We narrowed to 11,709 results for: chia
-
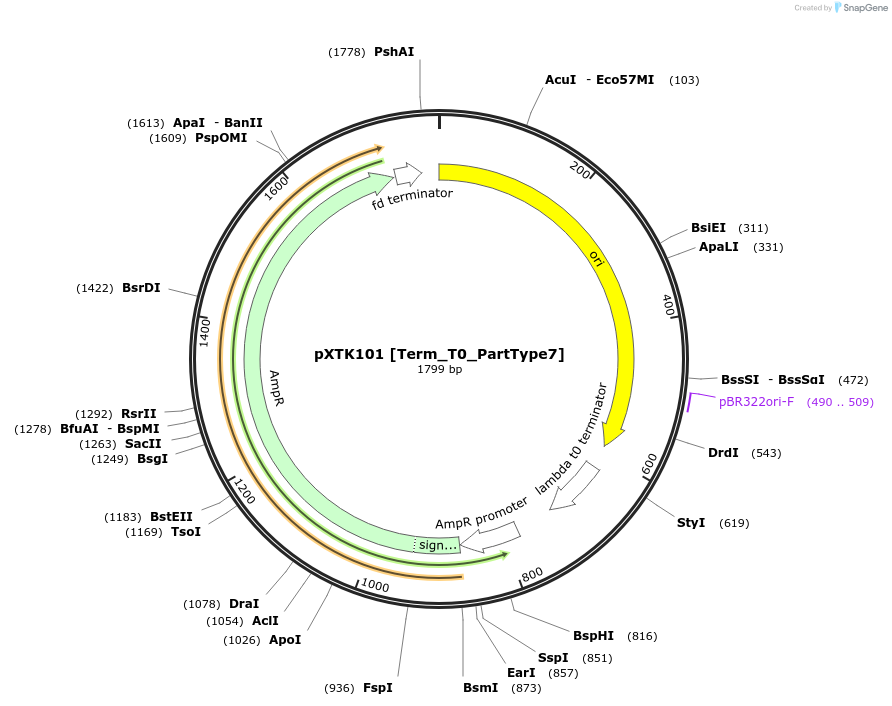
Plasmid#229338PurposeXanthoMoClo Golden Gate Part Plasmid for cloning, contains Term_T0_PartType7DepositorInsertT0 Terminator
ExpressionBacterialAvailable SinceMarch 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pXTK102 [Term_T1_PartType7]
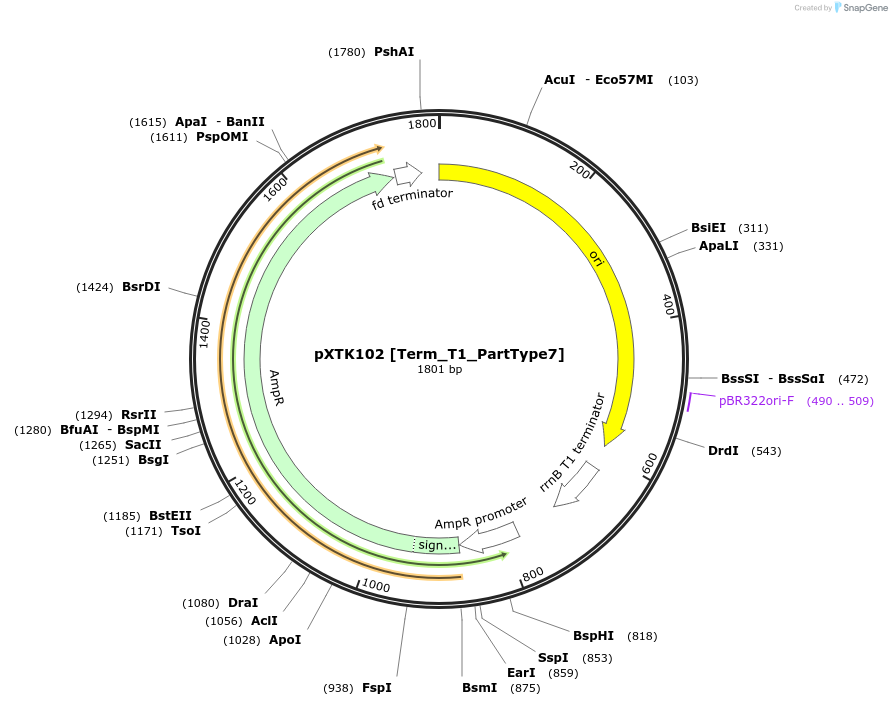
Plasmid#229339PurposeXanthoMoClo Golden Gate Part Plasmid for cloning, contains Term_T1_PartType7DepositorInsertT1 Terminator
ExpressionBacterialAvailable SinceMarch 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pXTK103 [Term_T7Te_PartType7]
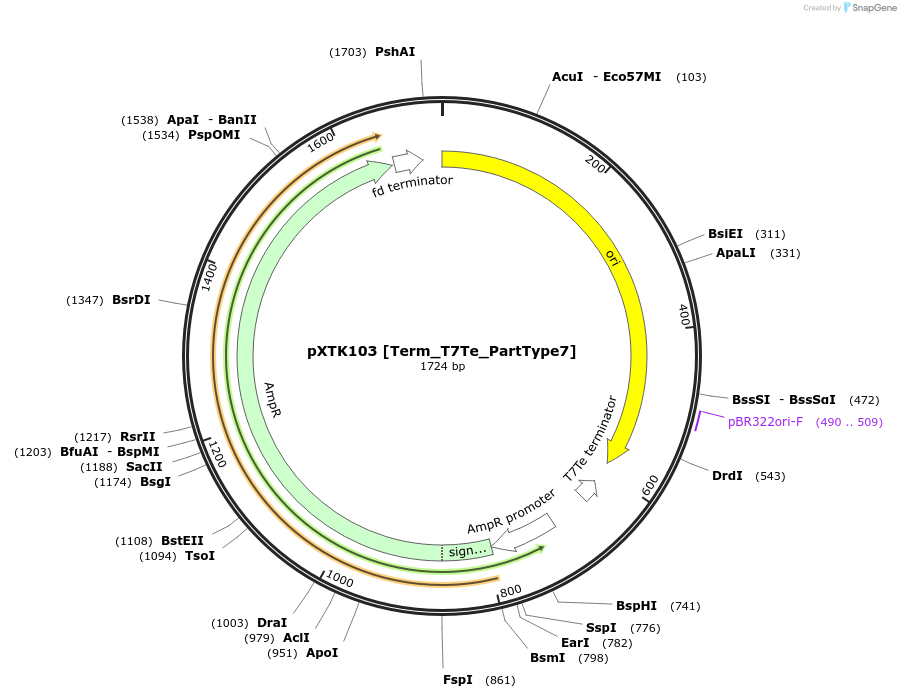
Plasmid#229340PurposeXanthoMoClo Golden Gate Part Plasmid for cloning, contains Term_T7Te_PartType7DepositorInsertT7Te Terminator
ExpressionBacterialAvailable SinceMarch 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pCAGGS-PHOSPHO2-TEV-Twin-Strep
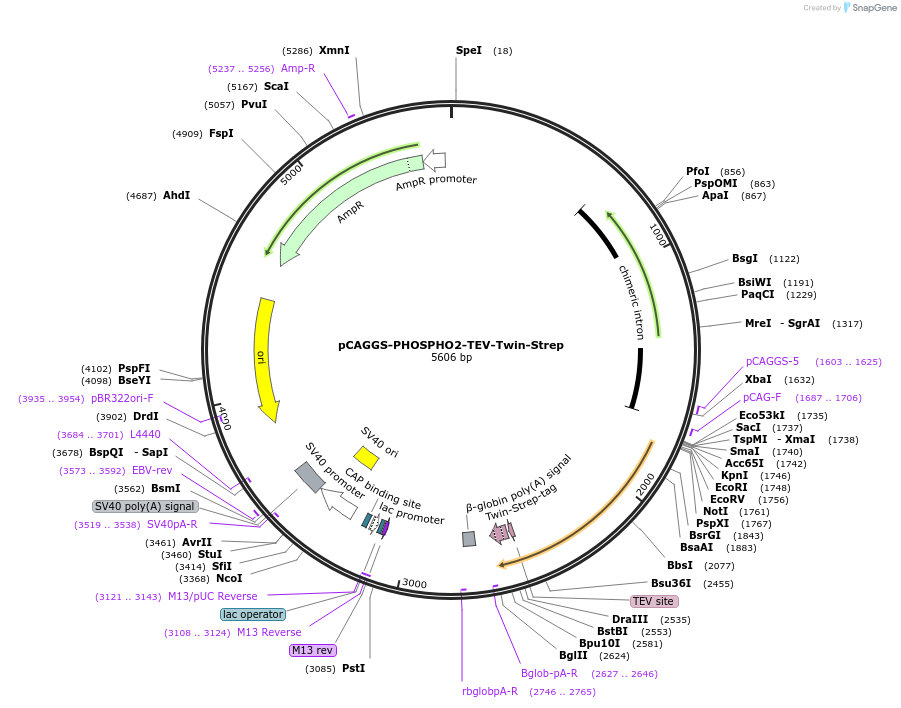
Plasmid#222016PurposeExpress human PHOSPHO2 protein (C-terminal Tobacco Etch Virus (TEV) protease cleavable Twin-Strep-fusion protein (ENLYFQGS-WSHPQFEK-(GGGS)2-GGSA-WSHPQFEK)) in mammalian cellDepositorInsertPHOSPHO2 (PHOSPHO2 Human)
TagsTEV cleavable site and Twin-Strep-tagExpressionMammalianMutationdeleted stop codon (TAA)Promoterchicken β-actin promoterAvailable SinceJan. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
pCAGGS-PHOSPHO2-TEV-Twin-Strep
Plasmid#222016PurposeExpress human PHOSPHO2 protein (C-terminal Tobacco Etch Virus (TEV) protease cleavable Twin-Strep-fusion protein (ENLYFQGS-WSHPQFEK-(GGGS)2-GGSA-WSHPQFEK)) in mammalian cellDepositorInsertPHOSPHO2 (PHOSPHO2 Human)
TagsTEV cleavable site and Twin-Strep-tagExpressionMammalianMutationdeleted stop codon (TAA)Promoterchicken β-actin promoterAvailable SinceJan. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
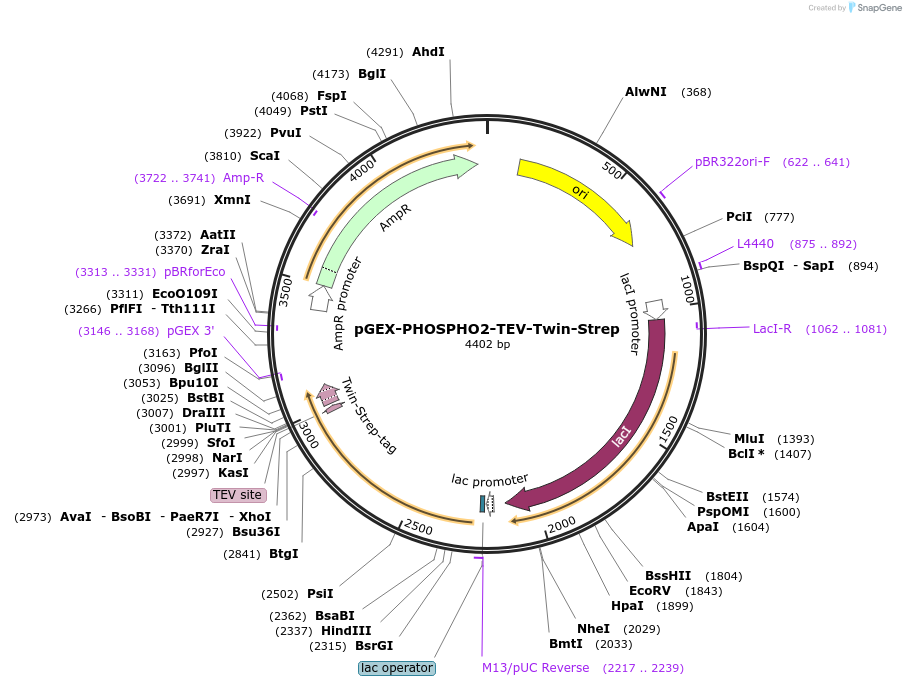
pGEX-PHOSPHO2-TEV-Twin-Strep
Plasmid#222017PurposeExpress human PHOSPHO2 protein (C-terminal Tobacco Etch Virus (TEV) protease cleavable Twin-Strep-fusion protein (ENLYFQGS-WSHPQFEK-(GGGS)2-GGSA-WSHPQFEK)) in E. coliDepositorInsertPHOSPHO2 (PHOSPHO2 Human)
TagsTEV cleavable site and Twin-Strep-tagExpressionBacterialMutationdeleted stop codon (TAA)Promoterlac promoterAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
pGEX-PHOSPHO2-TEV-Twin-Strep
Plasmid#222017PurposeExpress human PHOSPHO2 protein (C-terminal Tobacco Etch Virus (TEV) protease cleavable Twin-Strep-fusion protein (ENLYFQGS-WSHPQFEK-(GGGS)2-GGSA-WSHPQFEK)) in E. coliDepositorInsertPHOSPHO2 (PHOSPHO2 Human)
TagsTEV cleavable site and Twin-Strep-tagExpressionBacterialMutationdeleted stop codon (TAA)Promoterlac promoterAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
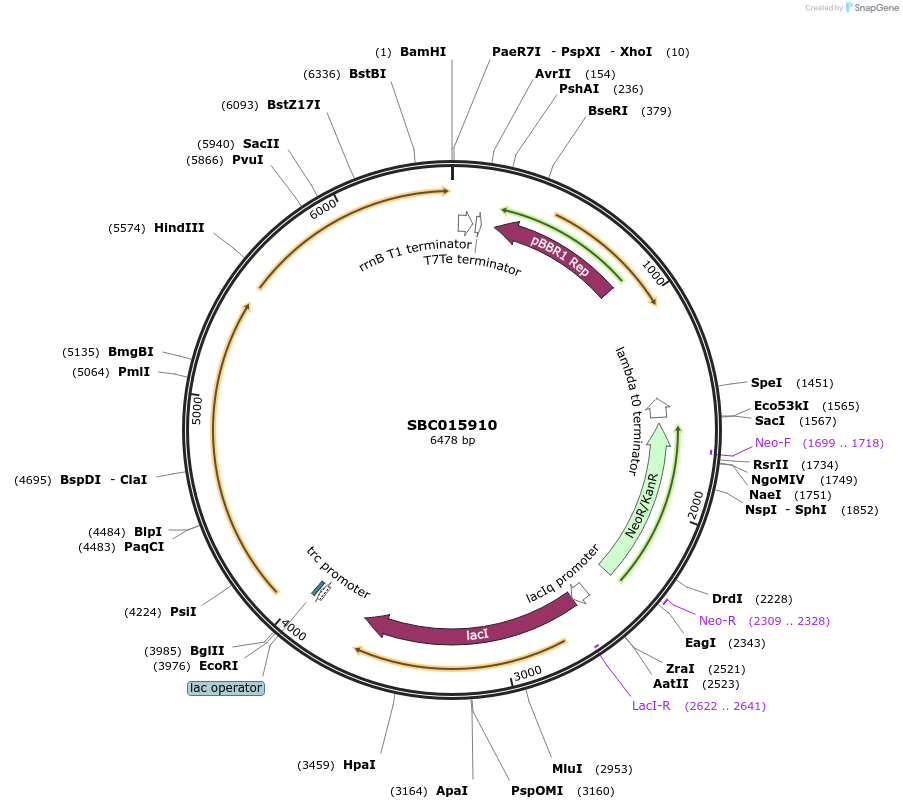
SBC015910
Plasmid#226287PurposeExpresses PhCFAT and PhIGS1 from trc promoter. Biosynthesis of isoeugenol from coniferyl alcohol.DepositorInsertsConiferyl alcohol acyltransferase
Isoeugenol synthase 1
ExpressionBacterialAvailable SinceNov. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
SBC015910
Plasmid#226287PurposeExpresses PhCFAT and PhIGS1 from trc promoter. Biosynthesis of isoeugenol from coniferyl alcohol.DepositorInsertsConiferyl alcohol acyltransferase
Isoeugenol synthase 1
ExpressionBacterialAvailable SinceNov. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
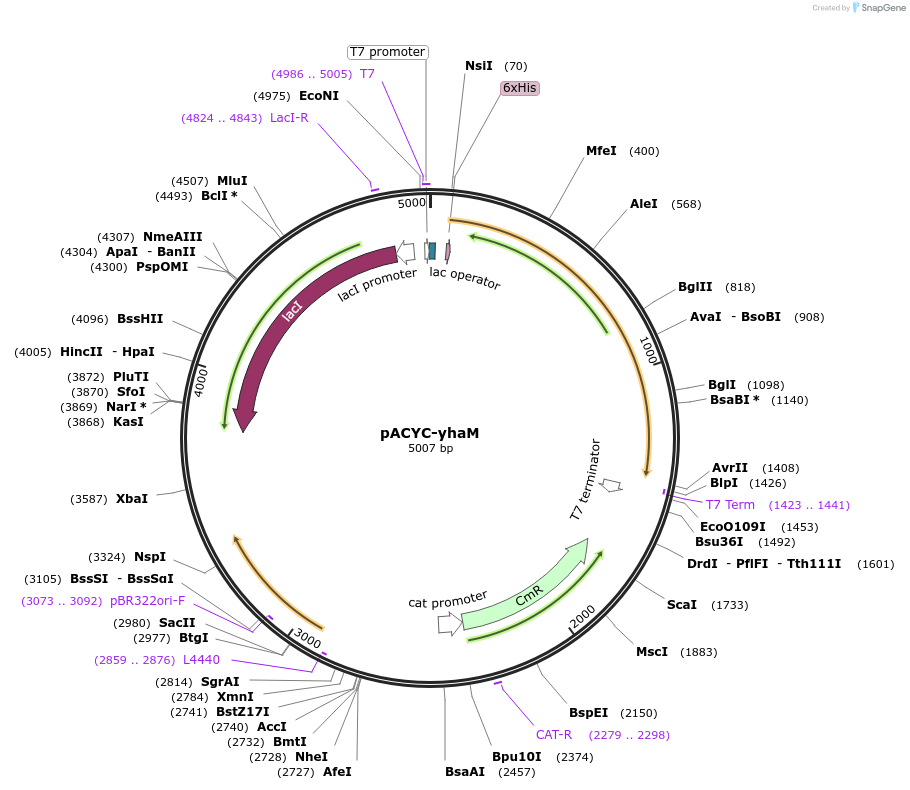
pACYC-yhaM
Plasmid#207650PurposeOverexpression of native L-cysteine desulfidase in E. coli MG1655DepositorInsertyhaM
Tags6xHisExpressionBacterialPromoterT7Available SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACYC-yhaM
Plasmid#207650PurposeOverexpression of native L-cysteine desulfidase in E. coli MG1655DepositorInsertyhaM
Tags6xHisExpressionBacterialPromoterT7Available SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
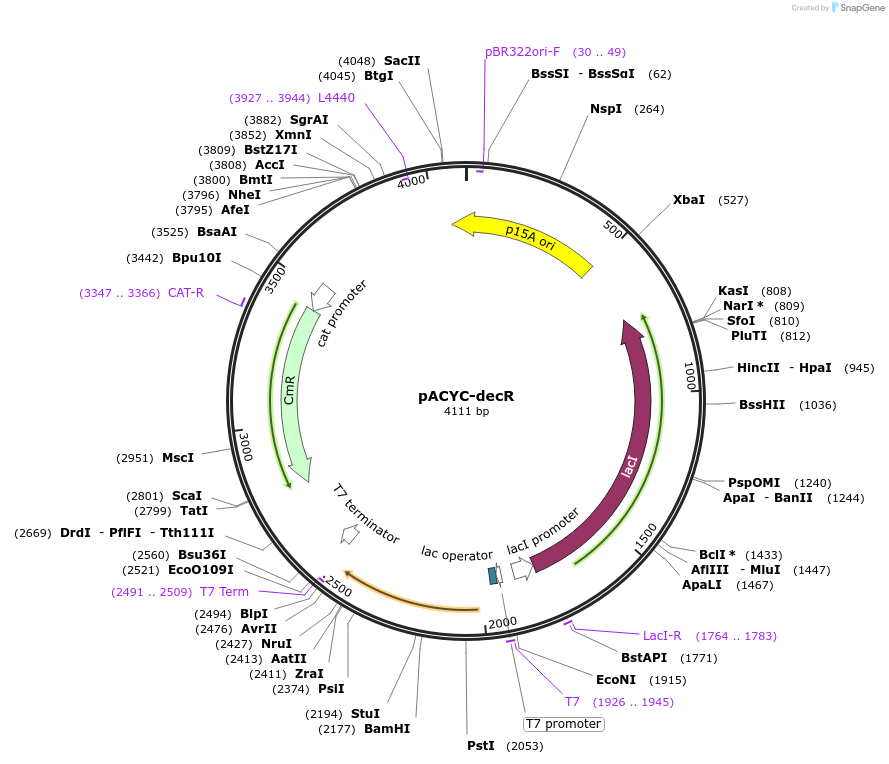
pACYC-decR
Plasmid#207653PurposeOverexpression of L-cysteine desulfidase transcriptional regulator in E. coli MG1655DepositorInsertdecR
ExpressionBacterialPromoterT7Available SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
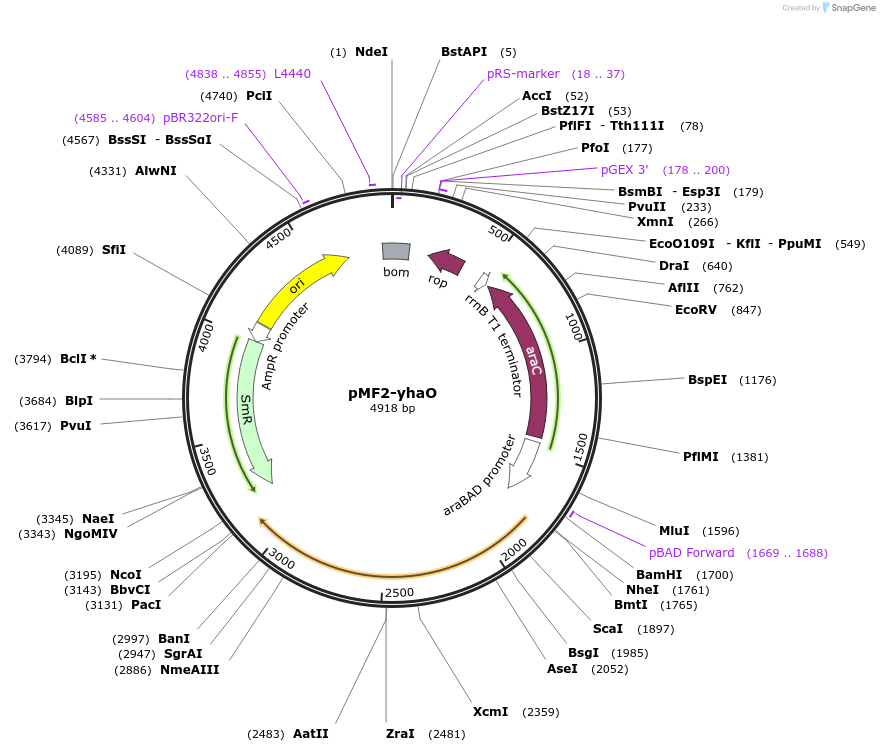
pMF2-yhaO
Plasmid#207654PurposeOverexpression of L-cysteine importer in E. coli MG1655DepositorInsertyhaO
Tags6xHisExpressionBacterialPromoteraraBpAvailable SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
pACYC-decR
Plasmid#207653PurposeOverexpression of L-cysteine desulfidase transcriptional regulator in E. coli MG1655DepositorInsertdecR
ExpressionBacterialPromoterT7Available SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
pMF2-yhaO
Plasmid#207654PurposeOverexpression of L-cysteine importer in E. coli MG1655DepositorInsertyhaO
Tags6xHisExpressionBacterialPromoteraraBpAvailable SinceOct. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
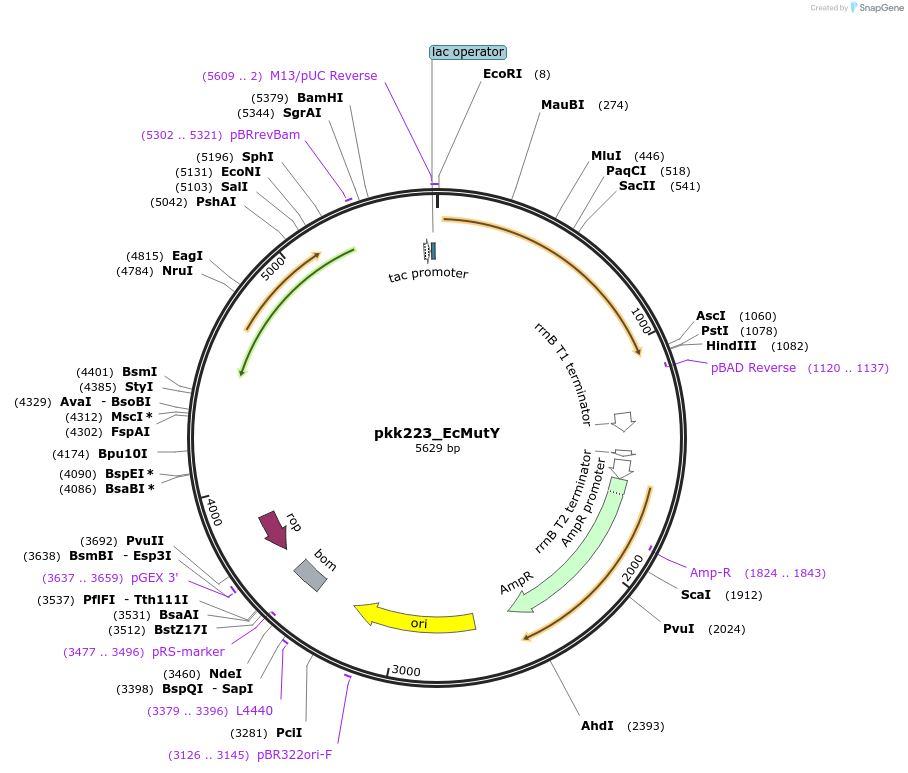
pkk223_EcMutY
Plasmid#213110Purposepkk223 with E coli MutY insertDepositorInsertMutY (adenine glycosylase) (mutY Escherichia coli)
ExpressionBacterialAvailable SinceApril 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
pkk223_EcMutY
Plasmid#213110Purposepkk223 with E coli MutY insertDepositorInsertMutY (adenine glycosylase) (mutY Escherichia coli)
ExpressionBacterialAvailable SinceApril 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
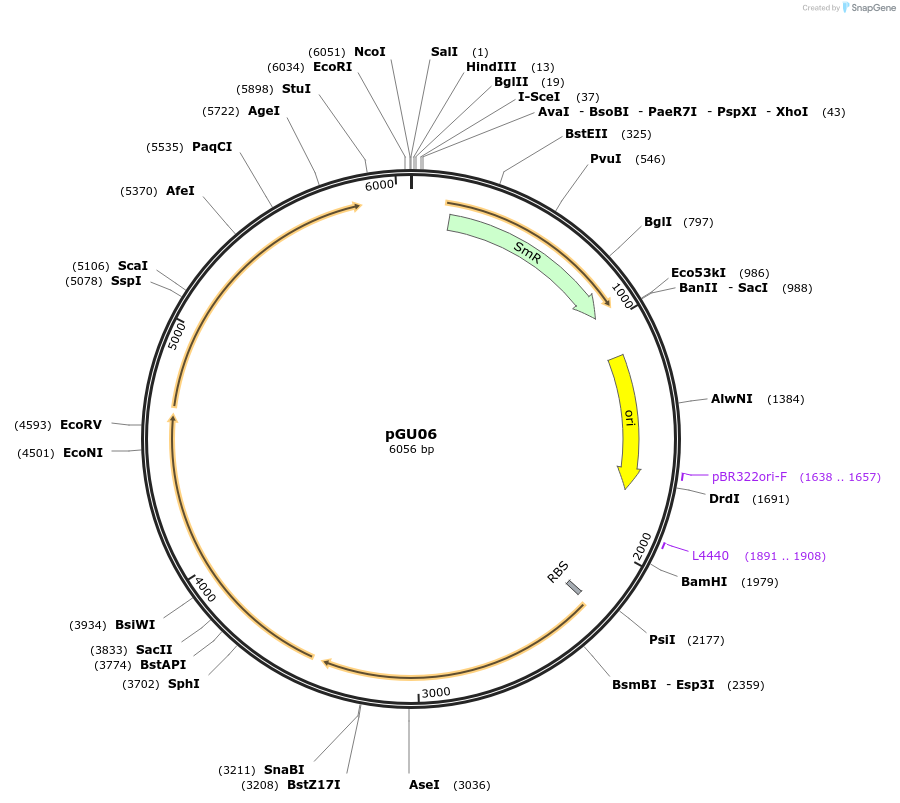
pGU06
Plasmid#212148PurposePlasmid expressing alanine-glyoxylate aminotransferase, glutamate-pyruvate aminotransferase and tyrosine aminotransferase under constitutive promoter PgyrA control.DepositorInsertExpressionBacterialPromoterPgyrAAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
pGU06
Plasmid#212148PurposePlasmid expressing alanine-glyoxylate aminotransferase, glutamate-pyruvate aminotransferase and tyrosine aminotransferase under constitutive promoter PgyrA control.DepositorInsertExpressionBacterialPromoterPgyrAAvailable SinceJan. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
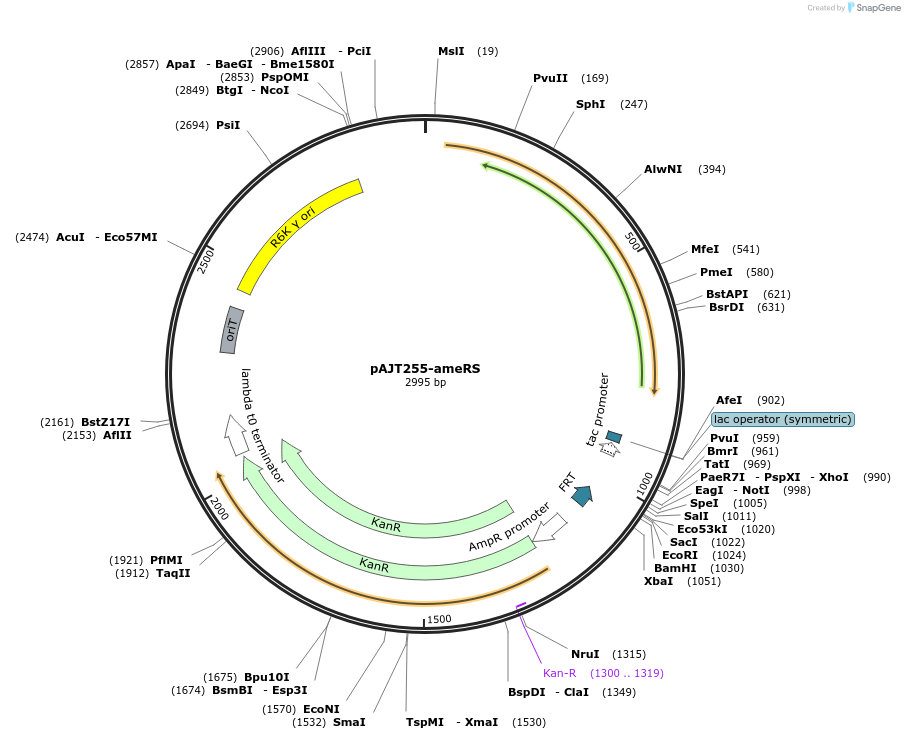
pAJT255-ameRS
Plasmid#202309PurposeameRS repressor plasmid for LPattB2 integrationDepositorInsertameRS
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT255-ameRS
Plasmid#202309PurposeameRS repressor plasmid for LPattB2 integrationDepositorInsertameRS
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
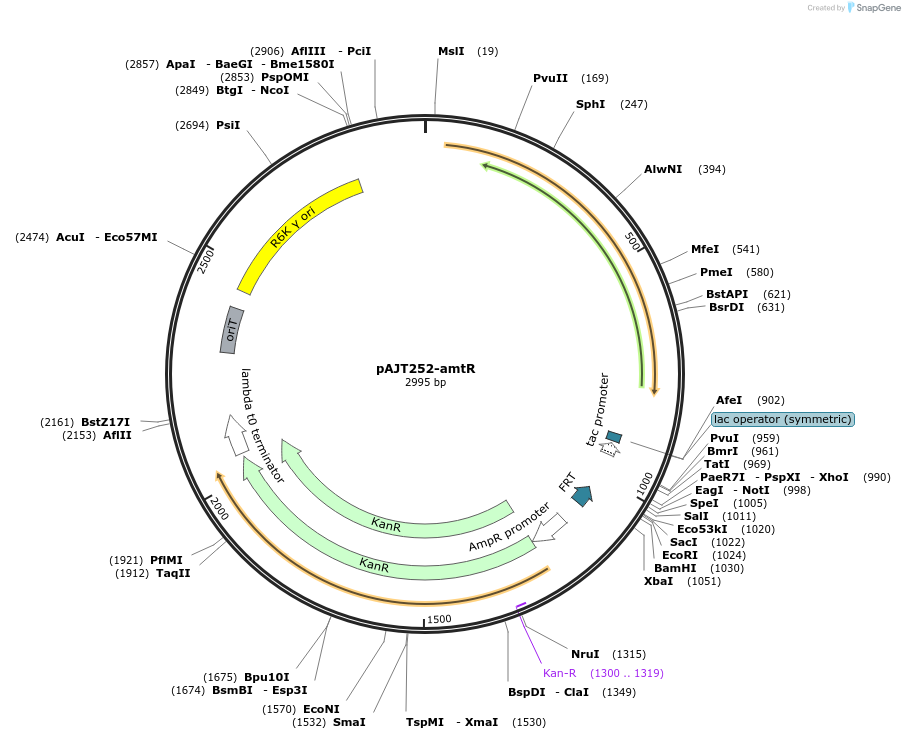
pAJT252-amtR
Plasmid#202307PurposeamtR repressor plasmid for LPattB2 integrationDepositorInsertamtR
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT252-amtR
Plasmid#202307PurposeamtR repressor plasmid for LPattB2 integrationDepositorInsertamtR
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
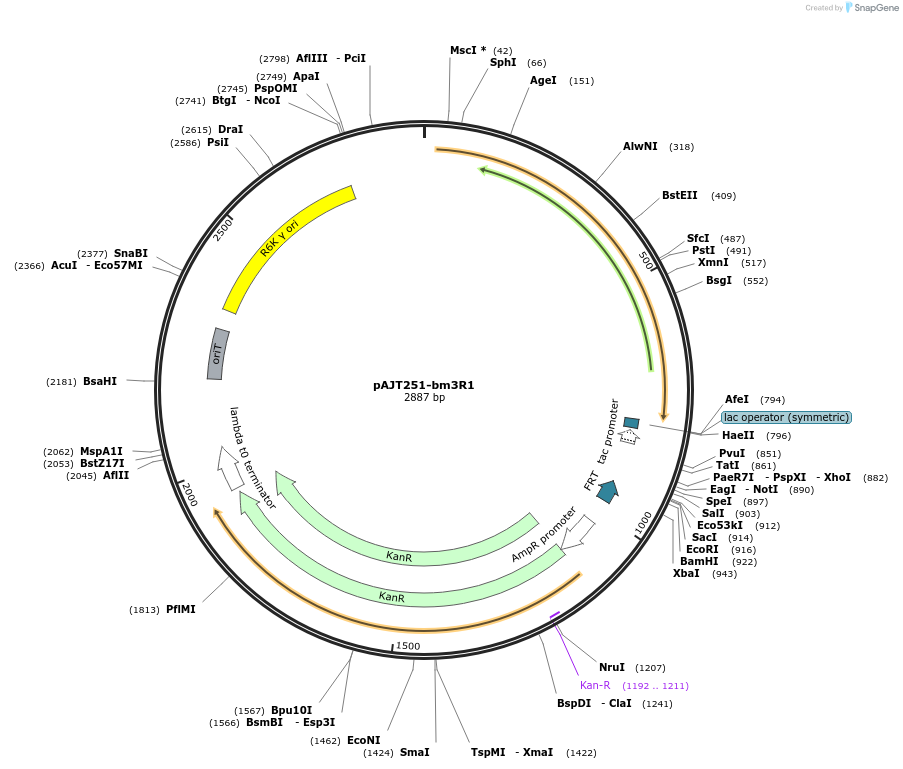
pAJT251-bm3R1
Plasmid#202306Purposebm3R1 repressor plasmid for LPattB2 integrationDepositorInsertbm3R1
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT251-bm3R1
Plasmid#202306Purposebm3R1 repressor plasmid for LPattB2 integrationDepositorInsertbm3R1
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
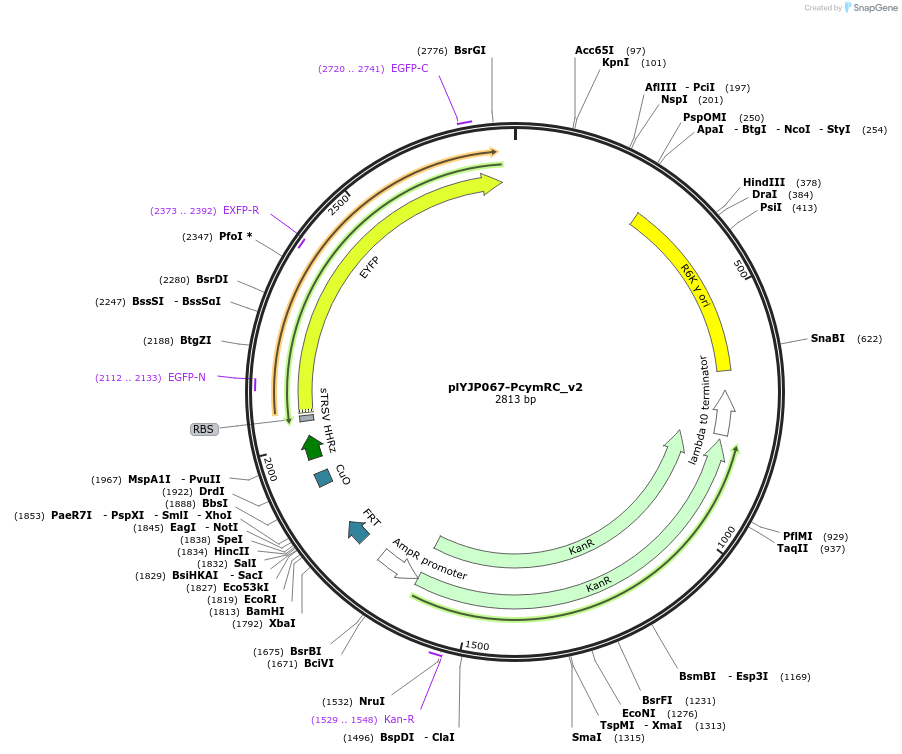
plYJP067-PcymRC_v2
Plasmid#202302PurposePCymRC_v2-YFP plasmid for LPattB2 integrationDepositorInsertPCymRC_v2-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
plYJP067-PcymRC_v2
Plasmid#202302PurposePCymRC_v2-YFP plasmid for LPattB2 integrationDepositorInsertPCymRC_v2-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
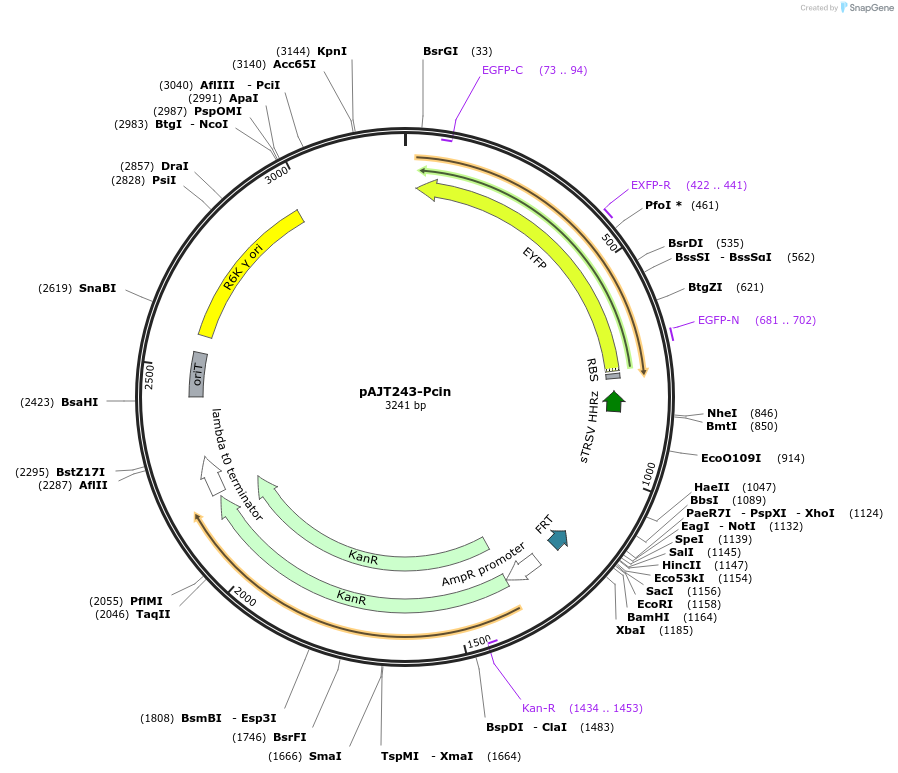
pAJT243-Pcin
Plasmid#202301PurposePCin-YFP plasmid for LPattB2 integrationDepositorInsertPcin-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT243-Pcin
Plasmid#202301PurposePCin-YFP plasmid for LPattB2 integrationDepositorInsertPcin-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
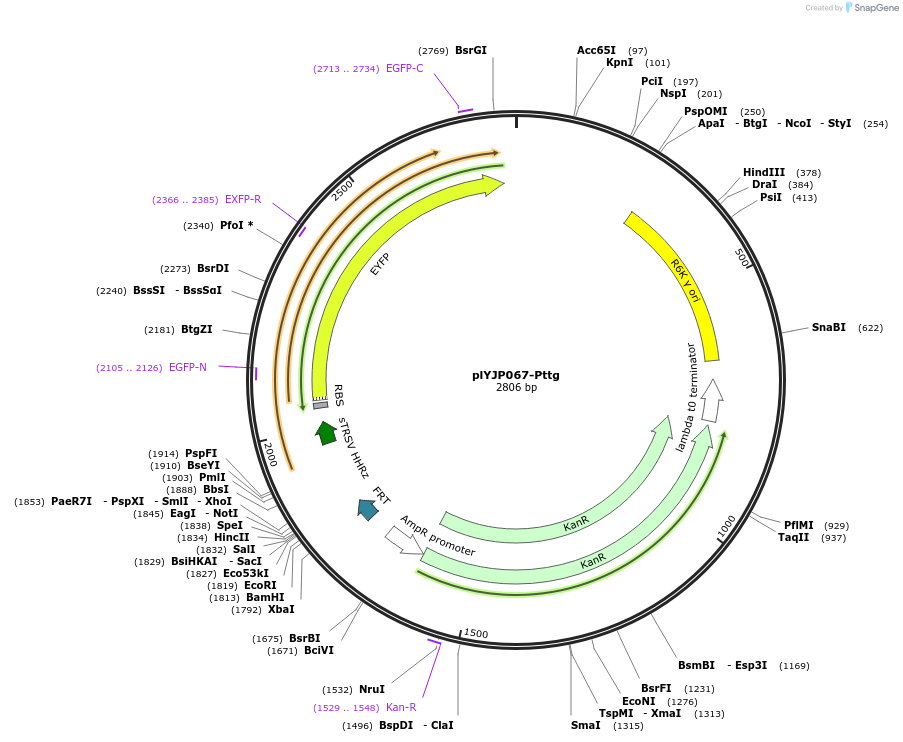
plYJP067-Pttg
Plasmid#202300PurposePTtg-YFP plasmid for LPattB2 integrationDepositorInsertPTtg-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
plYJP067-Pttg
Plasmid#202300PurposePTtg-YFP plasmid for LPattB2 integrationDepositorInsertPTtg-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
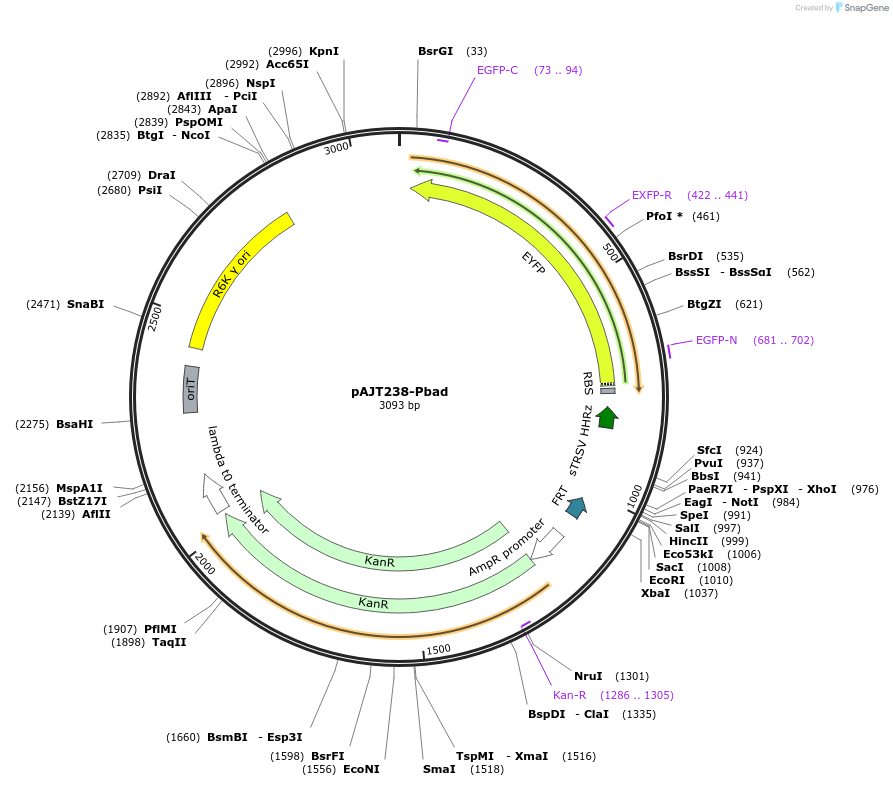
pAJT238-Pbad
Plasmid#202298PurposePBad-YFP plasmid for LPattB2 integrationDepositorInsertPbad-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT238-Pbad
Plasmid#202298PurposePBad-YFP plasmid for LPattB2 integrationDepositorInsertPbad-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
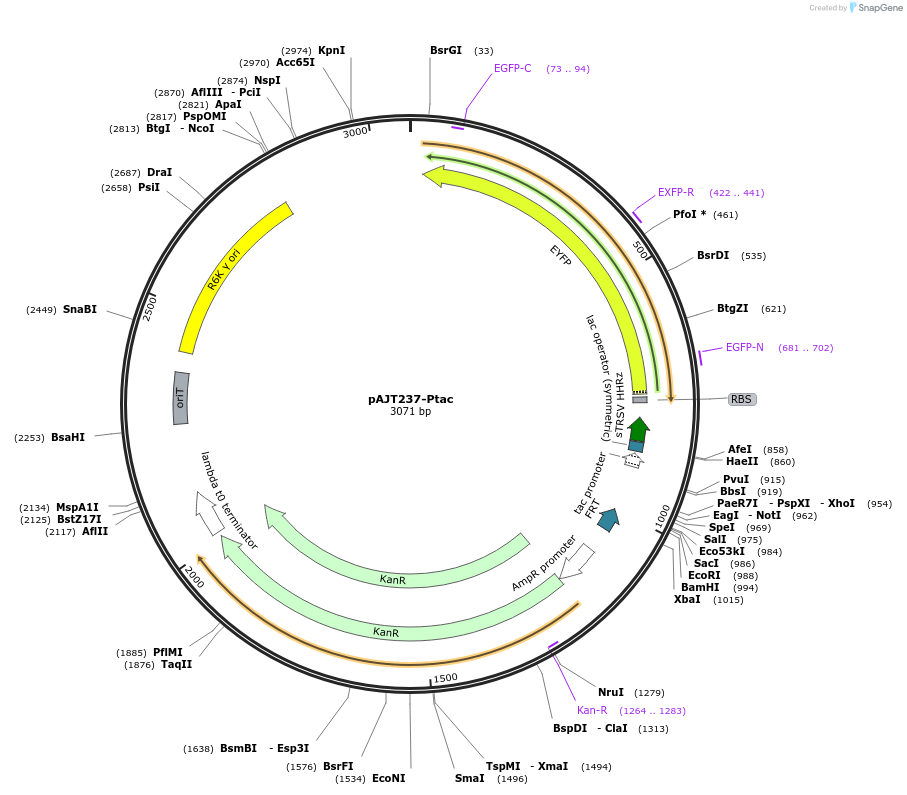
pAJT237-Ptac
Plasmid#202297PurposePTac-YFP plasmid for LPattB2 integrationDepositorInsertPTac-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT237-Ptac
Plasmid#202297PurposePTac-YFP plasmid for LPattB2 integrationDepositorInsertPTac-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
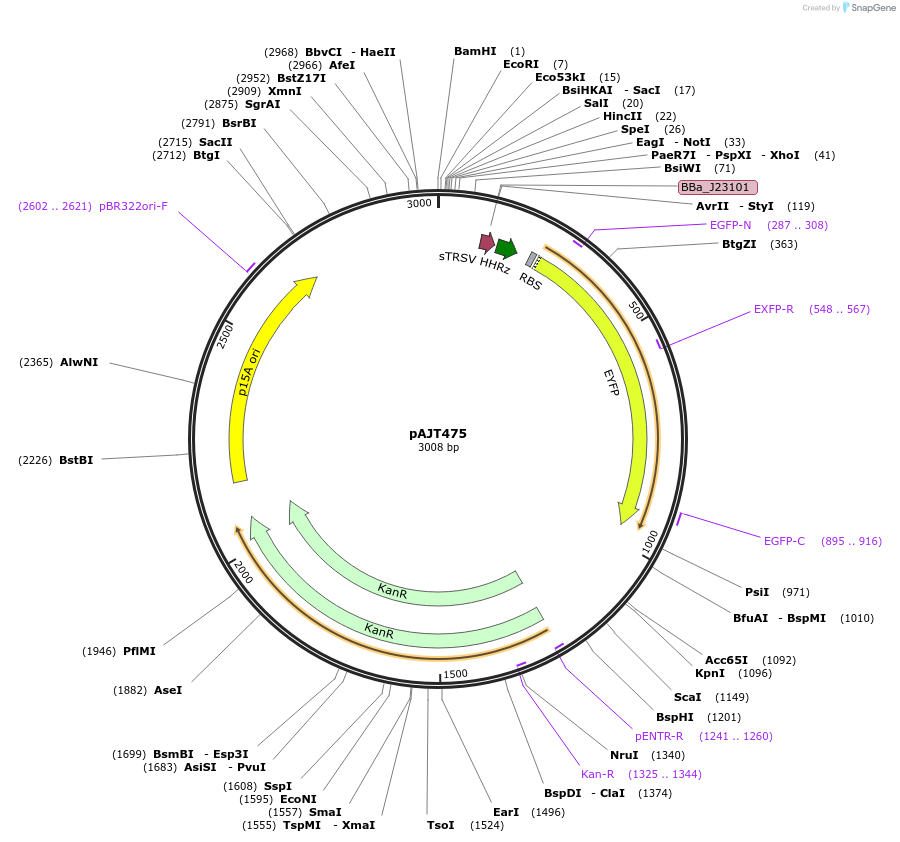
pAJT475
Plasmid#202292PurposeRPU plasmid w/ high RBS (p15a backbone)DepositorInsertJ23101-RiboJ00-B0034-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAJT475
Plasmid#202292PurposeRPU plasmid w/ high RBS (p15a backbone)DepositorInsertJ23101-RiboJ00-B0034-YFP
ExpressionBacterialAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
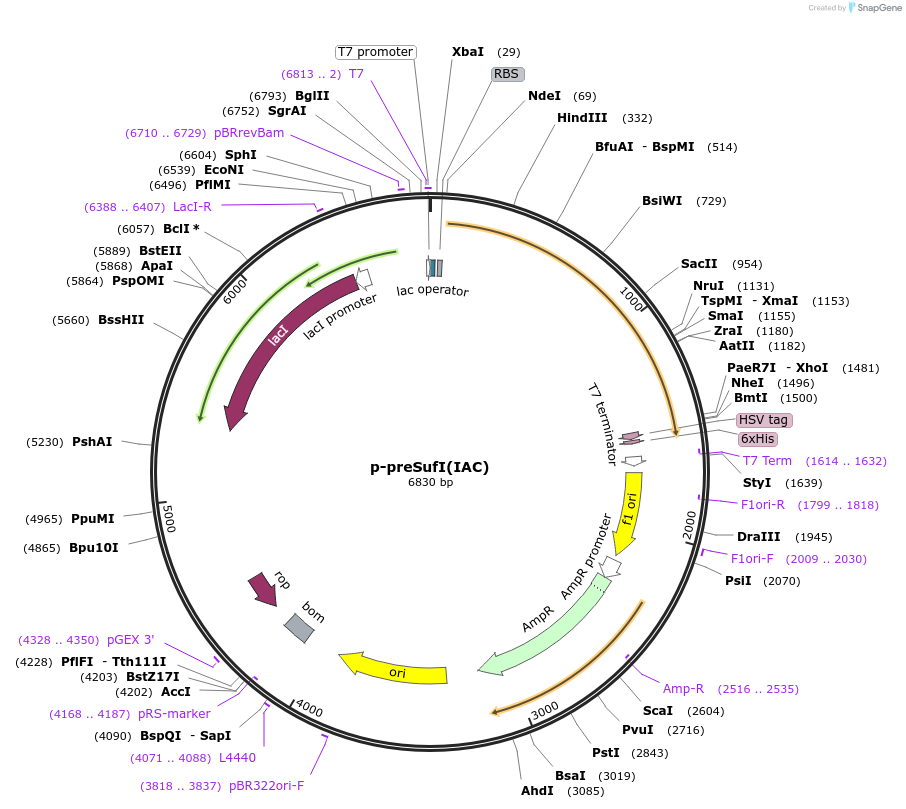
p-preSufI(IAC)
Plasmid#168516PurposeModified form of pre-SufI in pET25b. With C-terminal HHHHHHC (6xHisC) extension and mutations C17I and C295A.DepositorInsertpre-SufI(IAC)
Tags6xHisC tag and HSV tagExpressionBacterialMutationC17I and C295APromoterT7Available SinceSept. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
p-preSufI(IAC)
Plasmid#168516PurposeModified form of pre-SufI in pET25b. With C-terminal HHHHHHC (6xHisC) extension and mutations C17I and C295A.DepositorInsertpre-SufI(IAC)
Tags6xHisC tag and HSV tagExpressionBacterialMutationC17I and C295APromoterT7Available SinceSept. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
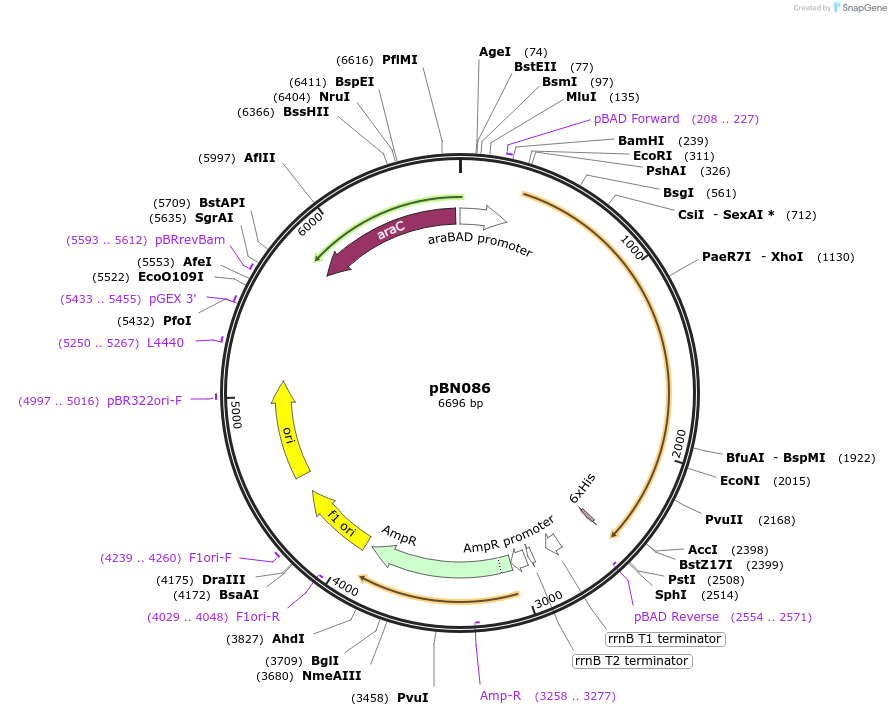
pBN086
Plasmid#173448Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y715EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y715E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y715EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
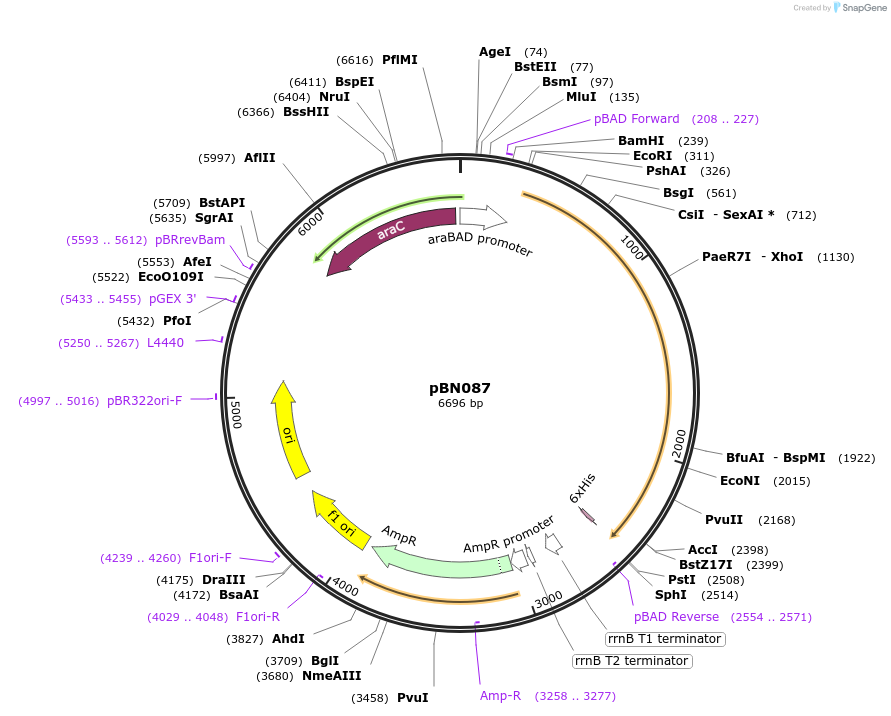
pBN087
Plasmid#173449Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y717EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y717E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y717EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
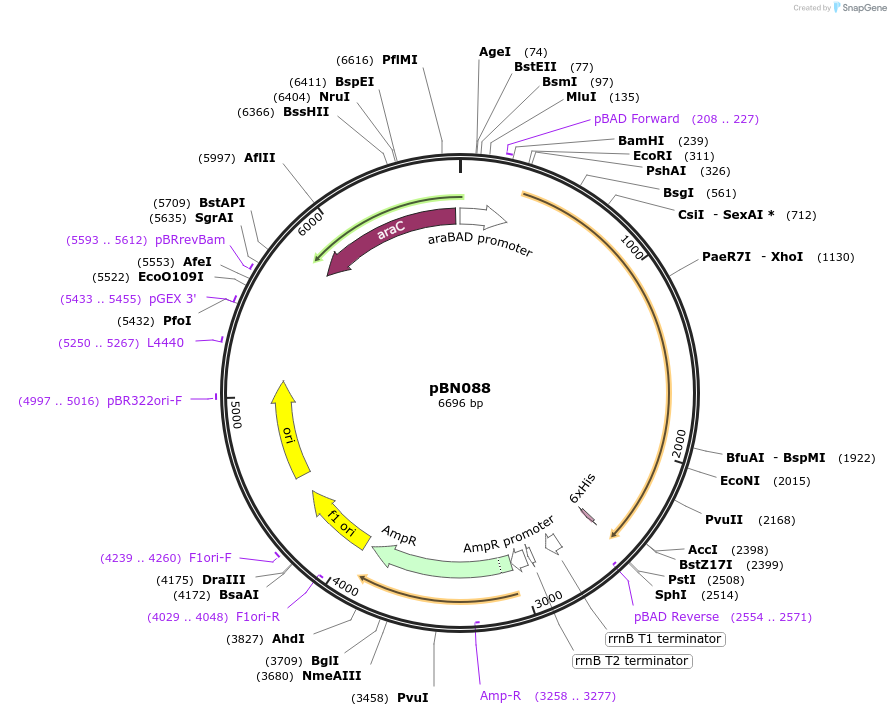
pBN088
Plasmid#173450Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y718EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y718E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y718EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
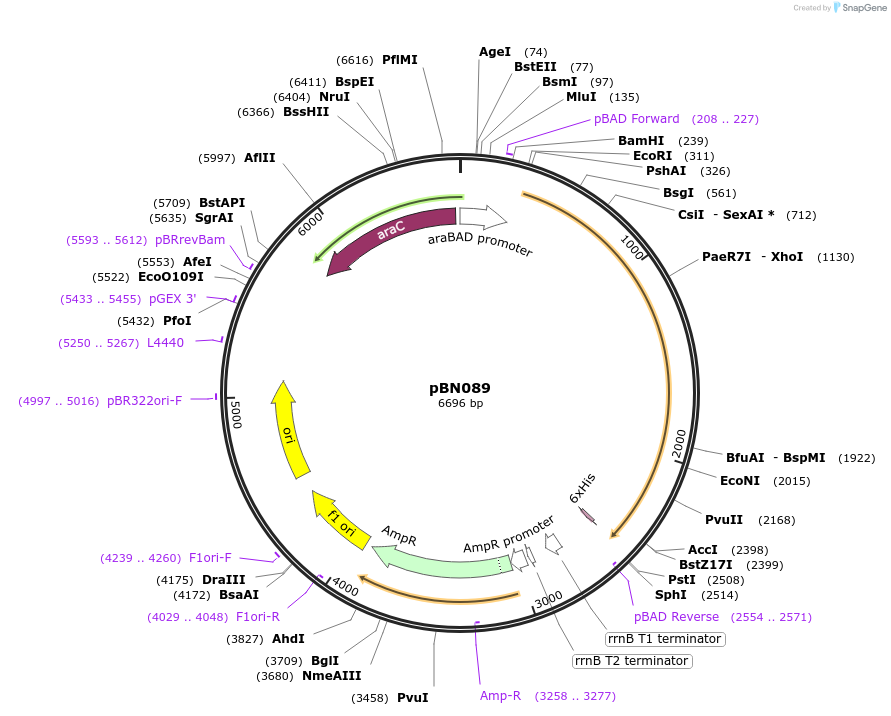
pBN089
Plasmid#173451Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y717E, Y718EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y717E, Y718E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y717E, Y718EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
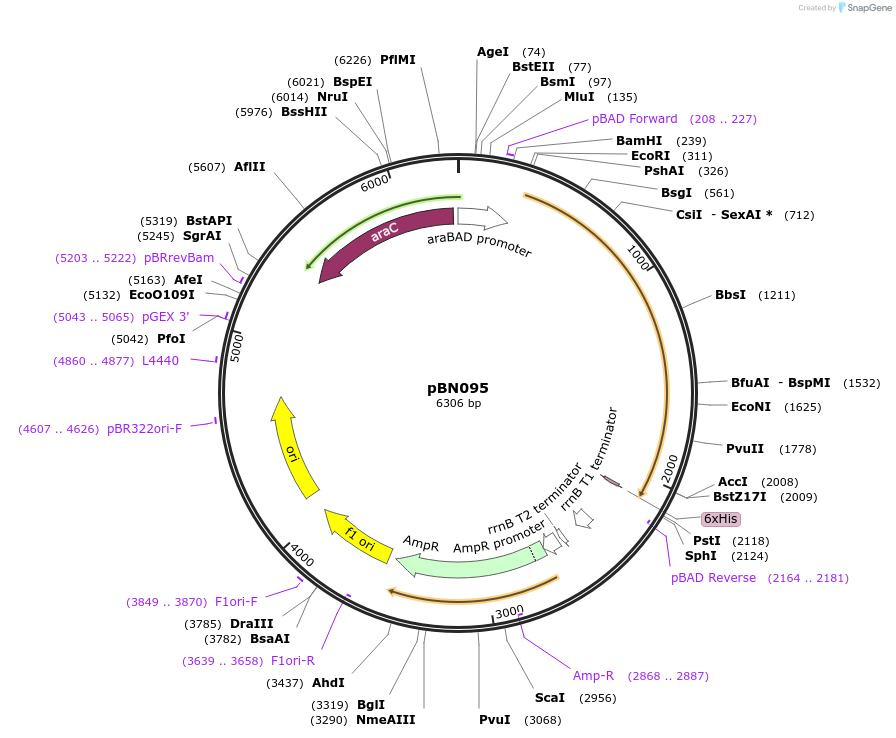
pBN095
Plasmid#173765Purposeexpress E. coli E69 (O9a:K30) Wzc with periplasmic motif 3 truncationDepositorInsertE. coli E69 (O9a:K30) Wzc with periplasmic motif 3 truncation
Tagshexa-histidine tagExpressionBacterialMutationdelete motif 3PromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBN086
Plasmid#173448Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y715EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y715E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y715EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBN087
Plasmid#173449Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y717EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y717E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y717EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBN088
Plasmid#173450Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y718EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y718E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y718EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBN089
Plasmid#173451Purposeexpress E. coli E69 (O9a:K30) Wzc_K540M with Y717E, Y718EDepositorInsertE. coli E69 (O9a:K30) Wzc_K540M with Y717E, Y718E
Tagshexa-histidine tagExpressionBacterialMutationK540M, Y717E, Y718EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBN095
Plasmid#173765Purposeexpress E. coli E69 (O9a:K30) Wzc with periplasmic motif 3 truncationDepositorInsertE. coli E69 (O9a:K30) Wzc with periplasmic motif 3 truncation
Tagshexa-histidine tagExpressionBacterialMutationdelete motif 3PromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
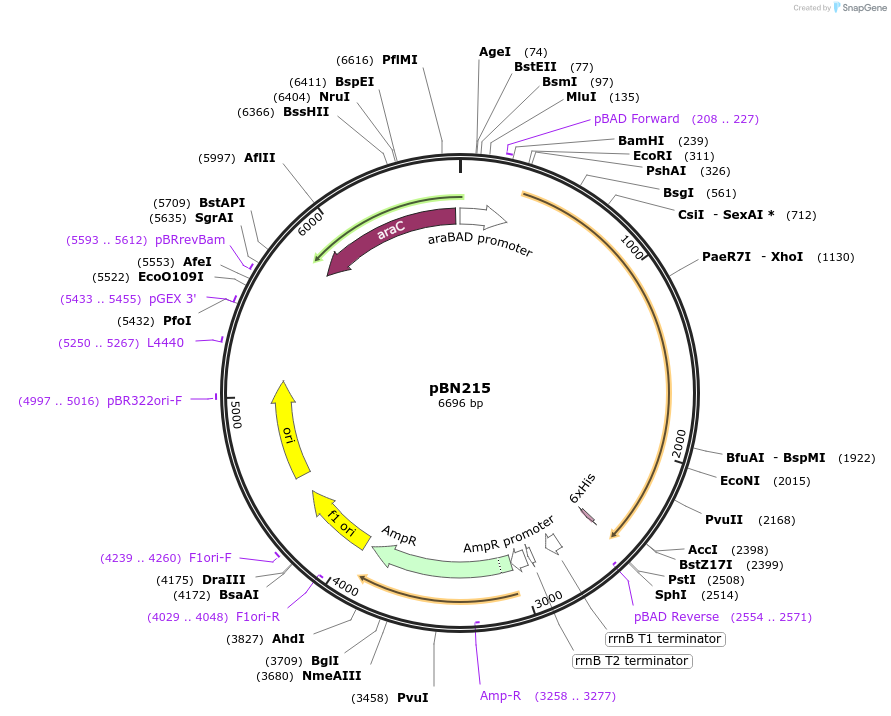
pBN215
Plasmid#167962Purposeexpress E. coli E69 (O9a:K30) Wzc with Y715E, Y717E, Y718EDepositorInsertE. coli E69 (O9a:K30) Wzc with Y715E, Y717E, Y718E
Tagshexa-histidine tagExpressionBacterialMutationY715E, Y717E, Y718EPromoteraraBADAvailable SinceOct. 7, 2021AvailabilityAcademic Institutions and Nonprofits only