4,457 results
-
Plasmid#246296PurposeEvaluation of CiU6.6 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.6 promoter
UseCRISPRExpressionPlantPromoterCiU6.6 promoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
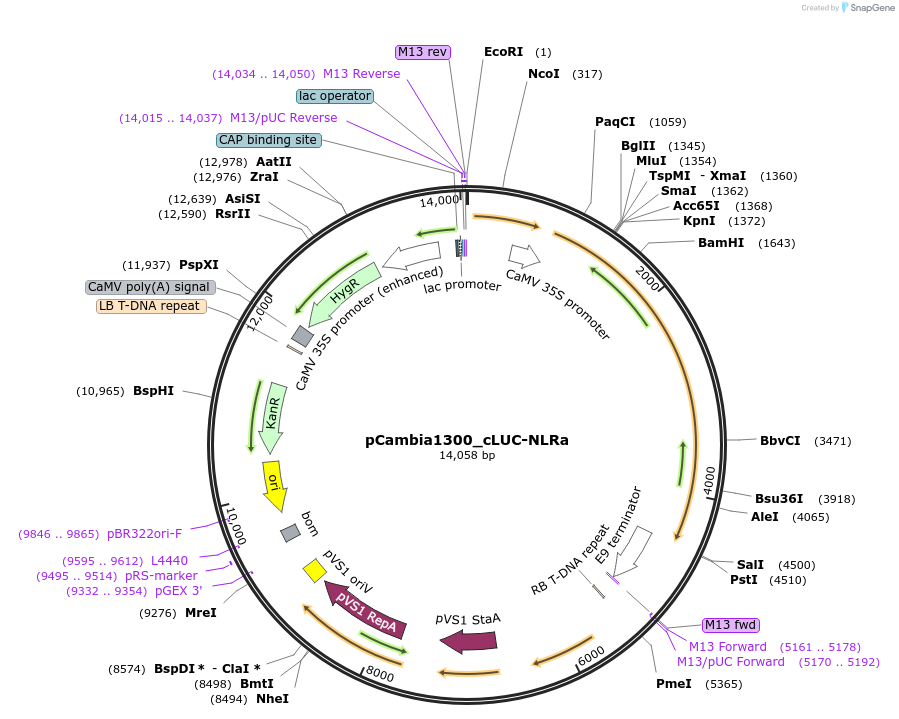
pCambia1300_cLUC-NLRa
Plasmid#233526PurposeExpresses split luciferase fusion cLUC-NLRa in plantaDepositorInsertSr62NLRa
TagsC-terminal fragment of LuciferaseExpressionPlantPromoter35SAvailable SinceDec. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
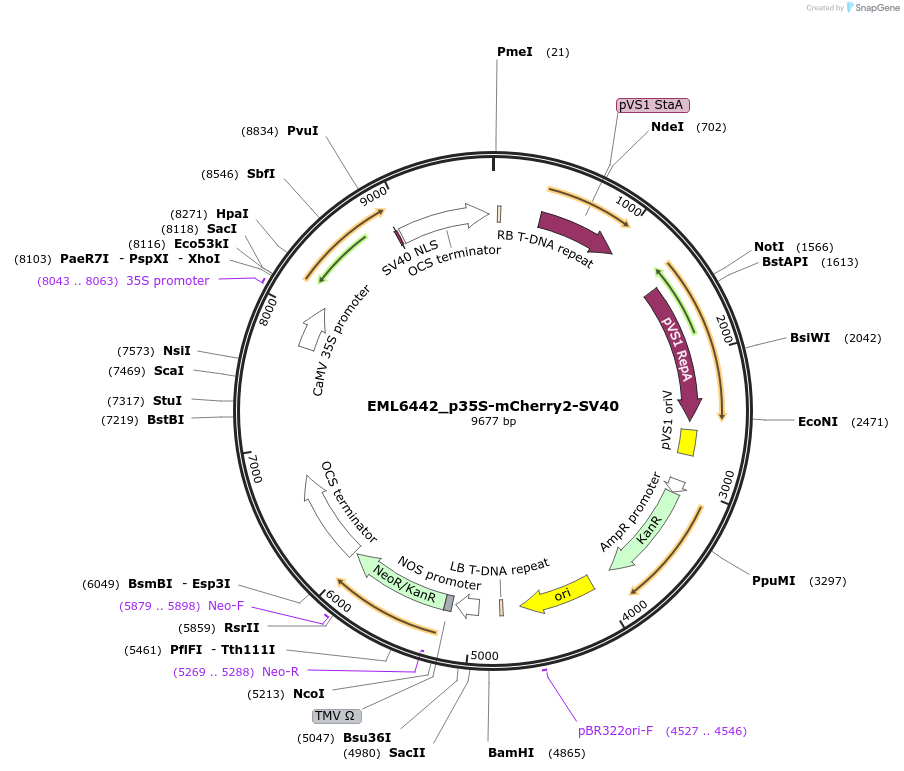
EML6442_p35S-mCherry2-SV40
Plasmid#234351PurposeExpressed mCherry2 florescent proteins in plant cells for tracing transformation eventsDepositorInsertSpeR
ExpressionPlantAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
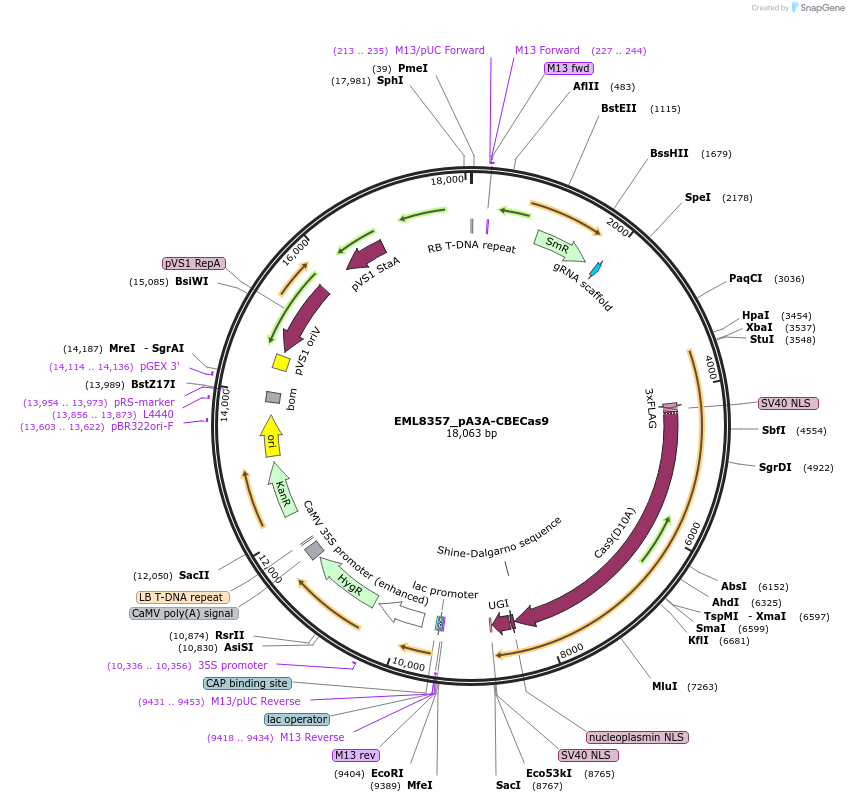
EML8357_pA3A-CBECas9
Plasmid#234352PurposeA base editor derived by egg-cell specific promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
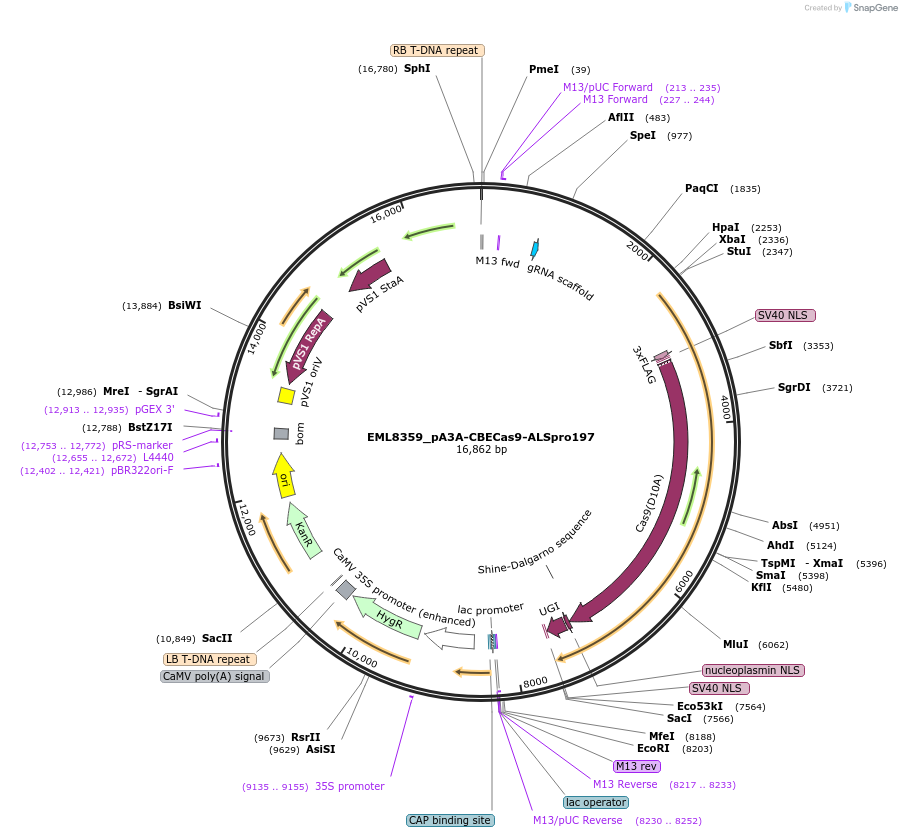
EML8359_pA3A-CBECas9-ALSpro197
Plasmid#234353PurposeBase editor targets to ALS Pro197 locusDepositorInsertALS
UseCRISPRExpressionPlantAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
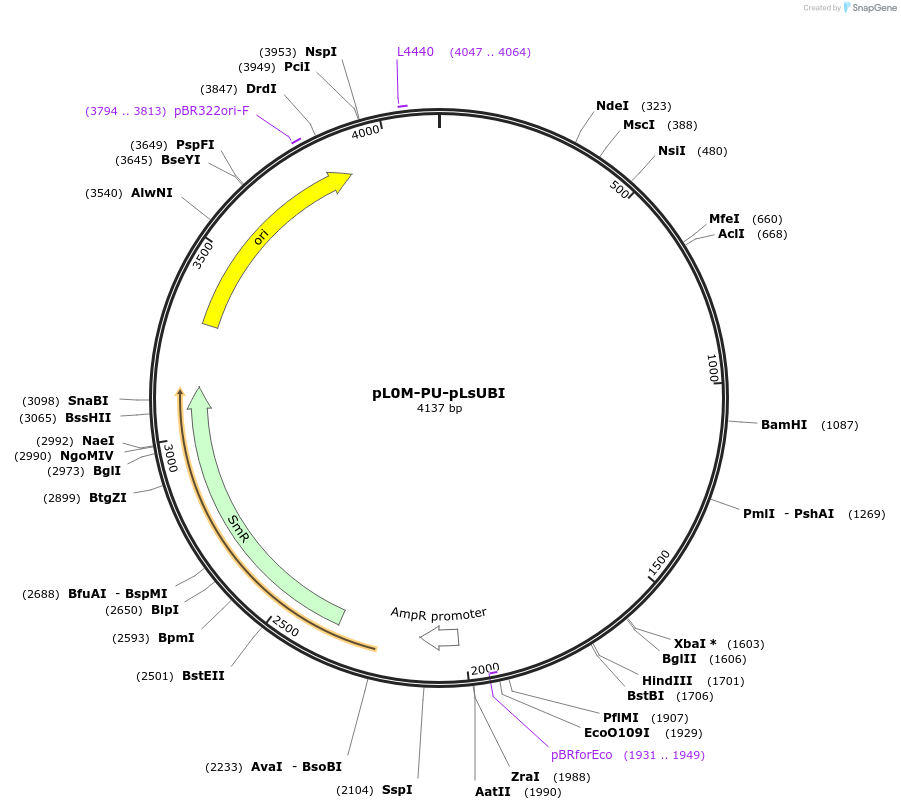
pL0M-PU-pLsUBI
Plasmid#244448PurposeLevel 0 Golden Gate "PU" module containing the domesticated sequence of the promoter of the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)DepositorInsertDomesticated sequence of the promoter for the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)
ExpressionPlantMutationThe promoter sequence 96336283-96337681 from NC_0…PromoterPolyubiquitin 4 gene (LOC111919935) from lettuceAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
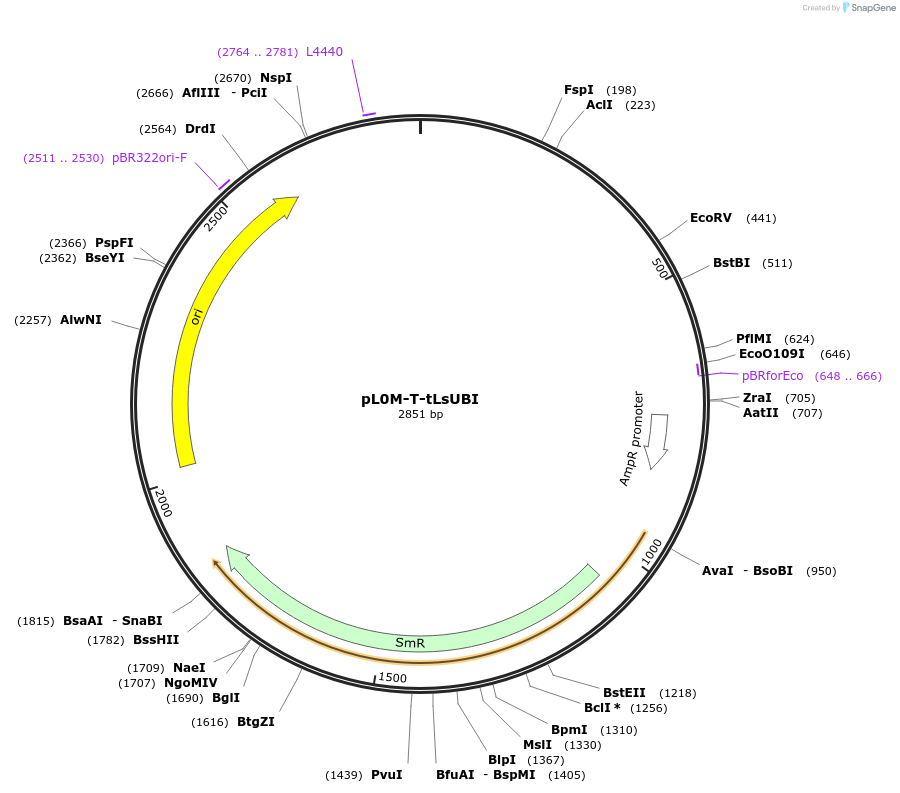
pL0M-T-tLsUBI
Plasmid#245484PurposeLevel 0 Golden Gate "T" module containing the sequence of the terminator of the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)DepositorInsertDomesticated sequence of the promoter for the polyubiquitin 4 gene (LOC111919935) from lettuce (Lactuca sativa)
ExpressionPlantMutationThe promoter sequence 96336283-96337681 from NC_0…Available SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
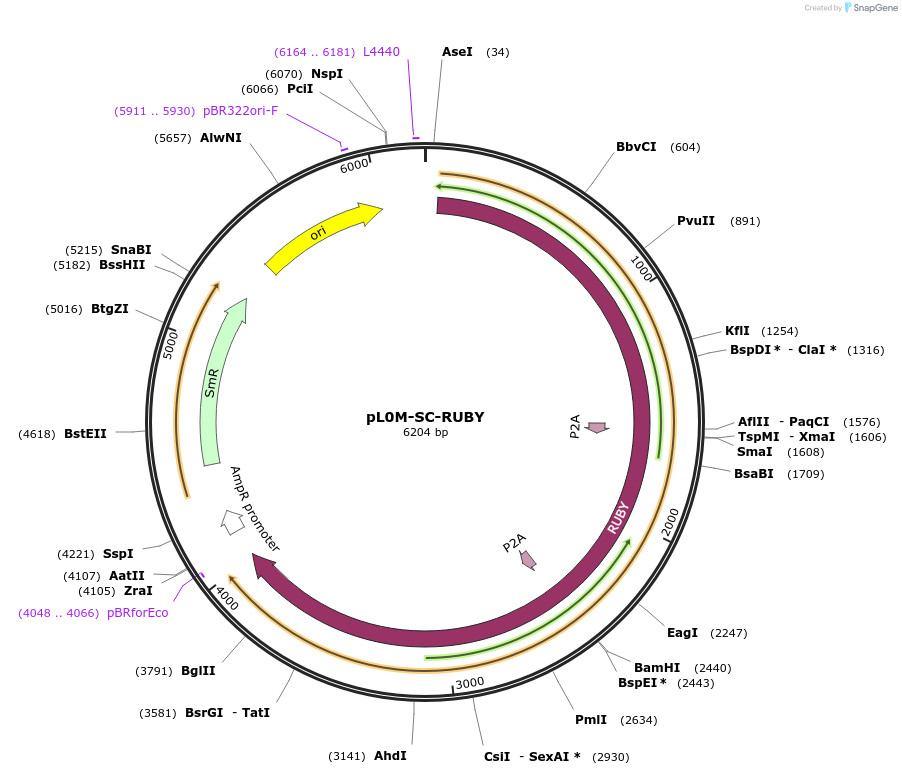
pL0M-SC-RUBY
Plasmid#245485PurposeGolden Gate Level 0 "SC" RUBY moduleDepositorInsertRUBY
ExpressionPlantAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
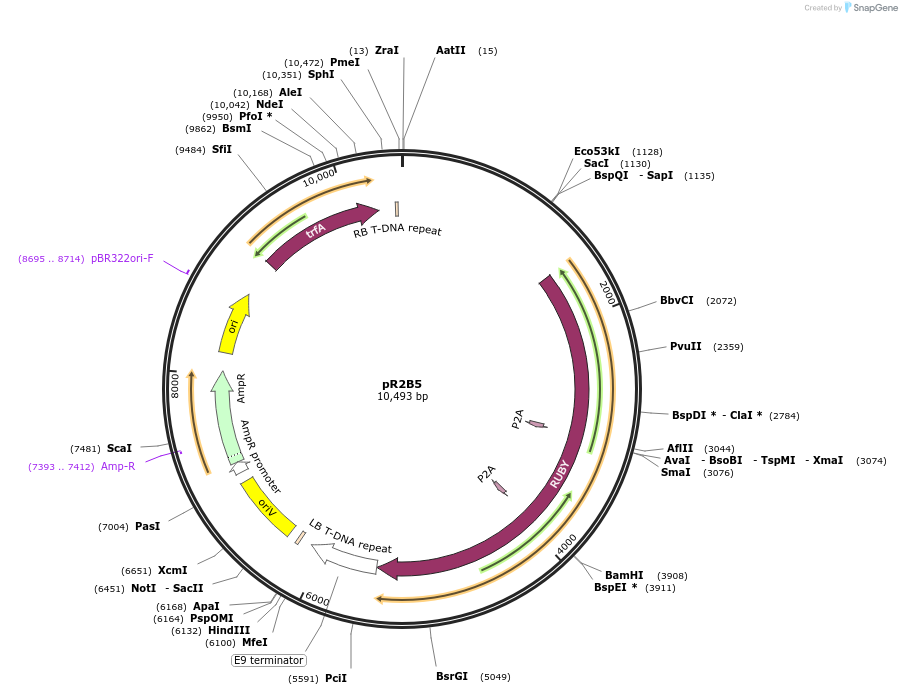
pR2B5
Plasmid#245486PurposepL1-R2-pAtUBI-RUBY-tRBCS (Golden Gate Level 1 module with a transcription unit expressing RUBY from the AtUBI promoter with the tRBC terminator for R2 position in Level 2 vector)DepositorInsertpAtUBI-RUBY-tRBCS
ExpressionPlantPromoterPolyubiquitin 10 gene from Arabidopsis thalianaAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
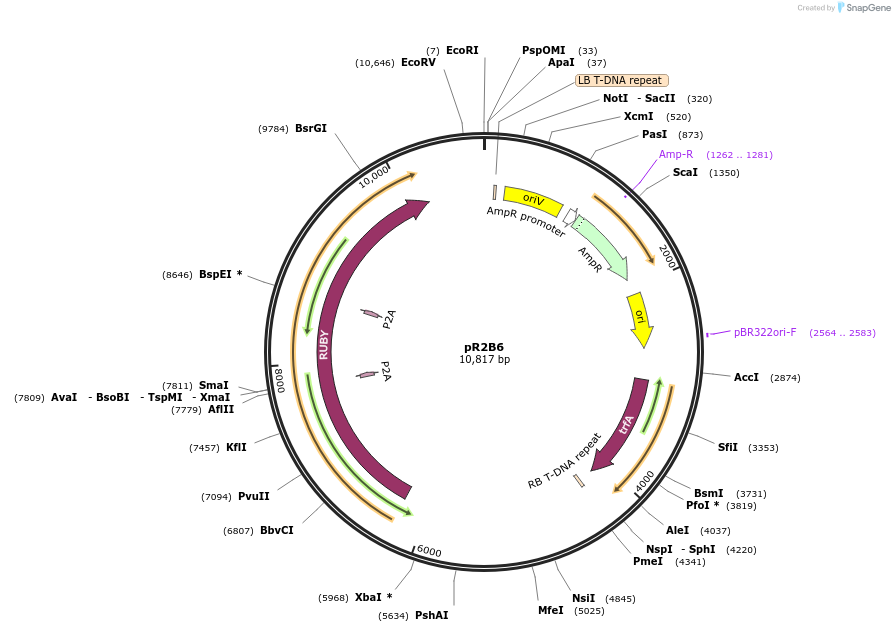
pR2B6
Plasmid#245487PurposepL1-R2-pLsUBI-RUBY-tLsUBI (Golden Gate Level 1 module with a transcription unit expressing RUBY from the LsUBI promoter with the tLsUBI terminator for R2 position in Level 2 vector)DepositorInsertpLsUBI-RUBY-tLSUBI
ExpressionPlantPromoterPolyubiquitin 4 gene (LOC111919935) from lettuceAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
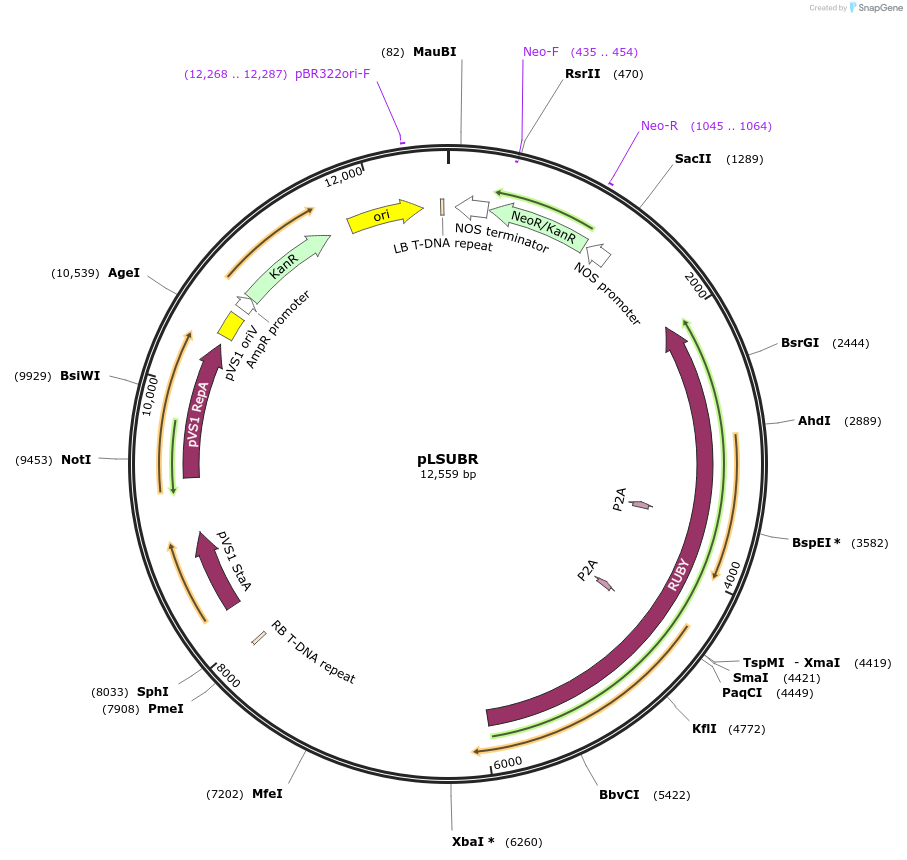
pLSUBR
Plasmid#245489PurposeBinary Plasmid for plant transformation expressing RUBY from the Lactuca sativa polyubiquitin 4 gene promoter and the terminator from the same gene. Kanamycin plant selectionDepositorInsertpLsUBI-RUBY-tLSUBI
ExpressionPlantPromoterPolyubiquitin 4 gene (LOC111919935) from lettuceAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
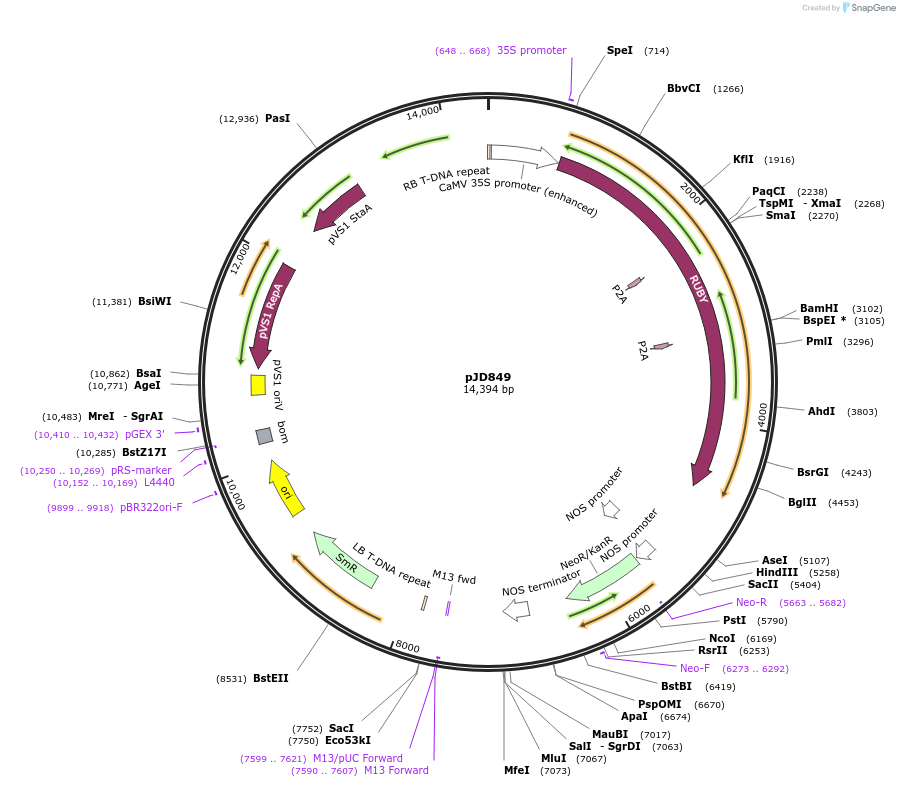
pJD849
Plasmid#245490PurposeBinary Plasmid for plant transformation expressing RUBY from the 35S promoter and the terminator from the Arabidopsis thaliana HSP gene. Kanamycin plant selectionDepositorInsertp35S-RUBY
ExpressionPlantPromoter35SAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
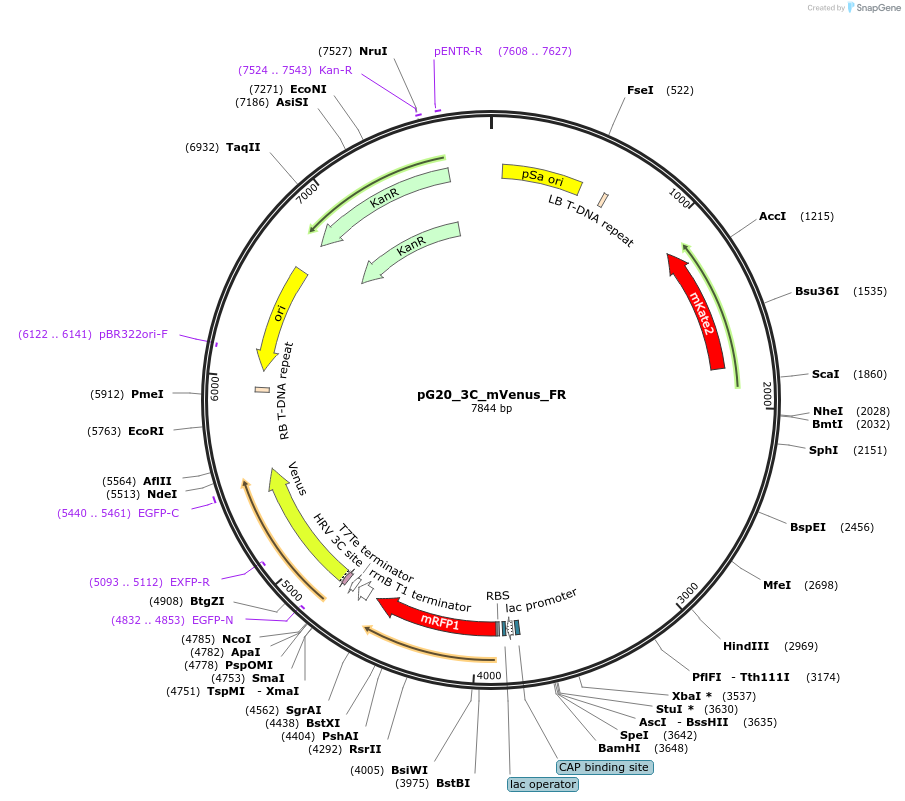
pG20_3C_mVenus_FR
Plasmid#240669PurposeExpression plasmid for 3C-mVenus-tagged proteins with FAST-Red transgenic seed selectionDepositorTypeEmpty backboneTags3C-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
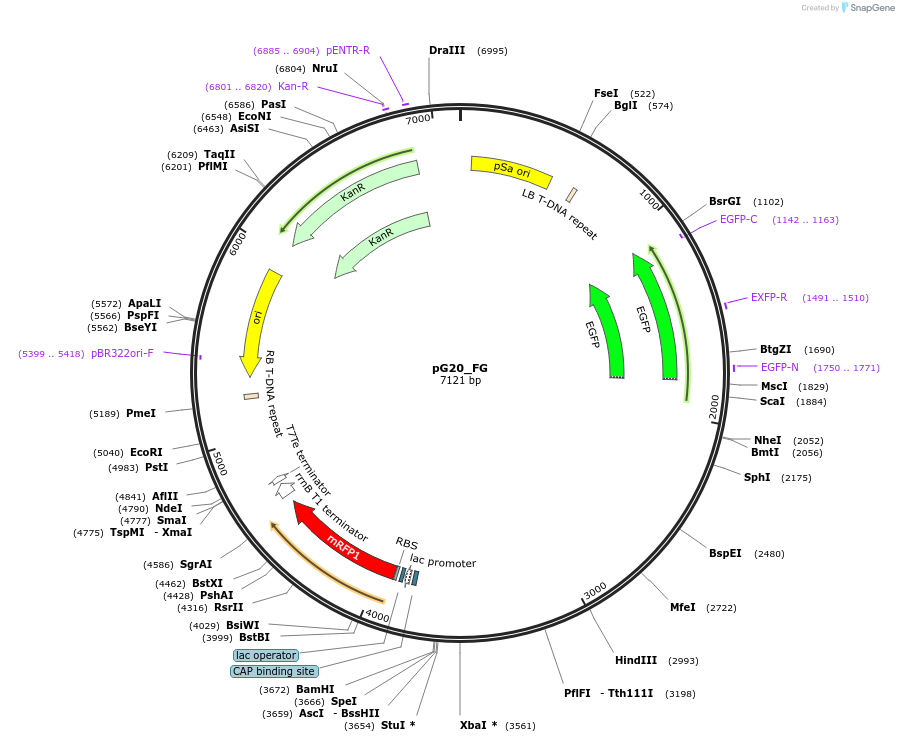
pG20_FG
Plasmid#240670PurposeExpression plasmid for un-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
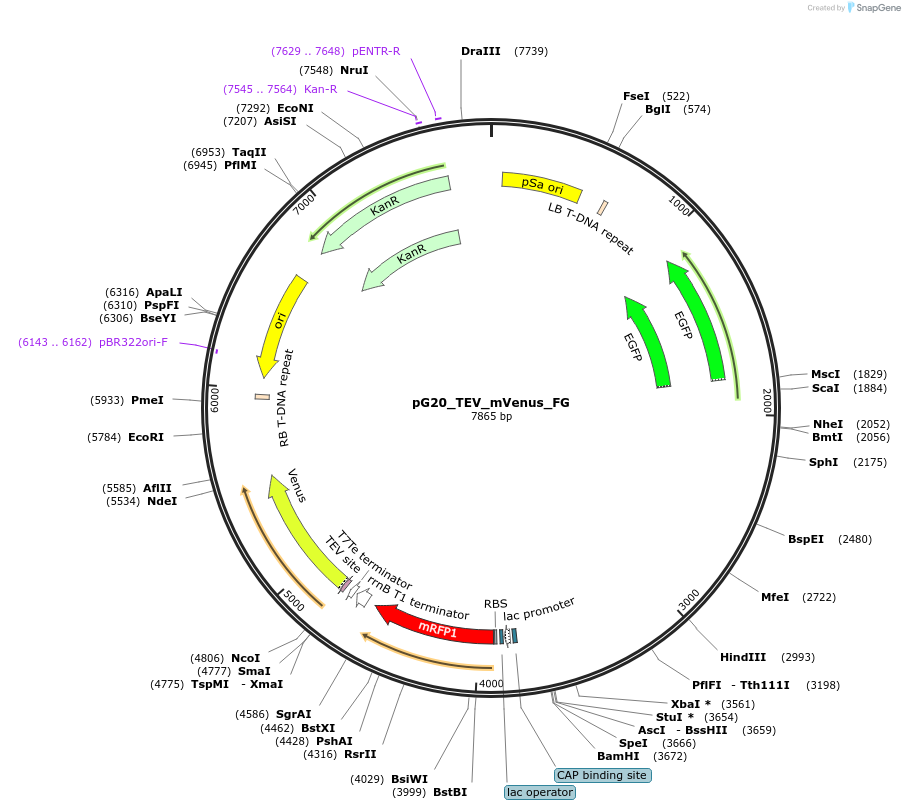
pG20_TEV_mVenus_FG
Plasmid#240671PurposeExpression plasmid for TEV-mVenus-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
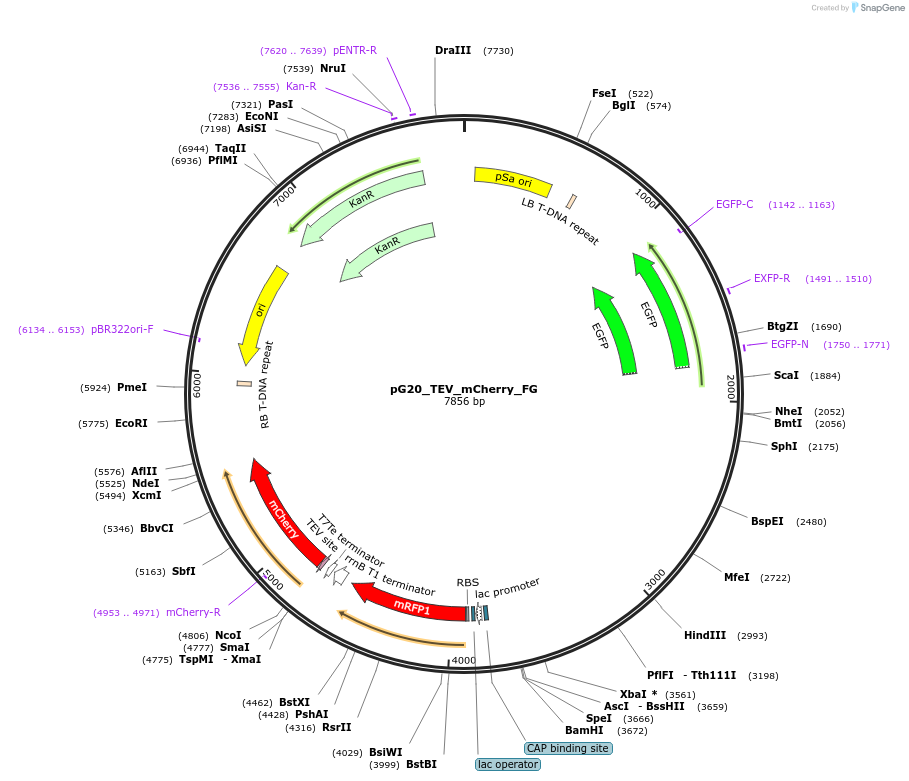
pG20_TEV_mCherry_FG
Plasmid#240672PurposeExpression plasmid for TEV-mCherry-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-mCherryExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
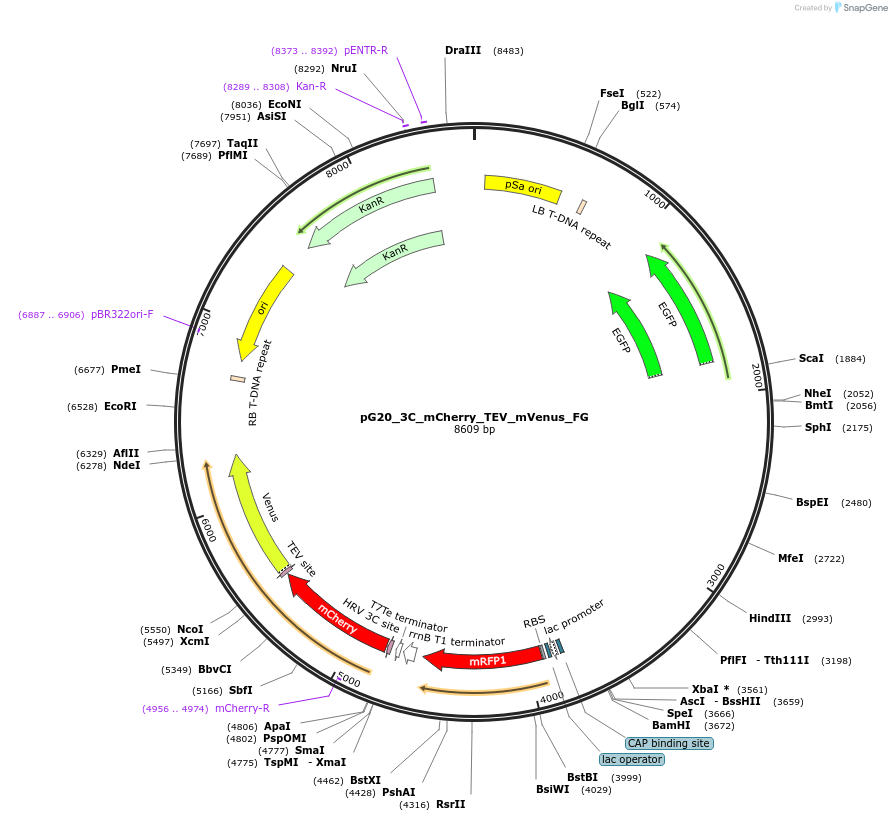
pG20_3C_mCherry_TEV_mVenus_FG
Plasmid#240674PurposeExpression plasmid for 3C-mCherry-TEV-mVenus-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTags3C-mCherry-TEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
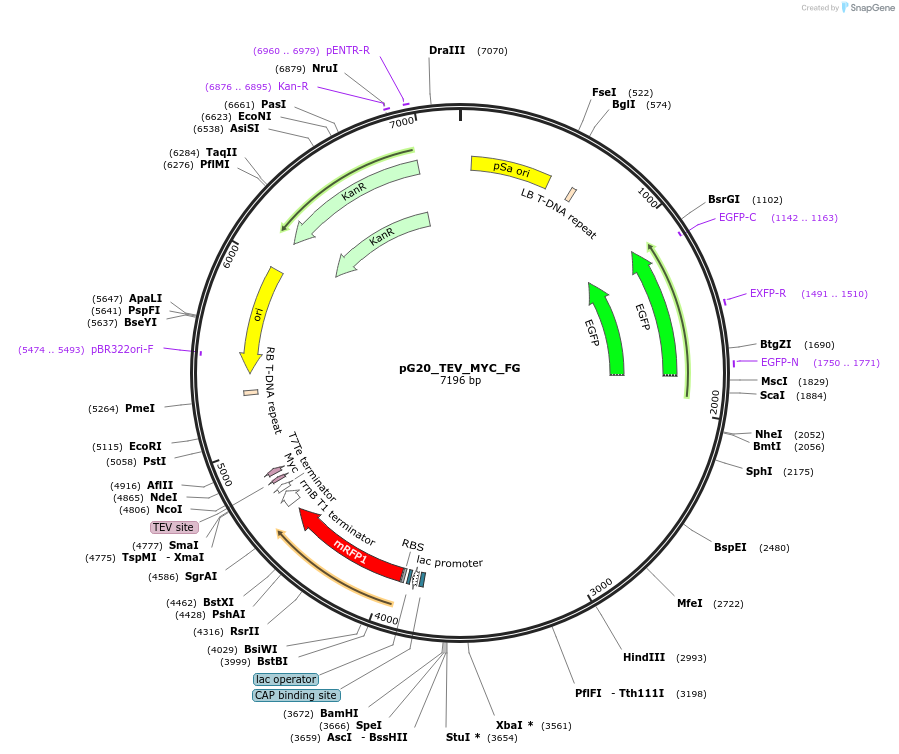
pG20_TEV_MYC_FG
Plasmid#240676PurposeExpression plasmid for TEV-MYC-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-MYCExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
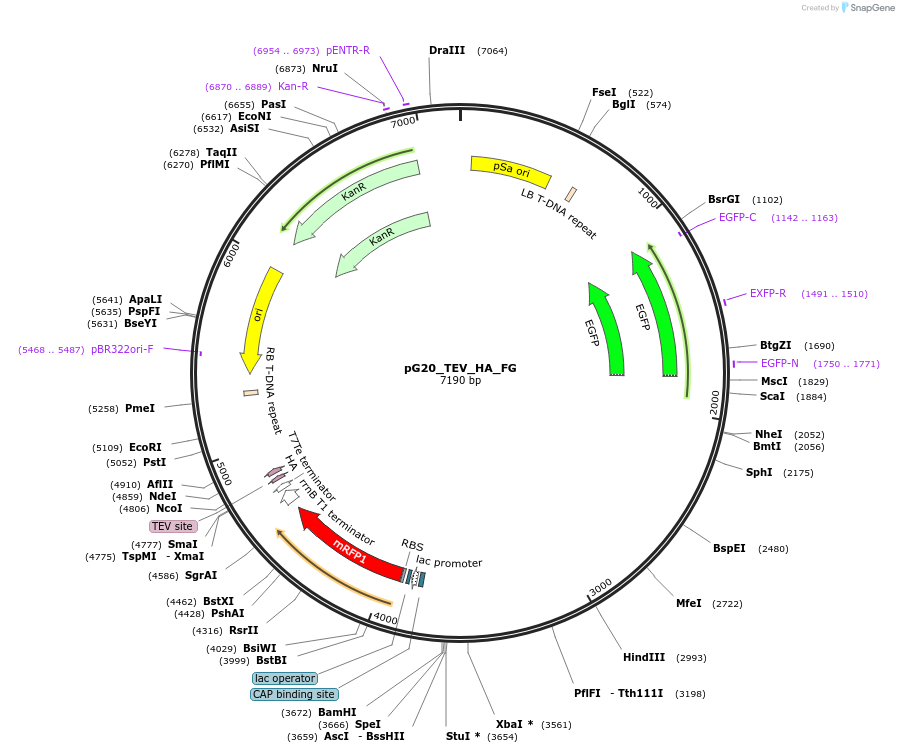
pG20_TEV_HA_FG
Plasmid#240677PurposeExpression plasmid for TEV-HA-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-HAExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
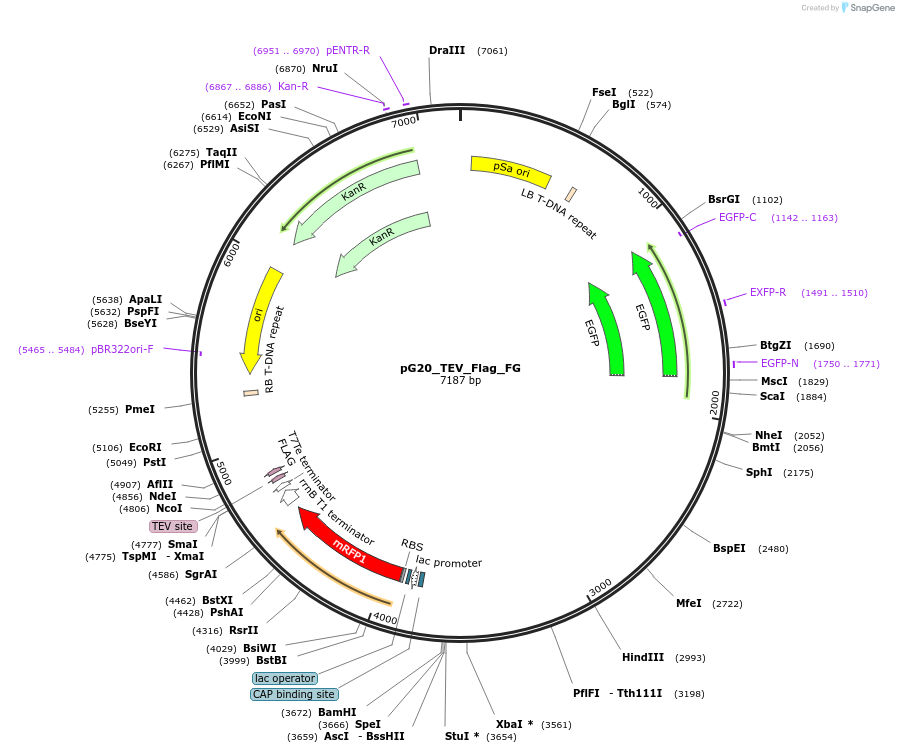
pG20_TEV_Flag_FG
Plasmid#240678PurposeExpression plasmid for TEV-Flag-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-FlagExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
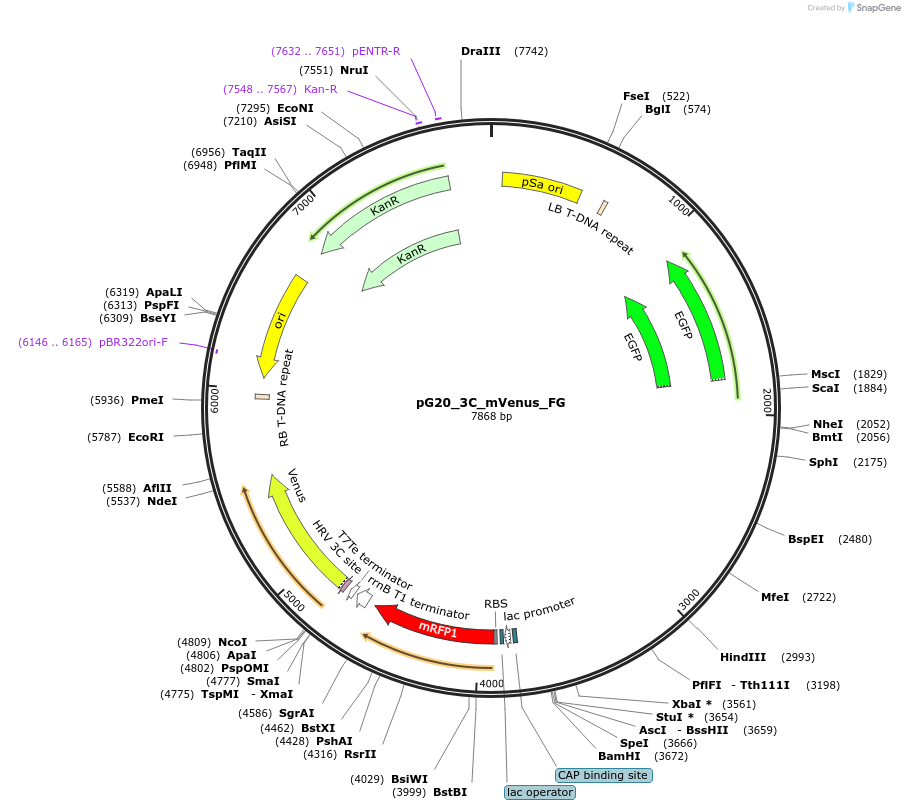
pG20_3C_mVenus_FG
Plasmid#240679PurposeExpression plasmid for 3C-mVenus-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTags3C-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
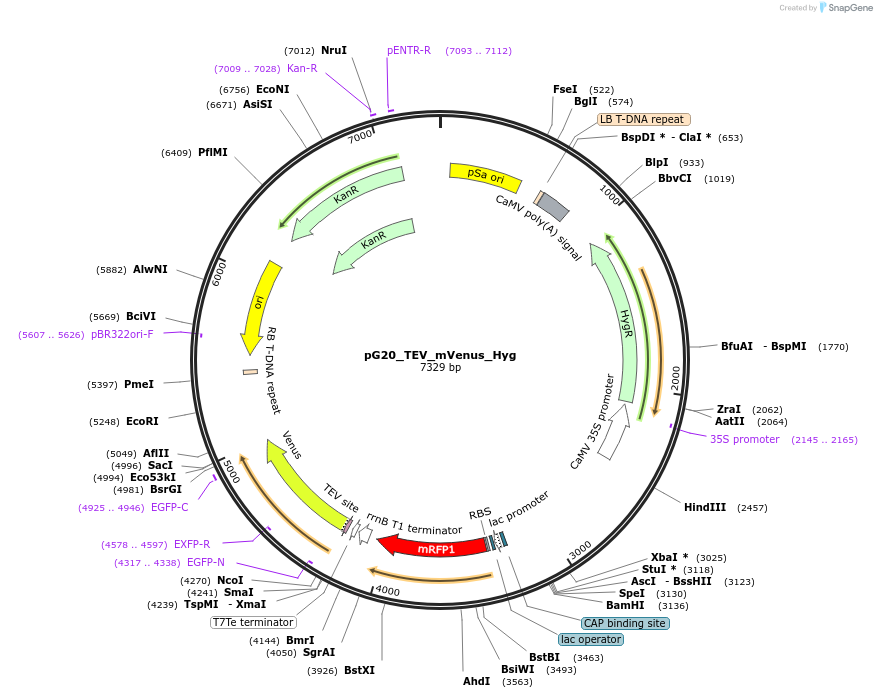
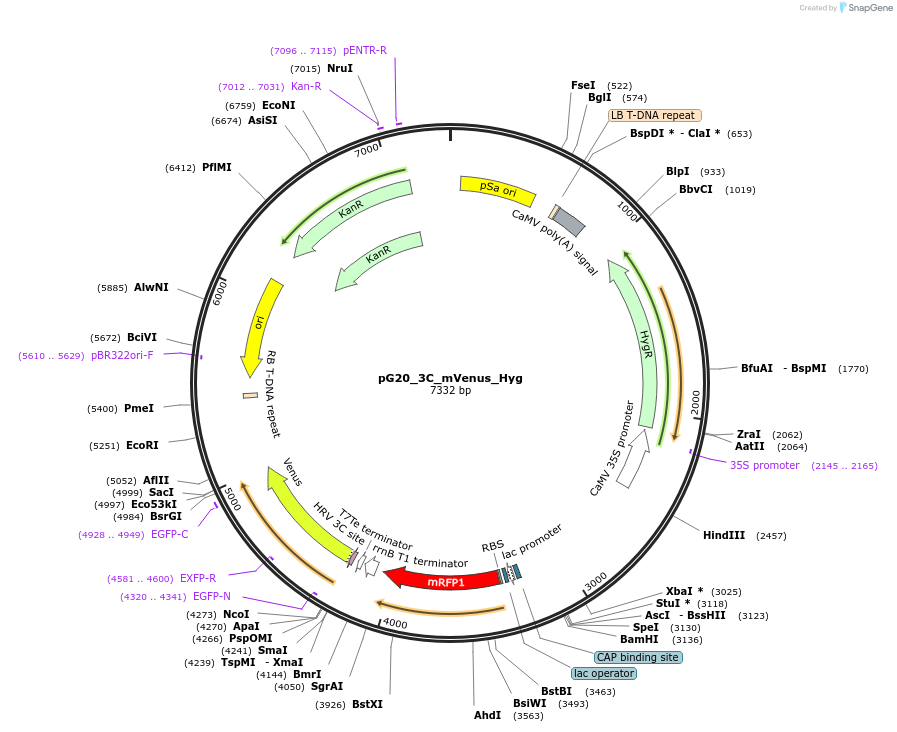
pG20_TEV_mVenus_Hyg
Plasmid#240651PurposeExpression plasmid for TEV-mVenus-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
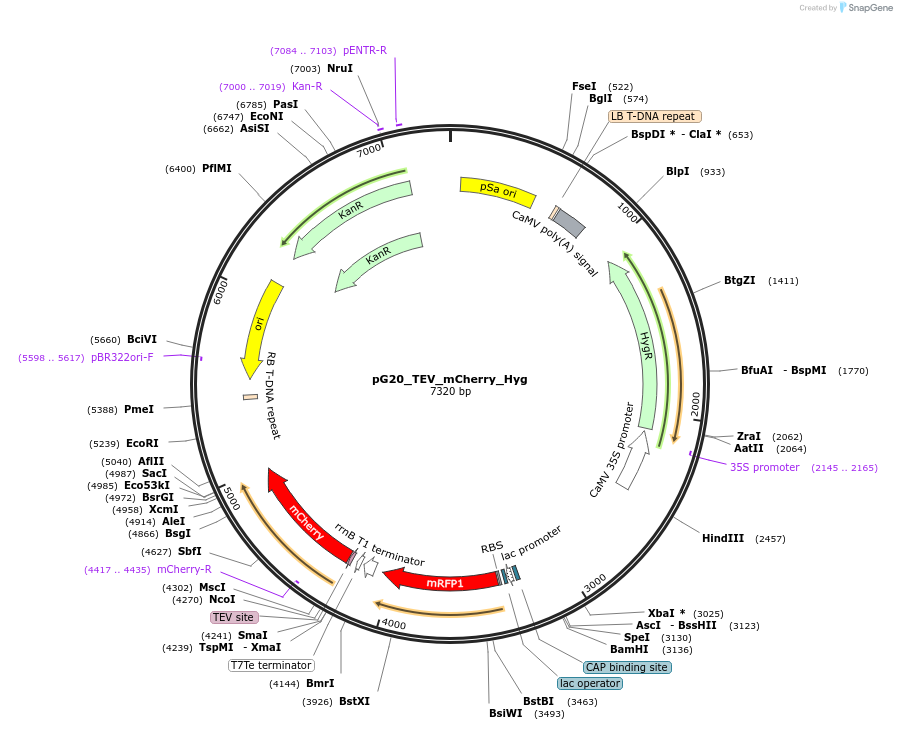
pG20_TEV_mCherry_Hyg
Plasmid#240652PurposeExpression plasmid for TEV-mCherry-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-mCherryExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
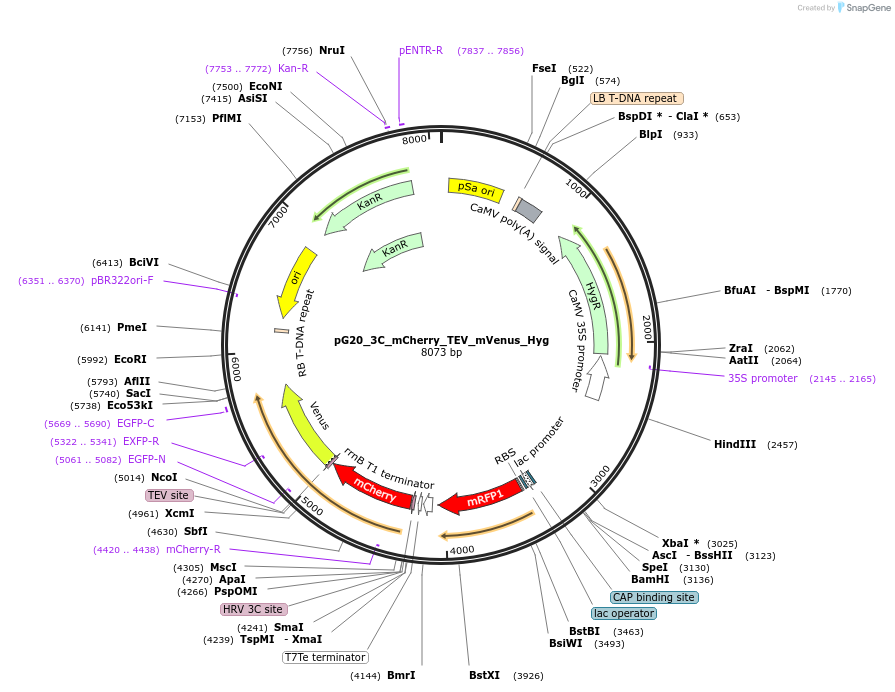
pG20_3C_mCherry_TEV_mVenus_Hyg
Plasmid#240654PurposeExpression plasmid for 3C-mCherry-TEV-mVenus-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTags3C-mCherry-TEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
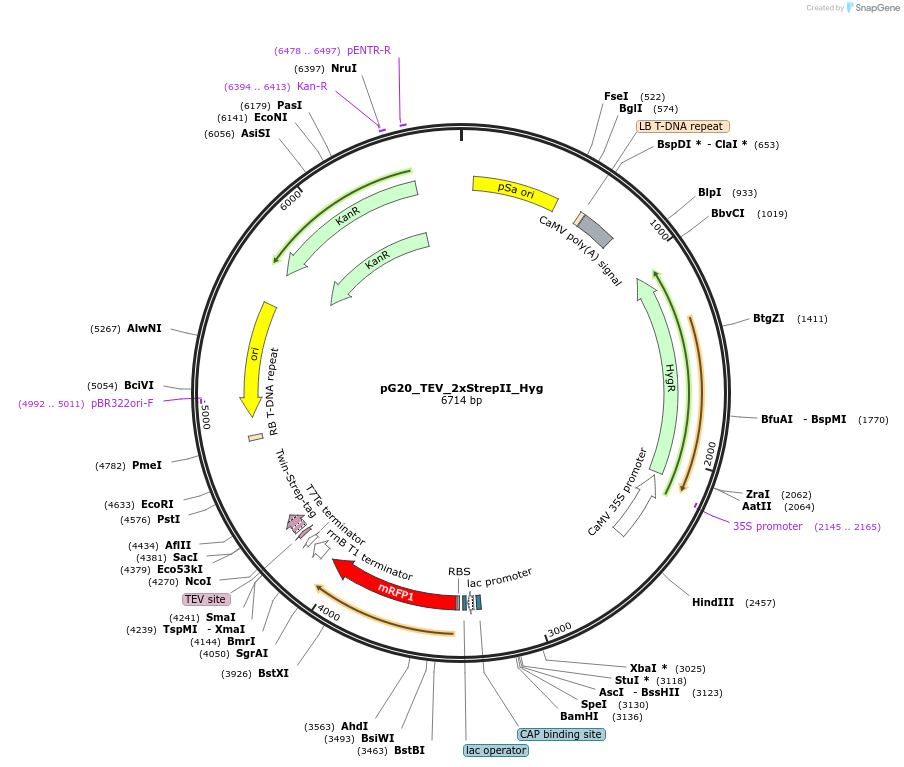
pG20_TEV_2xStrepII_Hyg
Plasmid#240655PurposeExpression plasmid for TEV-2xStrepII-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-2xStrepIIExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
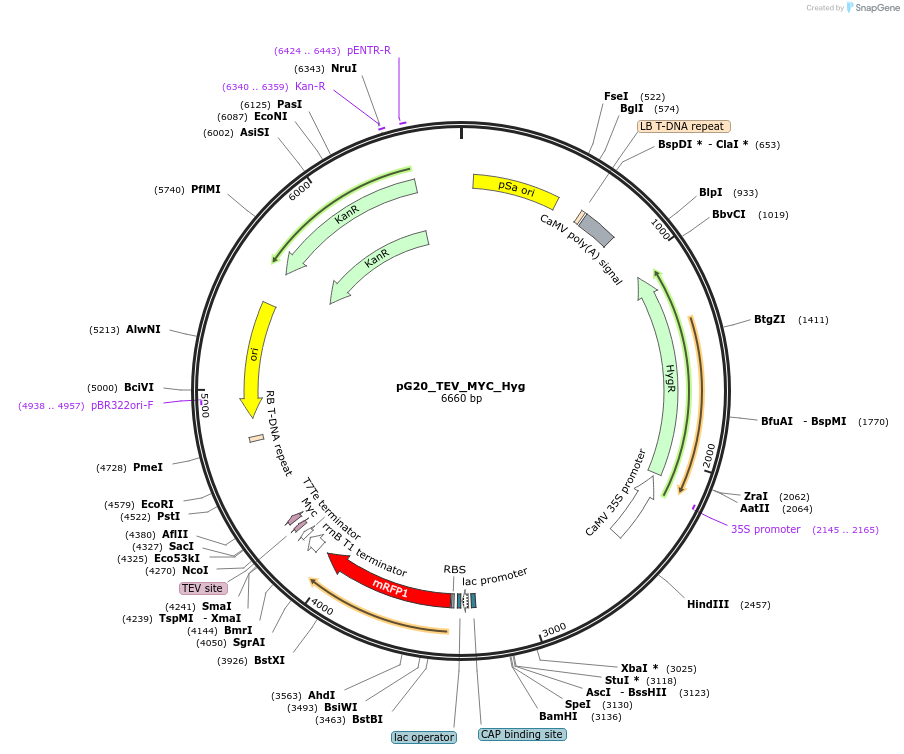
pG20_TEV_MYC_Hyg
Plasmid#240656PurposeExpression plasmid for TEV-MYC-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-MYCExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
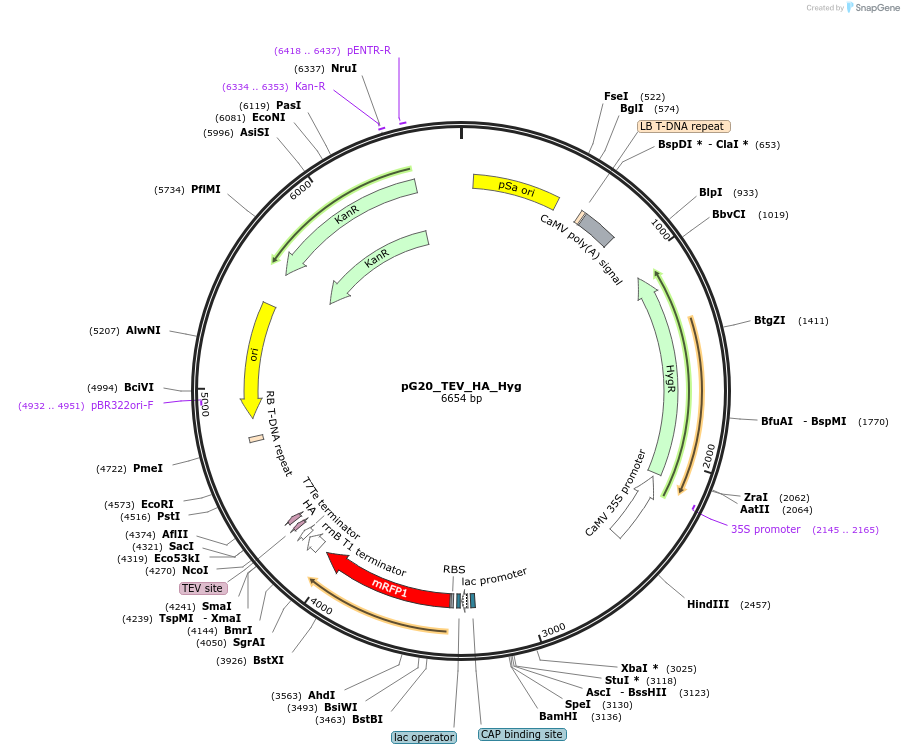
pG20_TEV_HA_Hyg
Plasmid#240657PurposeExpression plasmid for TEV-HA-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-HAExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
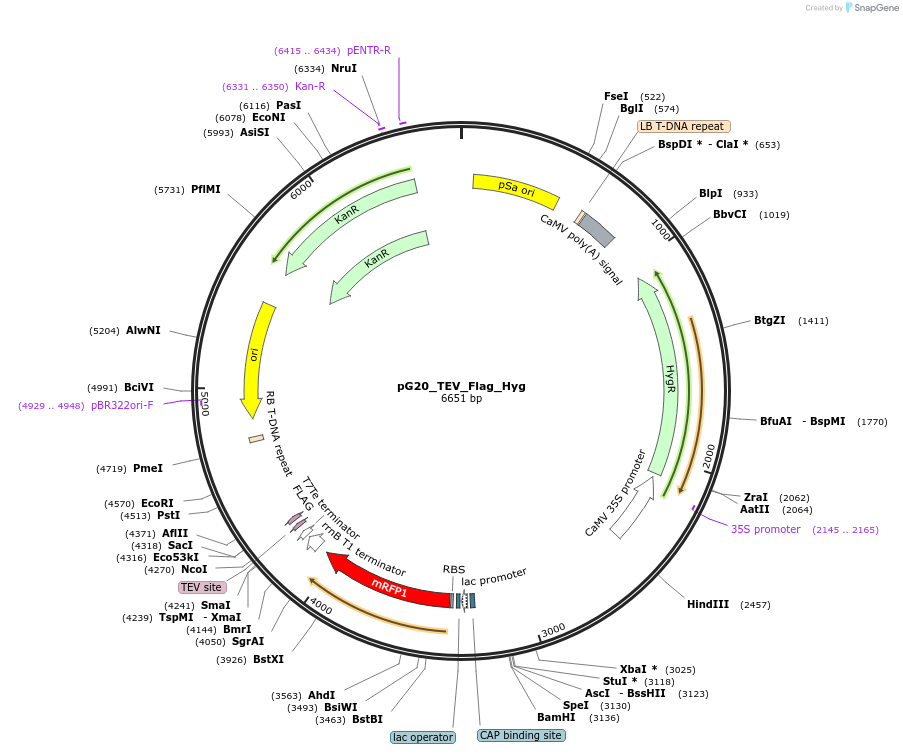
pG20_TEV_Flag_Hyg
Plasmid#240658PurposeExpression plasmid for TEV-Flag-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-FlagExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
pG20_3C_mVenus_Hyg
Plasmid#240659PurposeExpression plasmid for 3C-mVenus-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTags3C-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
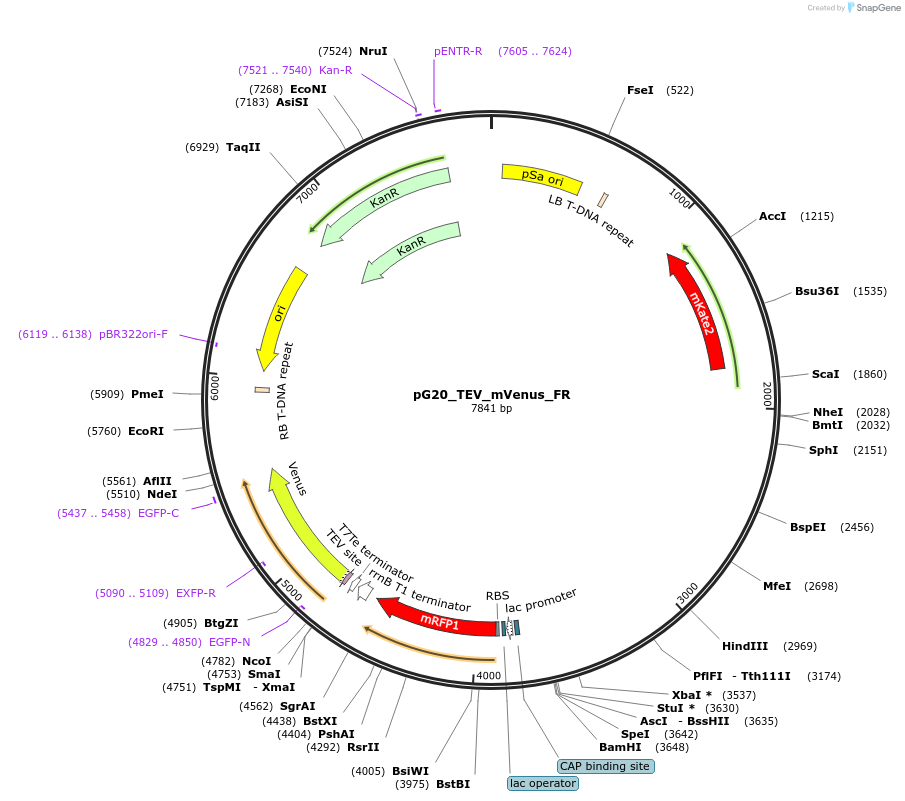
pG20_TEV_mVenus_FR
Plasmid#240661PurposeExpression plasmid for TEV-mVenus-tagged proteins with FAST-Red transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only