciferaseTagsLuciferaseExpressionMammalianMutationThe -181 G to A mutation is in a HNF1alpha bindin…Available SinceJan. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
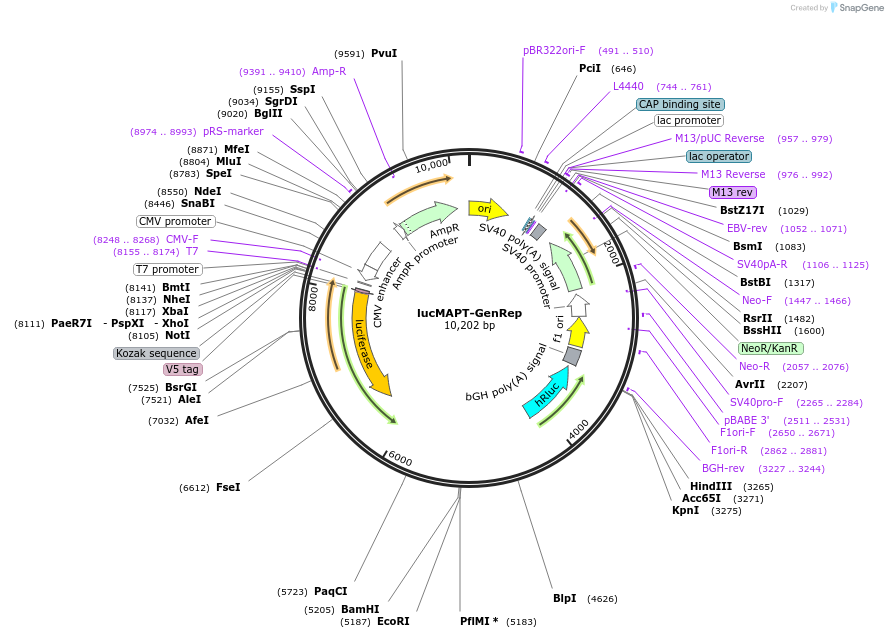
lucMAPT-GenRep
Plasmid#214674PurposeMAPT Exon 10 reporter, with Exon 10 and 100bp flanking intronic regions replaced with a BamHI/EcoRI cloning site for testing different exonsDepositorInsertMAPT (MAPT Human)
TagsFirefly Luciferase, Renilla Luciferase, and V5ExpressionMammalianMutationContains Exon 9 and Exon 11, with a BamHI EcoRI c…PromoterCMVAvailable SinceMarch 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
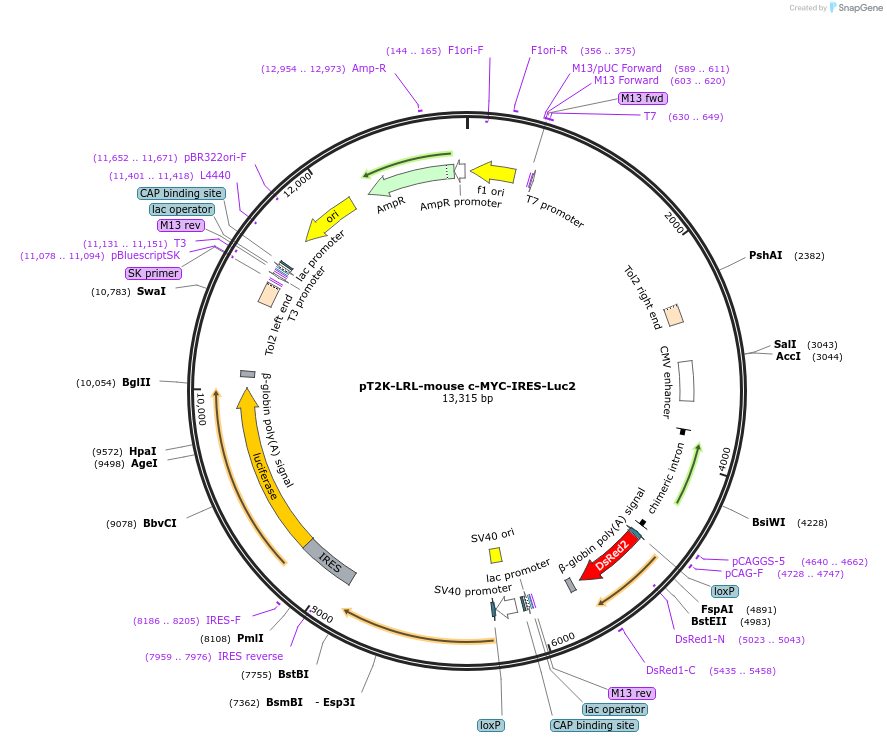
pT2K-LRL-mouse c-MYC-IRES-Luc2
Plasmid#109228PurposeFor Tol2 transposon-mediated stable expression of LRL, c-Myc and IRES-luciferaseDepositorAvailable SinceJune 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
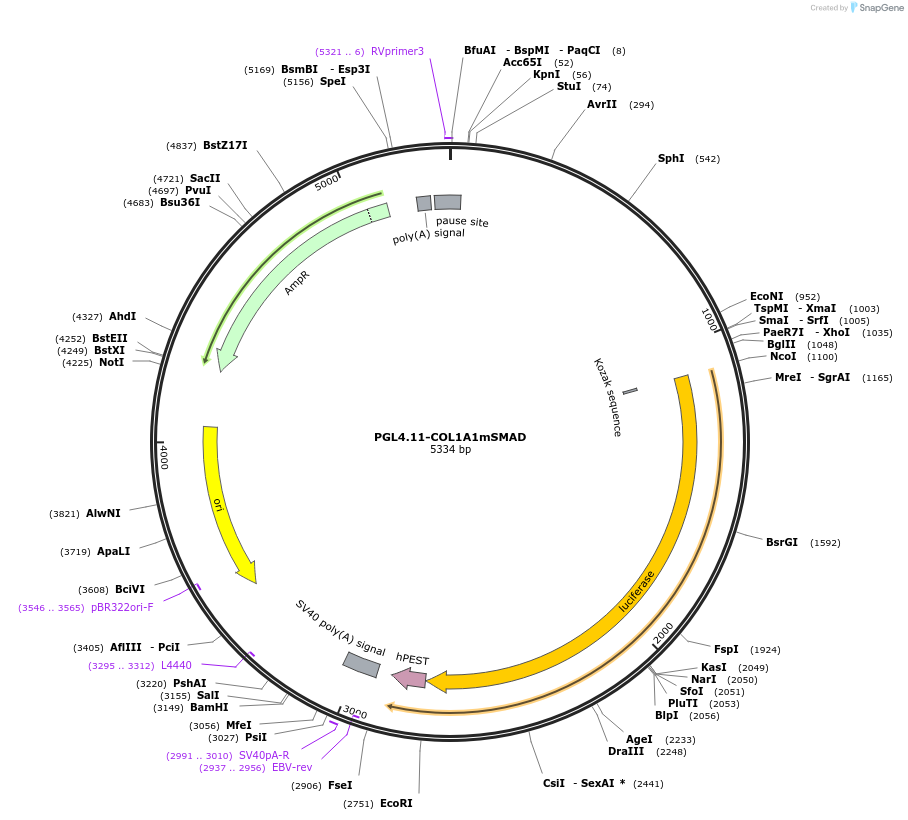
PGL4.11-COL1A1mSMAD
Plasmid#230917PurposeTested the COL1A1 promoter activity after abolition of the proximal SMAD binding sites in the promoter.DepositorInsertHuman COL1A1 promoter with mutant SMAD binding sequence (COL1A1 Human)
UseLuciferaseTagsLuciferase-luc2pExpressionBacterial and MammalianMutationCOL1A1 promoter with mutant potential SMAD bindin…PromoterCOL1A1Available SinceFeb. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
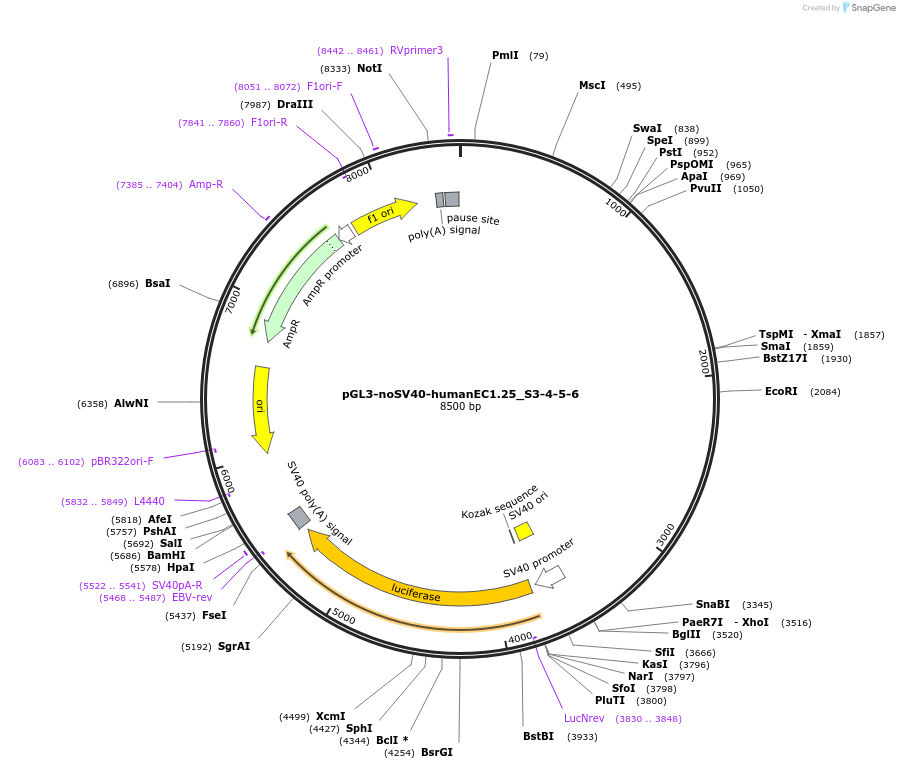
pGL3-noSV40-humanEC1.25_S3-4-5-6
Plasmid#173956PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer regions 3-6 (S3, S4, S5 and S6) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
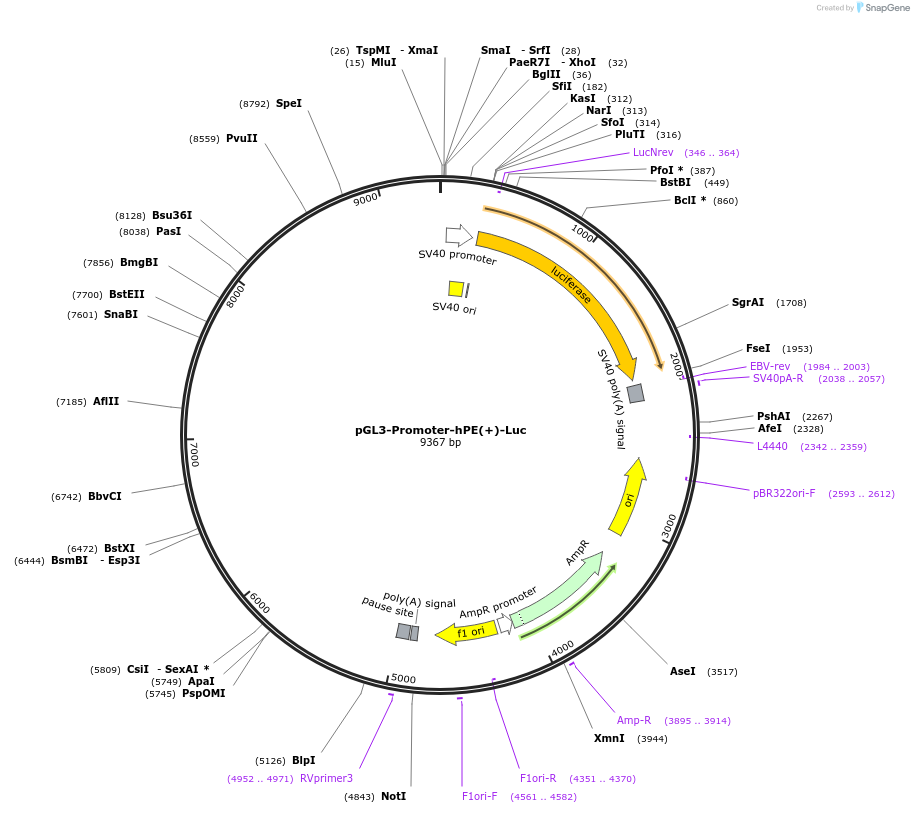
pGL3-Promoter-hPE(+)-Luc
Plasmid#172003PurposeLuciferase Reporter for Human PTEN Enhancer PE - Forward OrientationDepositorInsertHuman PE sequence (hPE)
UseLuciferaseExpressionBacterial and MammalianPromoterSV40 PromoterAvailable SinceJuly 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
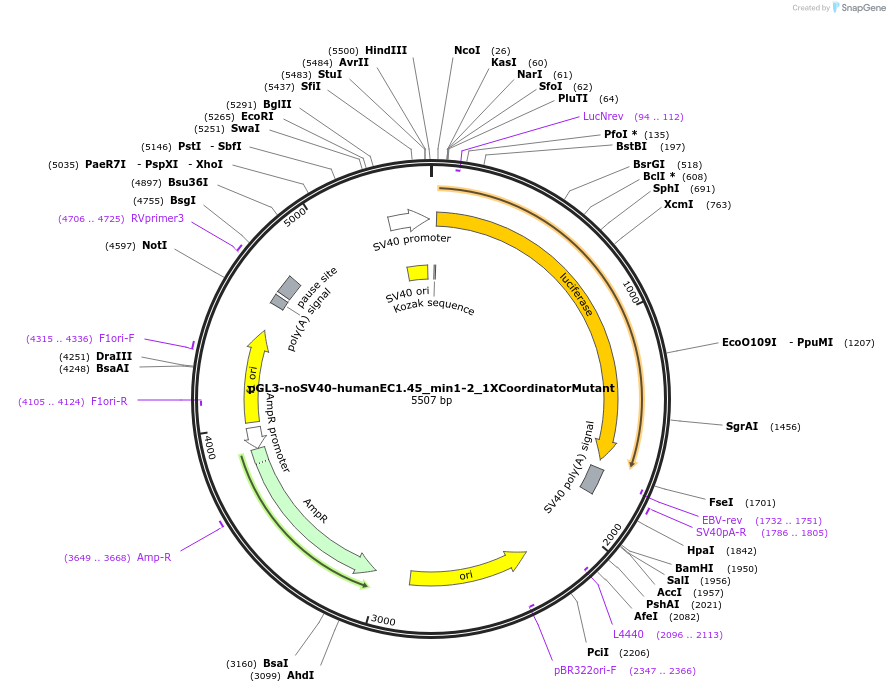
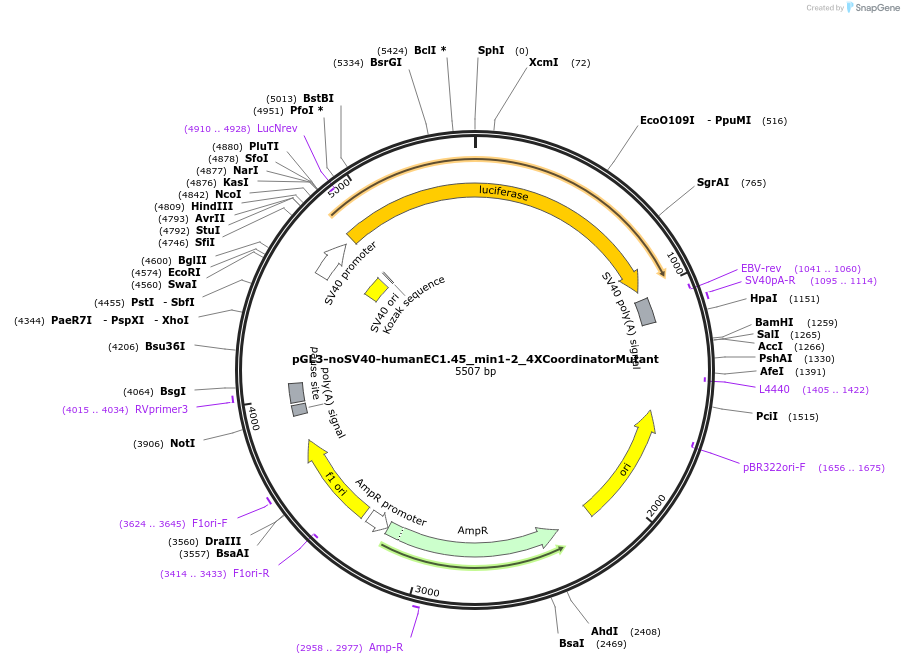
pGL3-noSV40-humanEC1.45_min1-2_1XCoordinatorMutant
Plasmid#173963PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer regions 1 and 2 (min1 and min2) from SOX9 enhancer cluster EC1.45 with 1 mutated Coordinator motif
UseLuciferaseMutation1 mutated Coordinator motifPromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
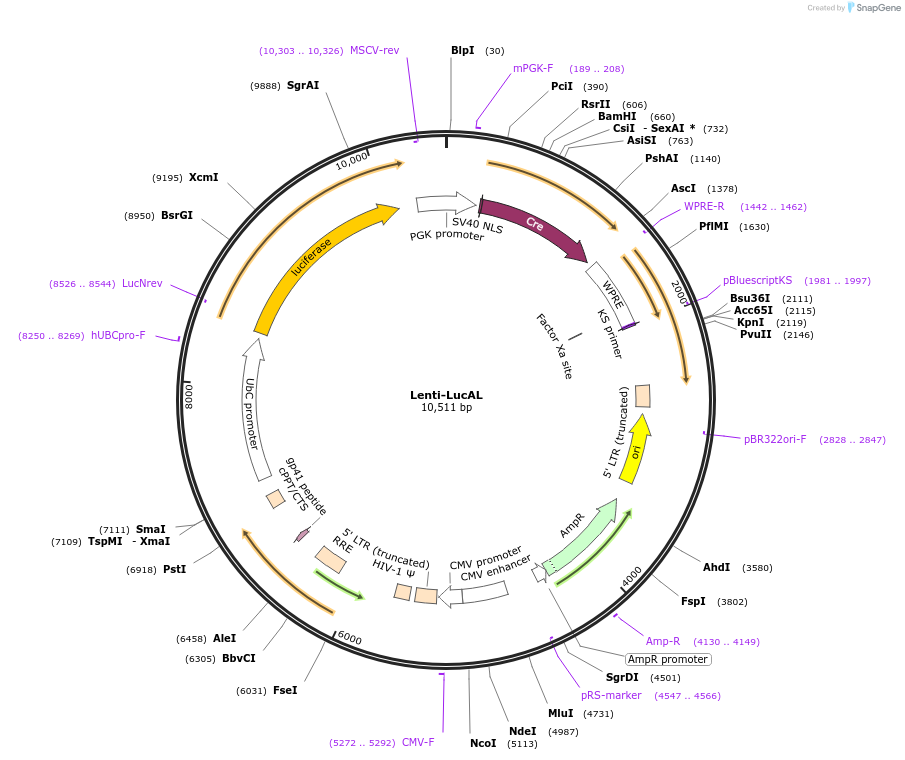
Lenti-LucAL
Plasmid#174049PurposeExpresses mALG8 (ITYTWTRL) and mLAMA4 (VGFNFRTL) antigens as a fusion to luciferase and Cre recombinaseDepositorInsertLucAL
UseCre/Lox, Lentiviral, and LuciferaseTagsLuciferaseExpressionMammalianMutationALG8 alanine 506 to threonine; LAMA4 glycine 1254…PromoterHuman Ubiquitin CAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
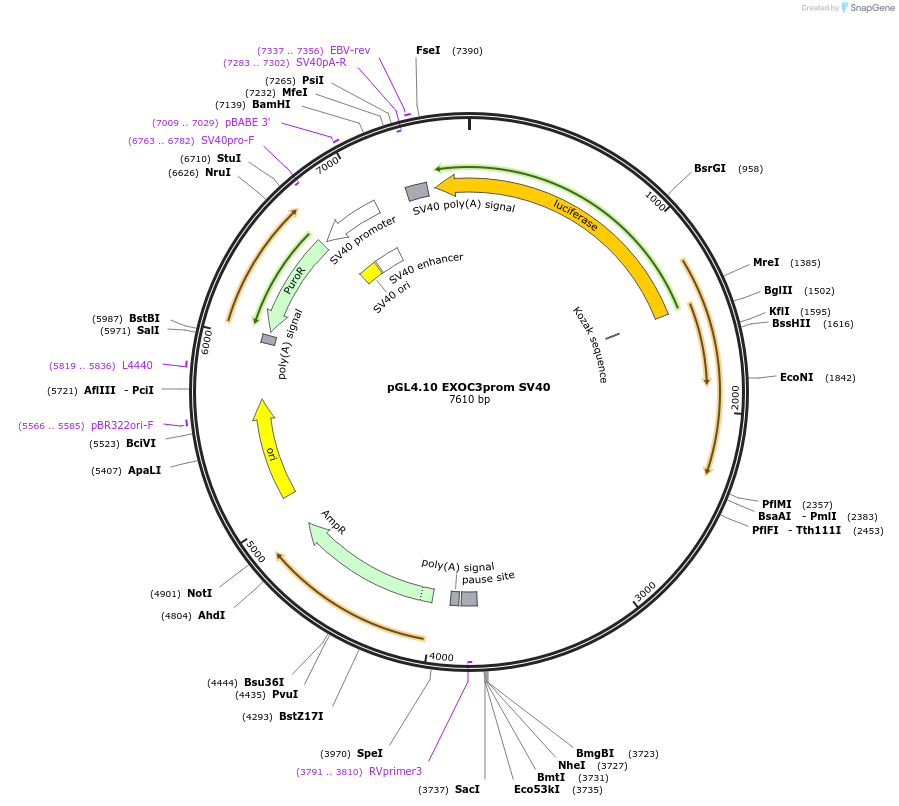
pGL4.10 EXOC3prom SV40
Plasmid#246713PurposeTo investigate the influence of a universal enhancer (SV40) on EXOC3 promoter activity (measured by luciferase expression)DepositorAvailable SinceNov. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
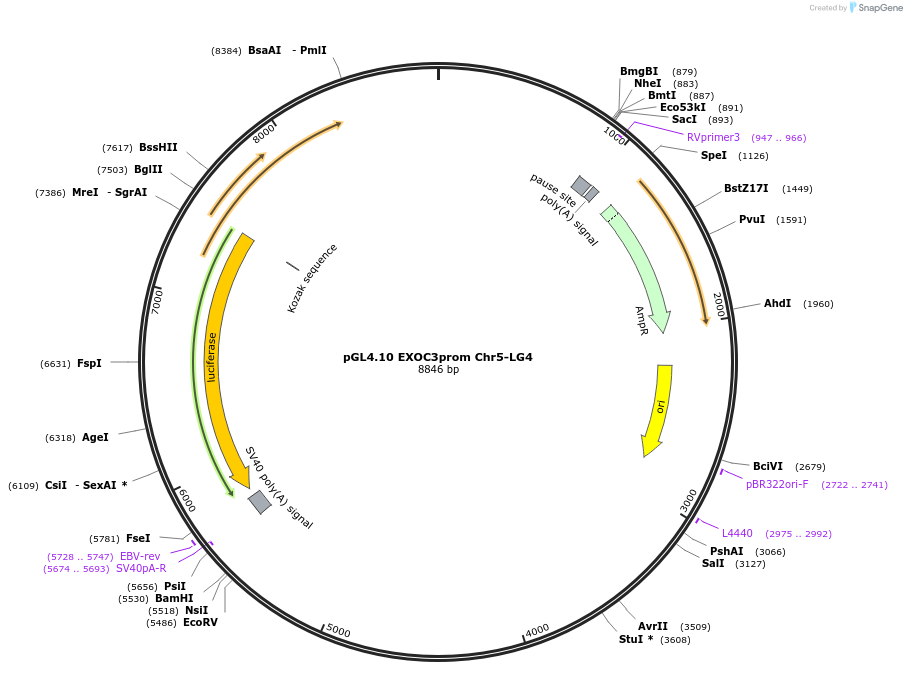
pGL4.10 EXOC3prom Chr5-LG4
Plasmid#246712PurposeTo investigate the influence of an LG4 enhancer (chr5 LG4) on EXOC3 promoter activity (measured by luciferase expression)DepositorAvailable SinceNov. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
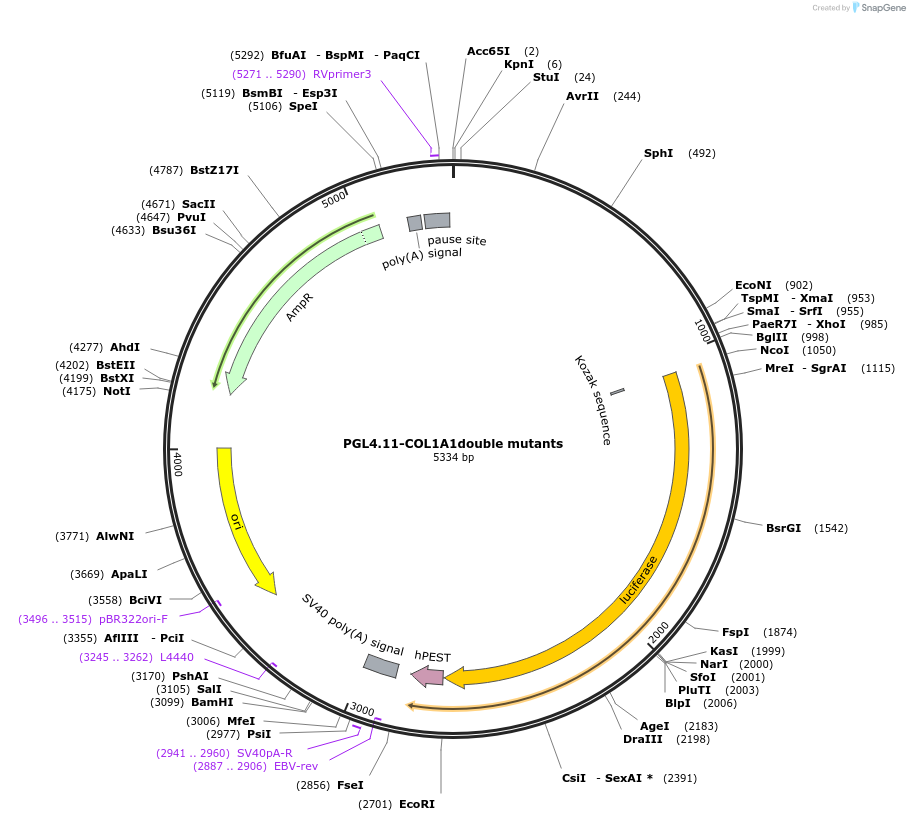
PGL4.11-COL1A1double mutants
Plasmid#230918PurposeDouble mutation of TEAD and SMAD binding elements in COL1A1 promoter; check the promoter activity.DepositorInsertHuman CLO1A1 promoter including SMAD and TEAD mutant elements (COL1A1 Human)
UseLuciferaseTagsLuciferase-luc2pExpressionBacterial and MammalianMutationCOL1A1 promoter with mutant SMAD and TEAD binding…PromoterCOL1A1 with mutant SMAD and TEAD elementsAvailable SinceFeb. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
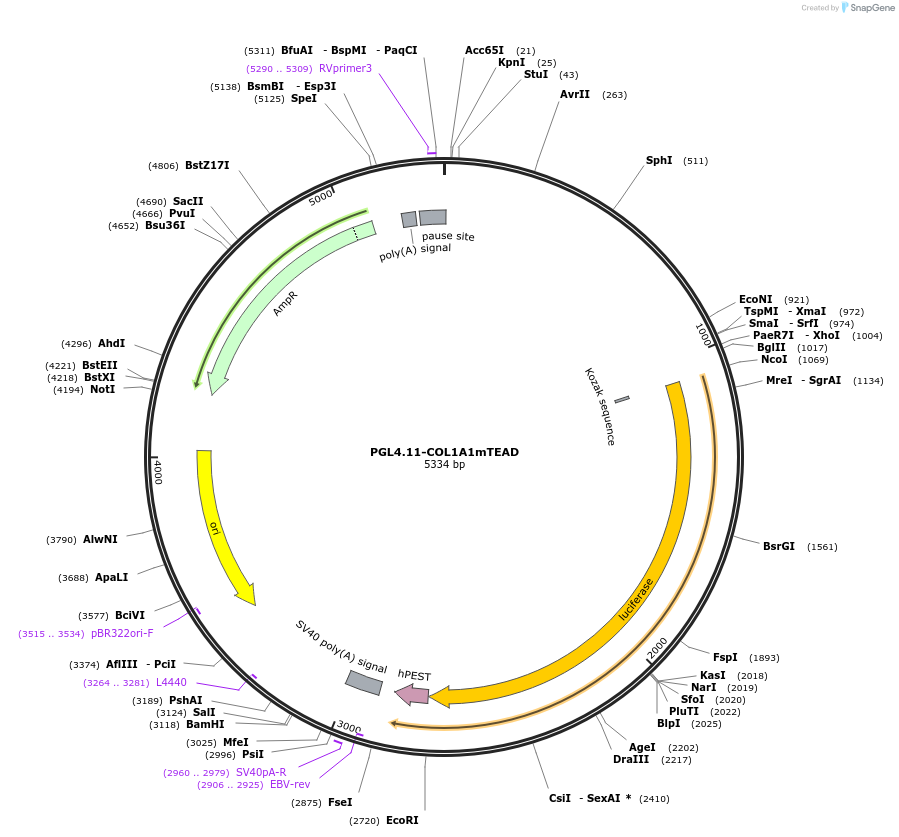
PGL4.11-COL1A1mTEAD
Plasmid#230916PurposeComparing with wild type COL1A1 promoter, tested whether mutant TEAD binding sites changed the promoter activity.DepositorInsertHuman COL1A1 promoter with mutant TEAD elements (COL1A1 Human)
UseLuciferaseTagsLuciferase- luc2pExpressionBacterial and MammalianMutationmutant potential TEAD bind elements AGGAAT to CTG…PromoterCOL1A1Available SinceFeb. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
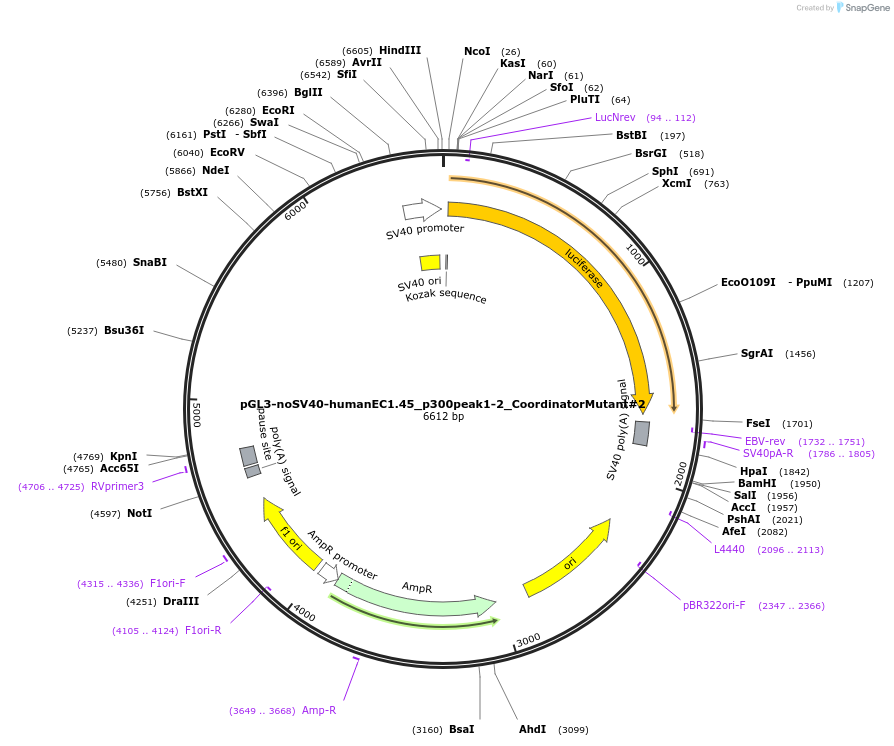
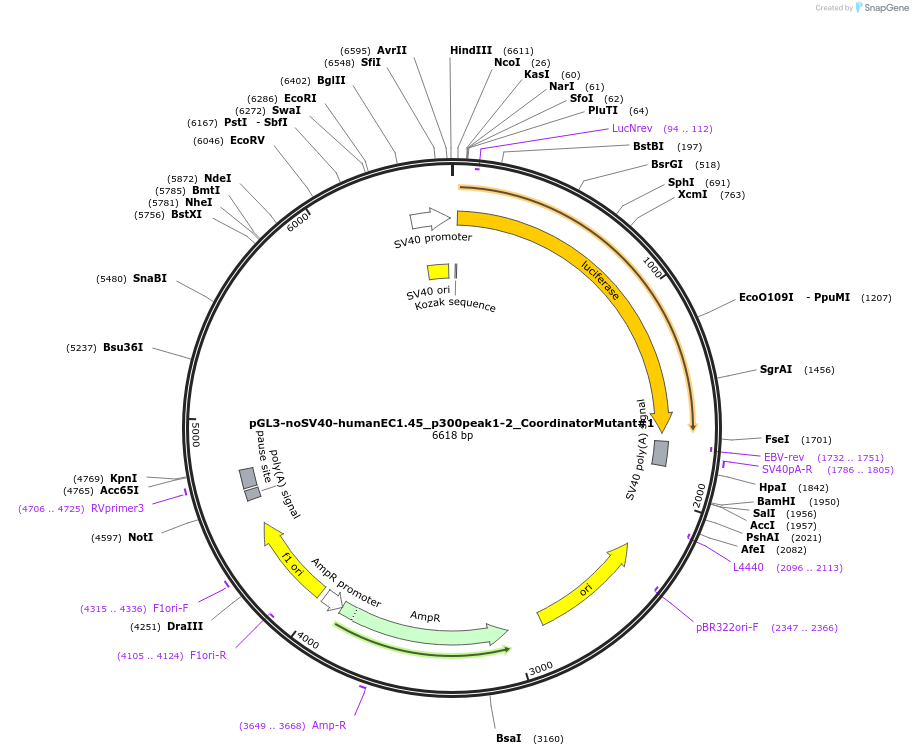
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#2
Plasmid#173986PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #2 mutated
UseLuciferaseMutationCoordinator motif mutation #2PromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
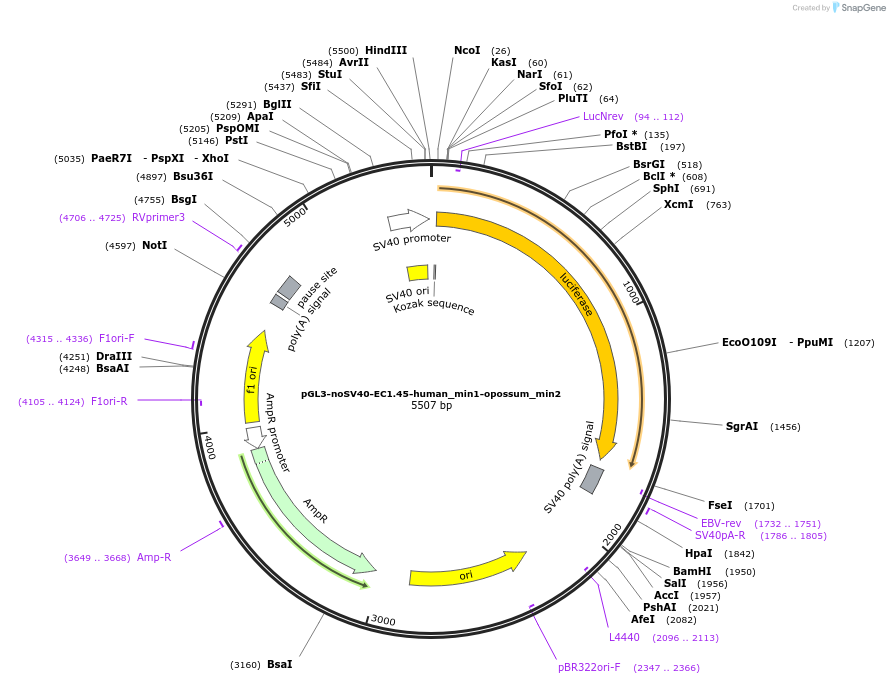
pGL3-noSV40-EC1.45-human_min1-opossum_min2
Plasmid#173966PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and opossum minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationSee Depositor Comments BelowPromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
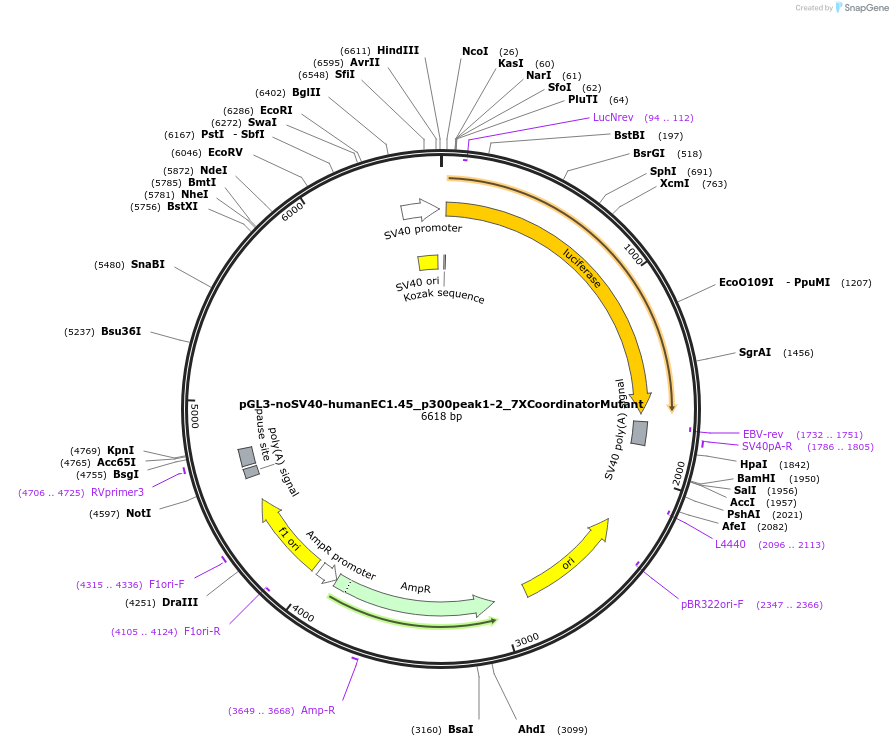
pGL3-noSV40-humanEC1.45_p300peak1-2_7XCoordinatorMutant
Plasmid#173994PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with all 7 Coordinator motifs mutated
UseLuciferaseMutation7X Coordinator motif mutation #1-2-3-4-5-6-7PromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_min1-2_4XCoordinatorMutant
Plasmid#173962PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer regions 1 and 2 (min1 and min2) from SOX9 enhancer cluster EC1.45 with 4 mutated Coordinator motifs
UseLuciferaseMutation4 mutated Coordinator motifsPromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#1
Plasmid#173985PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #1 mutated
UseLuciferaseMutationCoordinator motif mutation #1PromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only