8,173 results
-
Plasmid#190042PurposeBackbone vector for generation of BiFC plasmidsDepositorTypeEmpty backboneExpressionYeastPromoterMet3Available SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only
-
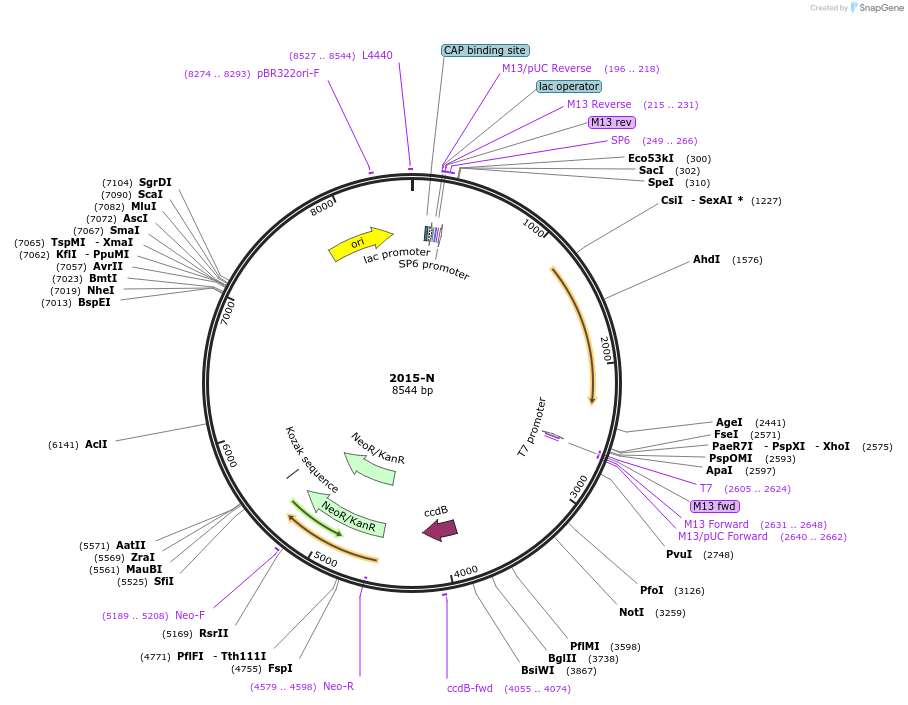
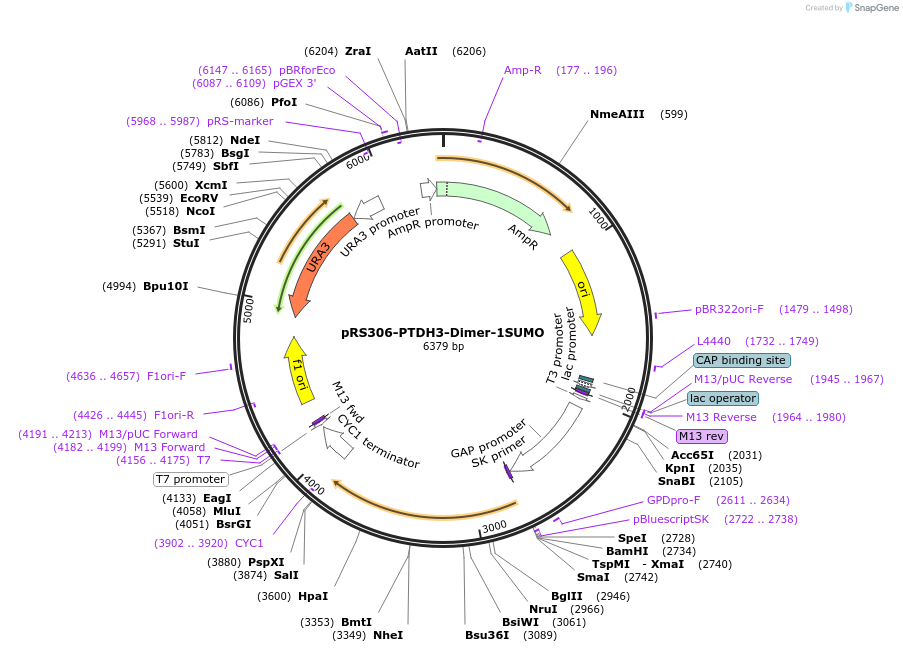
pRS306-PTDH3-Dimer-1SUMO
Plasmid#190249PurposeExpression dimer-1SUMO component in yeast cellsDepositorInsertdimer-SUMO
ExpressionYeastAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
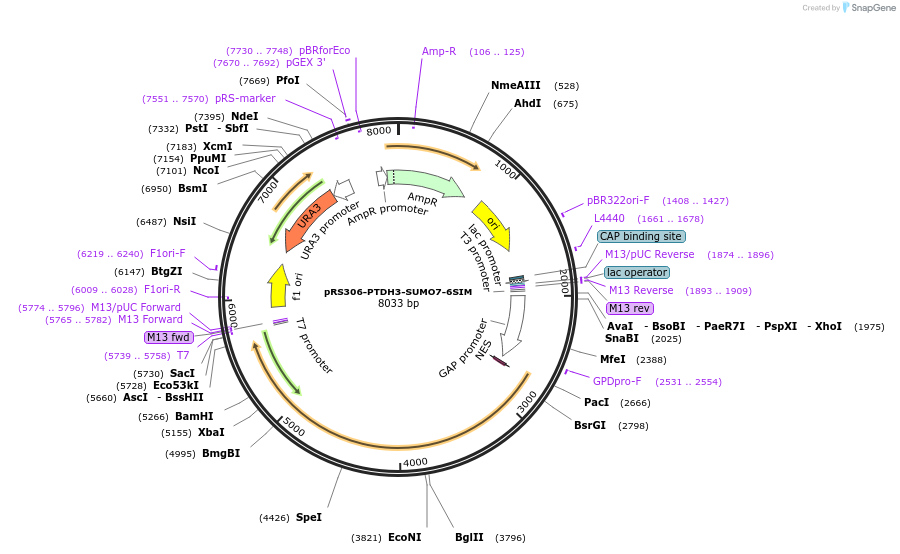
pRS306-PTDH3-SUMO7-6SIM
Plasmid#190250PurposeExpression SUMO7-SIM6 in yeast cellsDepositorInsert7SUMO-6SIM
ExpressionYeastAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
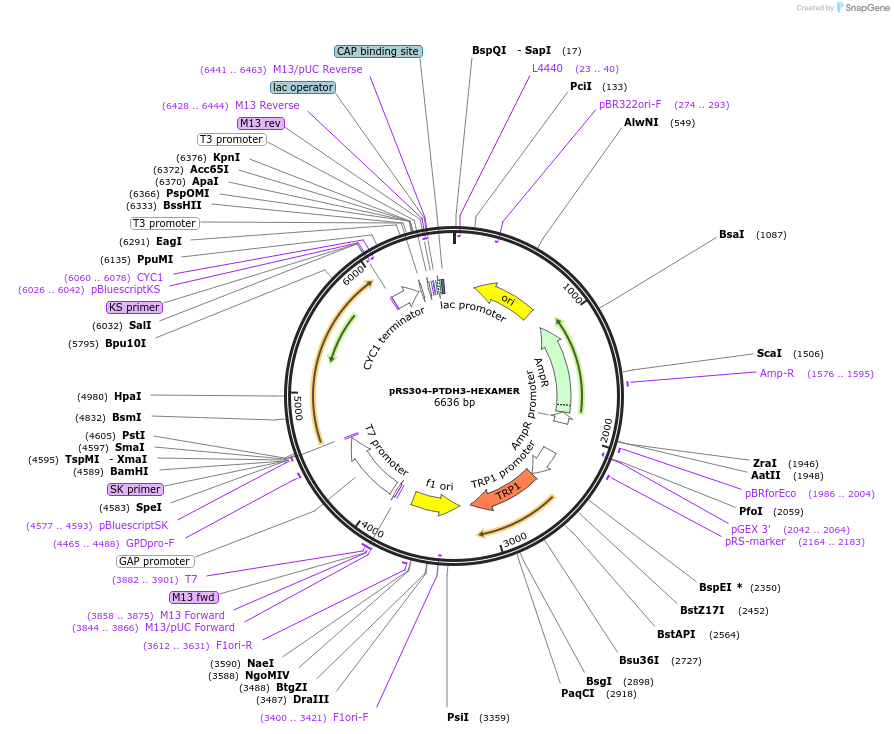
pRS304-PTDH3-HEXAMER
Plasmid#190251PurposeExpression hexamer component in yeastDepositorInserthexamer
TagsmCherryExpressionYeastAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
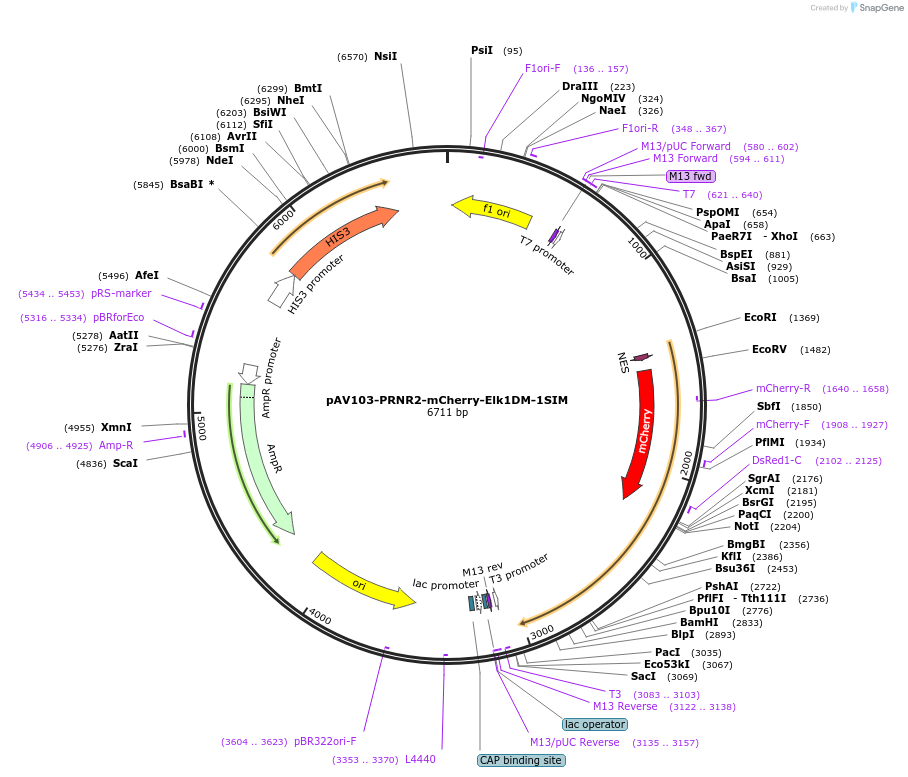
pAV103-PRNR2-mCherry-Elk1DM-1SIM
Plasmid#190252PurposeExpression mCherry-Elk1DM in yeast cellsDepositorInsertELK1DM-SIM
TagsmCherryExpressionYeastAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
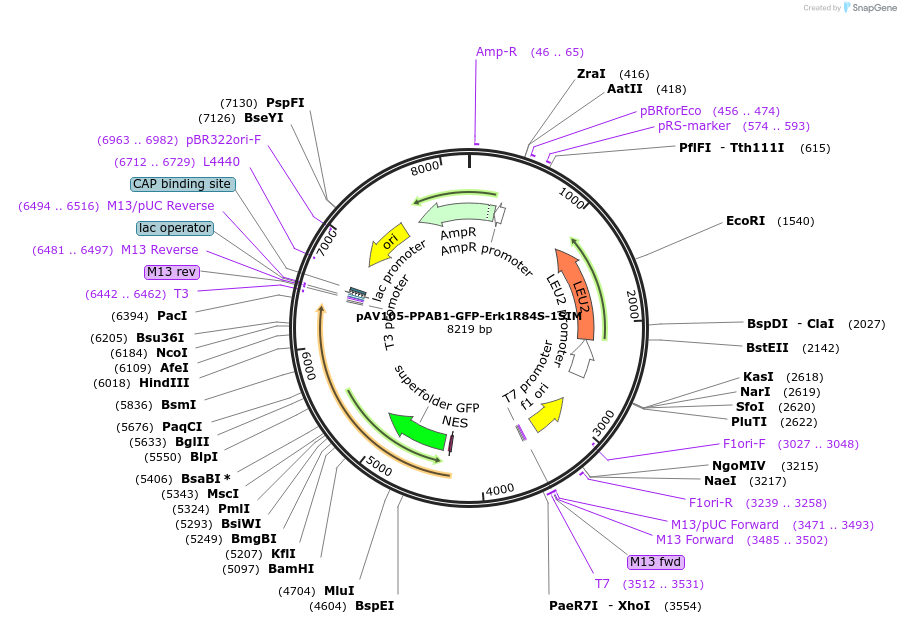
pAV105-PPAB1-GFP-Erk1R84S-1SIM
Plasmid#190253PurposeExpression Erk1-GFP-1SIM in yeast cellsDepositorInsertERK1R84S-SIM
TagsGFPExpressionYeastMutationERK1R84SAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
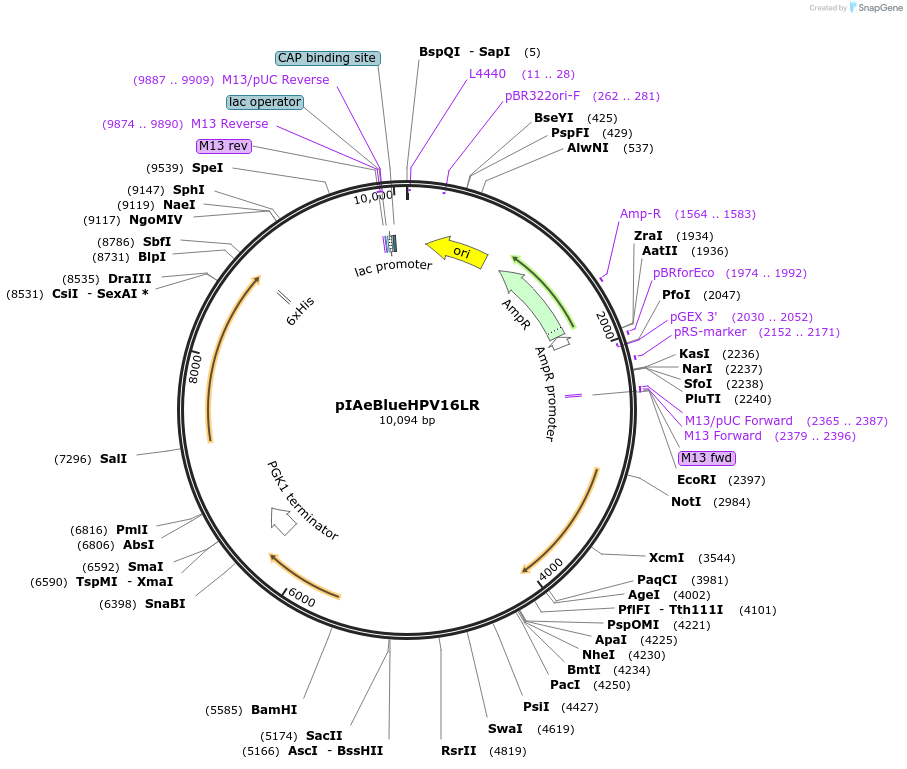
pIAeBlueHPV16LR
Plasmid#185894PurposeIn vivo gene amplification (HamAmp) of AeBlue + HPV16 L1 expression module at RPL25 locusDepositorInsertPRPL25(Arm1)>Kl.LEU2>TKl.LEU2-TRPL25(Arm3)-ARS305-PALD6>EforRed>TPGK1-PSe.GAL2>HPV16-L1_C-6*H>TRPL41B-PBTS1>RPL25(partial;Arm2)
ExpressionYeastMutationNoneAvailable SinceNov. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
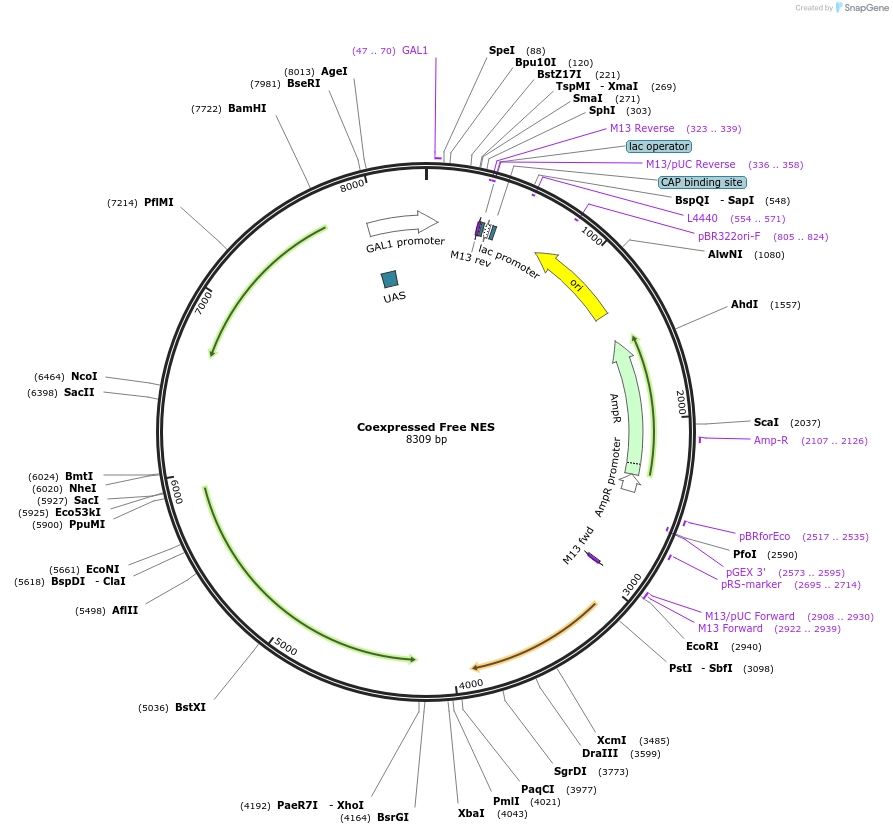
Coexpressed Free NES
Plasmid#182482PurposeYeast integrative plasmid for expressing ERG20 (GAL10 promoter) and nerolidol synthase from Actinidia chinensis (AcNES1; GAL7 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertsFPPS
NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10 and GAL7Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
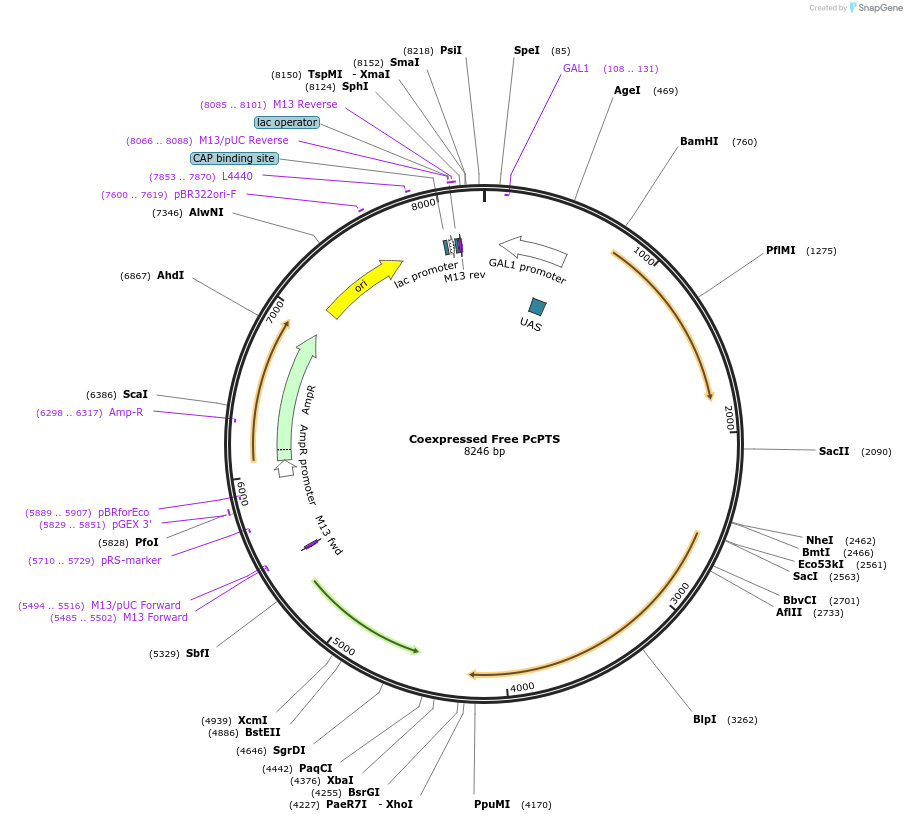
Coexpressed Free PcPTS
Plasmid#182483PurposeYeast integrative plasmid for expressing ERG20 (GAL10 promoter) and patchoulol synthase from Pogostemon cablin (PcPTS; GAL7 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertsFPPS
PTS
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10 and GAL7Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
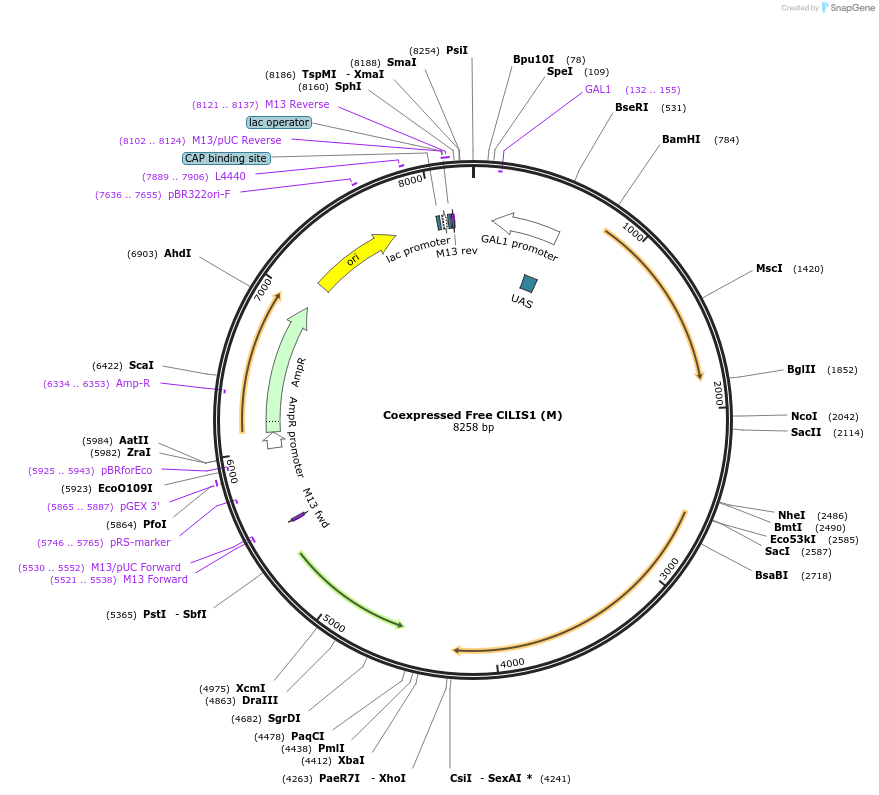
Coexpressed Free ClLIS1 (M)
Plasmid#182484PurposeYeast integrative plasmid for expressing ERG20(F96W-N127W) (GAL10 promoter) and limonene synthase from Citrus limon (ClLIS1; GAL7 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertsFPPS(M)
LS
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10 and GAL7Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
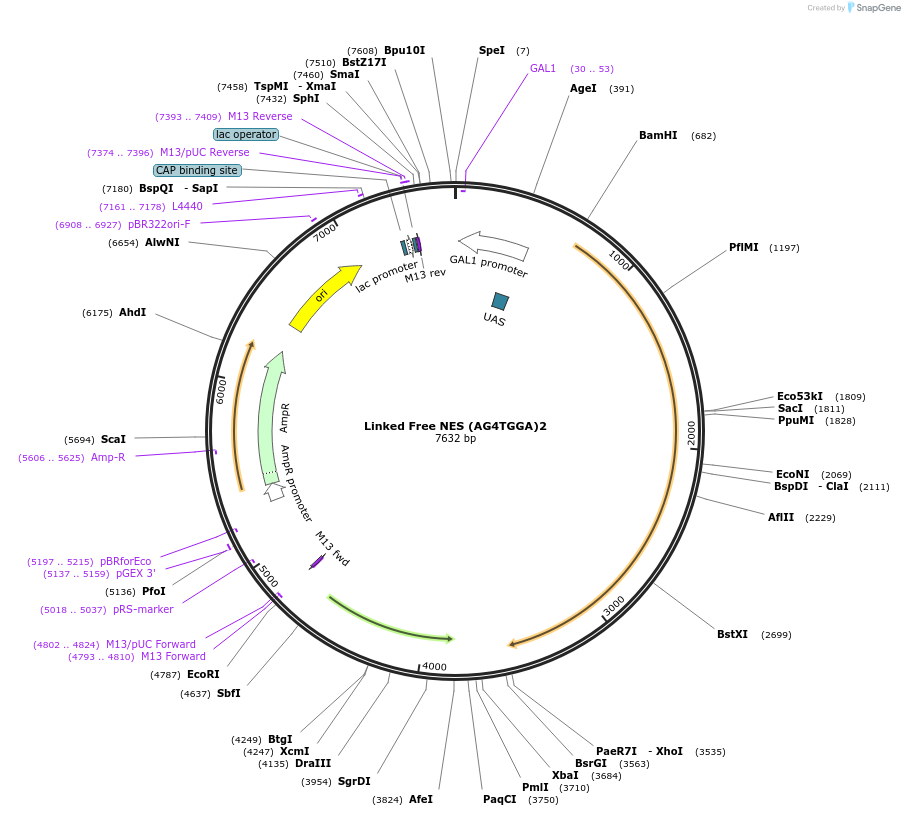
Linked Free NES (AG4TGGA)2
Plasmid#182488PurposeYeast integrative plasmid for expressing fusion protein ERG20-AcNES1, with linker sequence (AG4TGGA)2 (GAL10 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertFPPS-NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
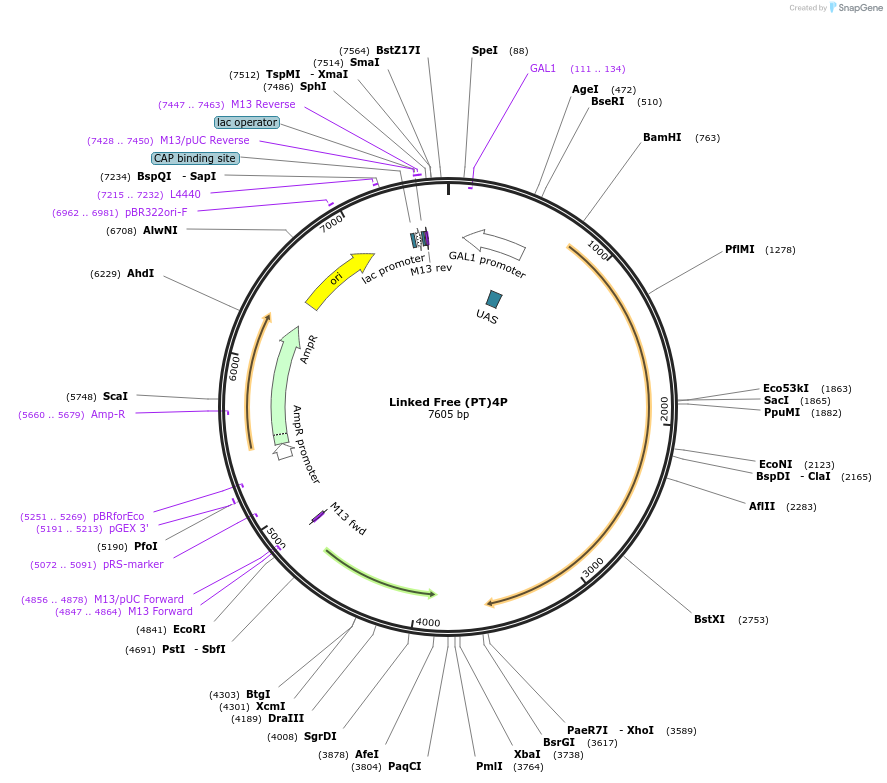
Linked Free (PT)4P
Plasmid#182490PurposeYeast integrative plasmid for expressing fusion protein ERG20-AcNES1, with linker sequence PTPTPTPTP (GAL10 promoter). Contains uracil auxotrophic marker (K. lactis URA3).DepositorInsertFPPS-NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
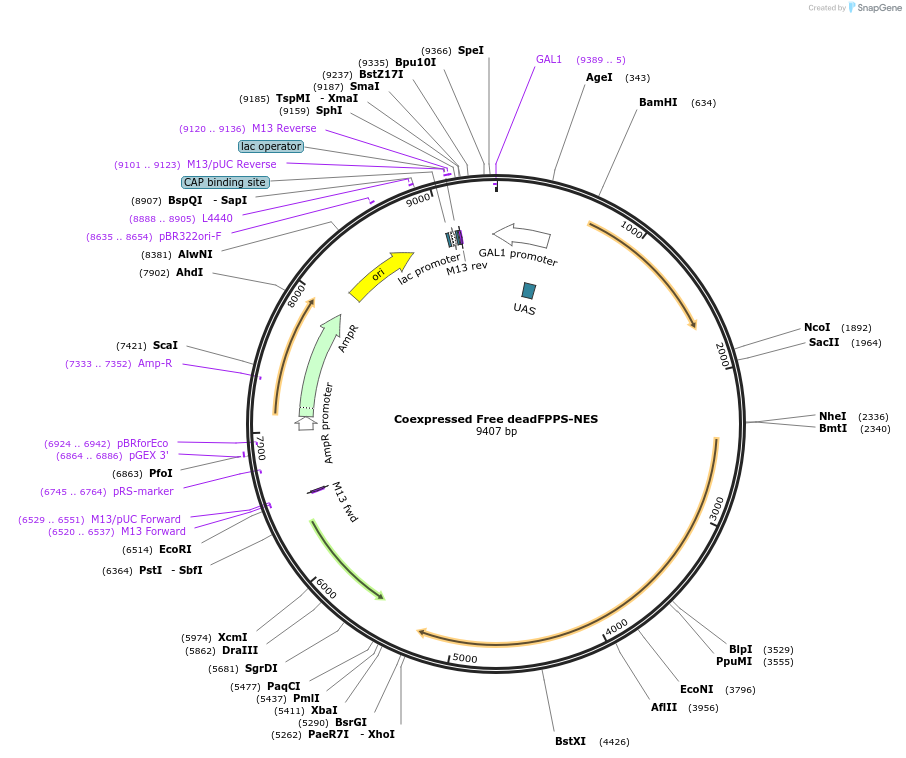
Coexpressed Free deadFPPS-NES
Plasmid#182492PurposeYeast integrative plasmid for expressing ERG20 (GAL10 promoter) and fusion protein deadFPPS-AcNES1 (GAL7 promoter). deadFPPS is ERG20(K197G-K254A), an inactive mutant of ERG20.DepositorInsertsFPPS
deadFPPS-NES
UseSynthetic Biology; Metabolic engineeringExpressionYeastPromoterGAL10 and GAL7Available SinceNov. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
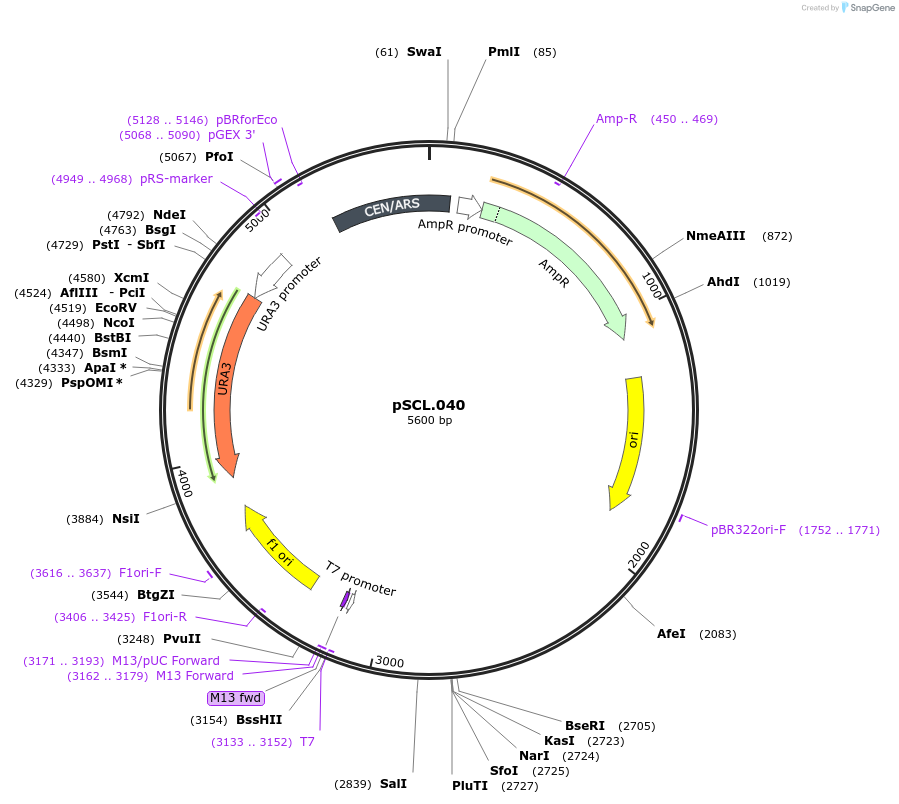
pSCL.040
Plasmid#184974PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v2
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
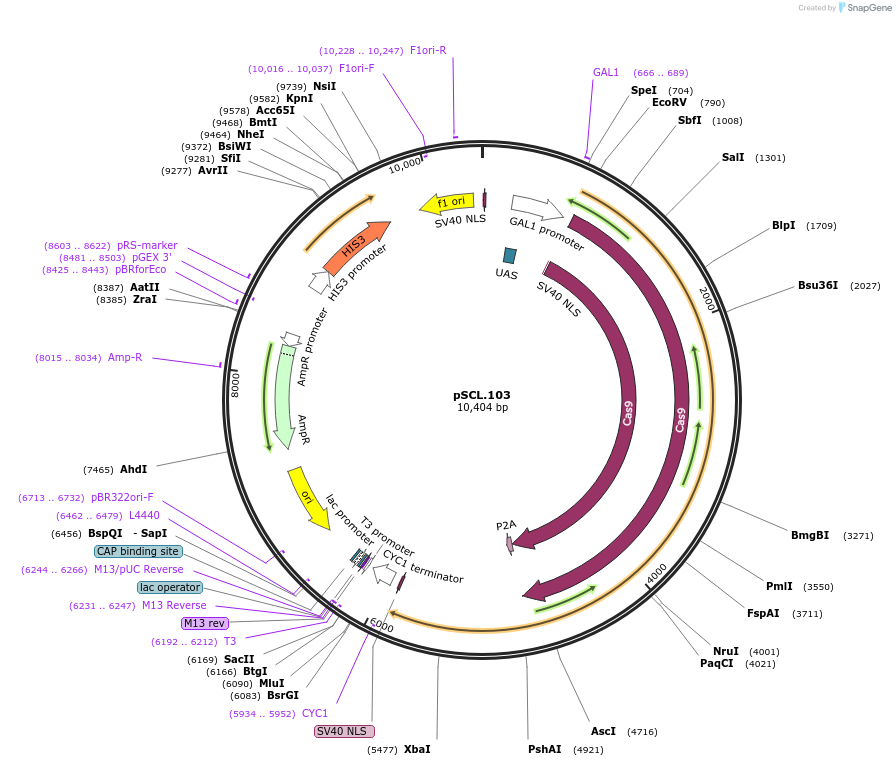
pSCL.103
Plasmid#184988PurposeExpress Cas9-P2A-Eco1RTDepositorInsertCas9-P2A-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
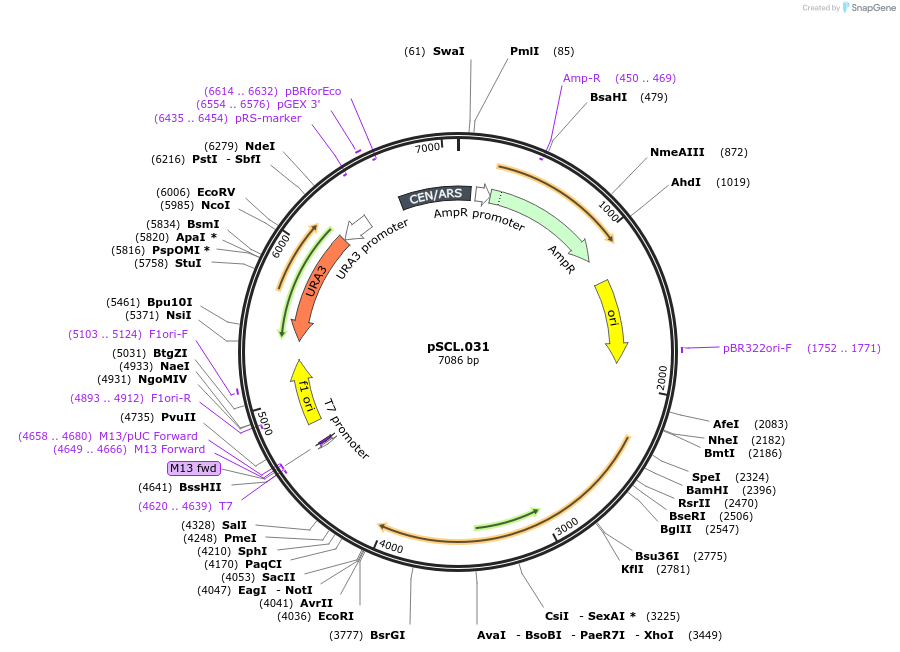
pSCL.031
Plasmid#184968PurposeTest effect of extending -Eco2 a1/a2 in yeastDepositorInsertEco2: RT and ncRNA(extended), a1/a2 length: 29
ExpressionYeastMutationhuman codon optimized RT, a1/a2 length extended t…PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
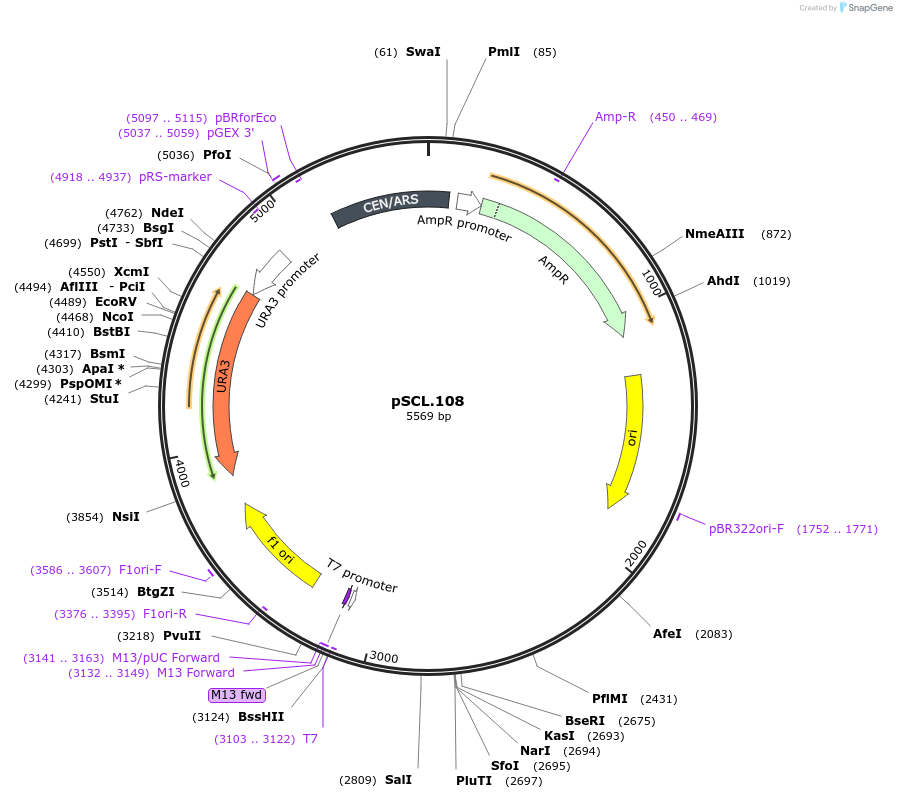
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
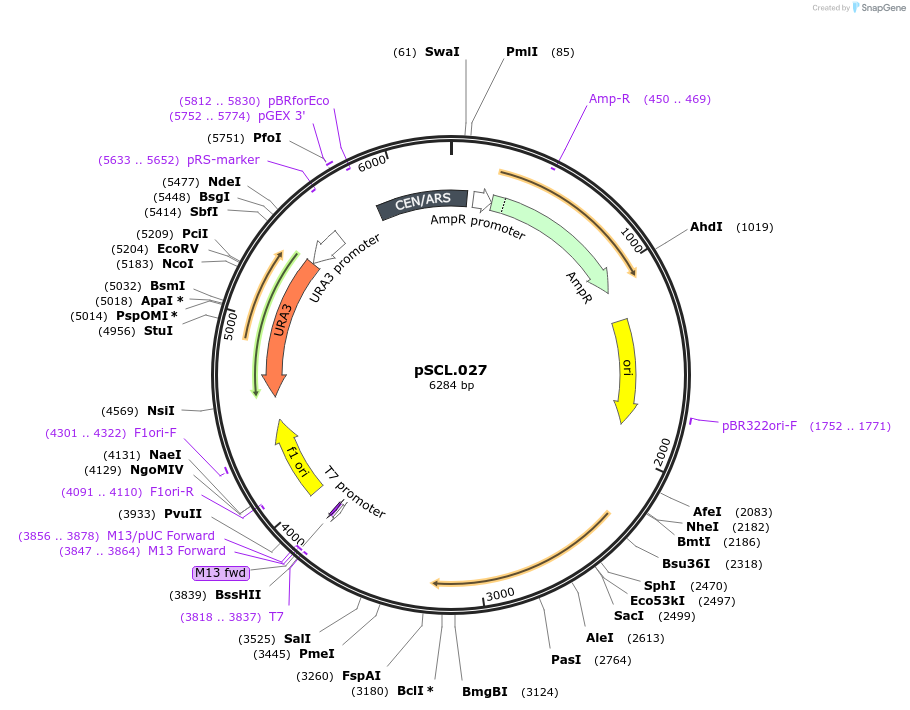
pSCL.027
Plasmid#184964PurposeExpress -Eco1 RT and ncRNA in yeastDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12
ExpressionYeastMutationhuman codon optimized RTPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
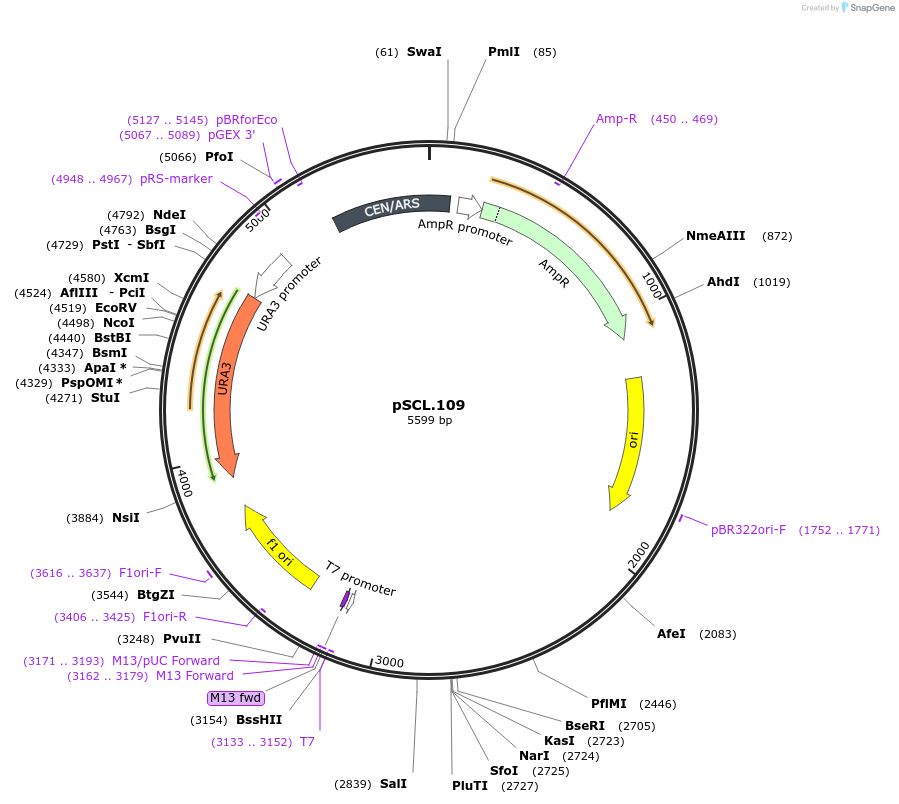
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
ExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
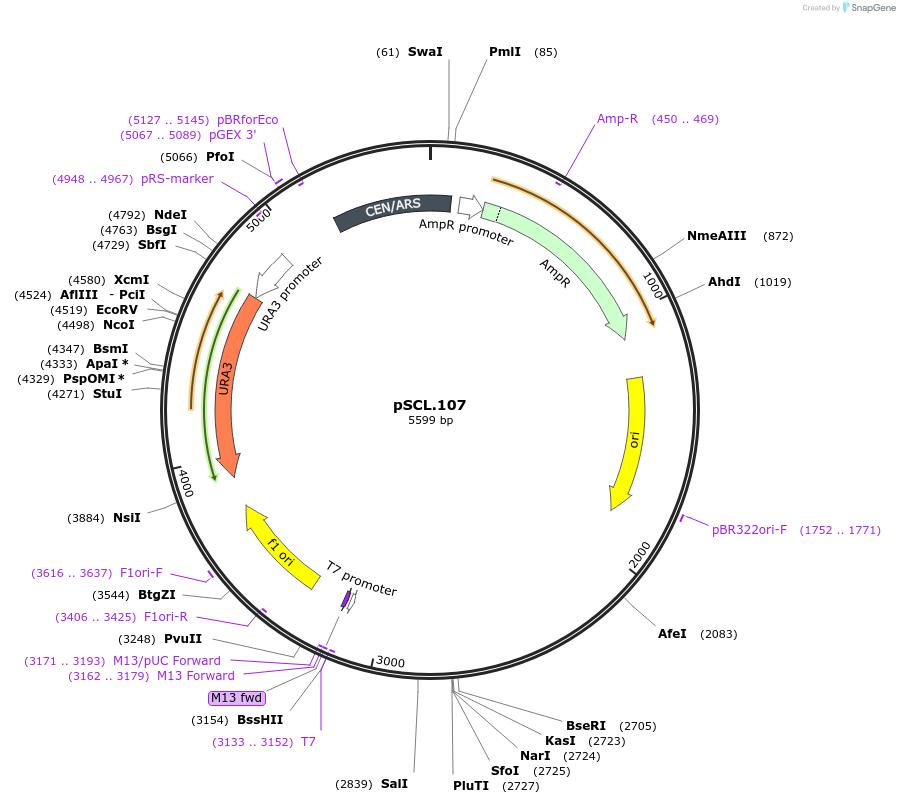
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
ExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
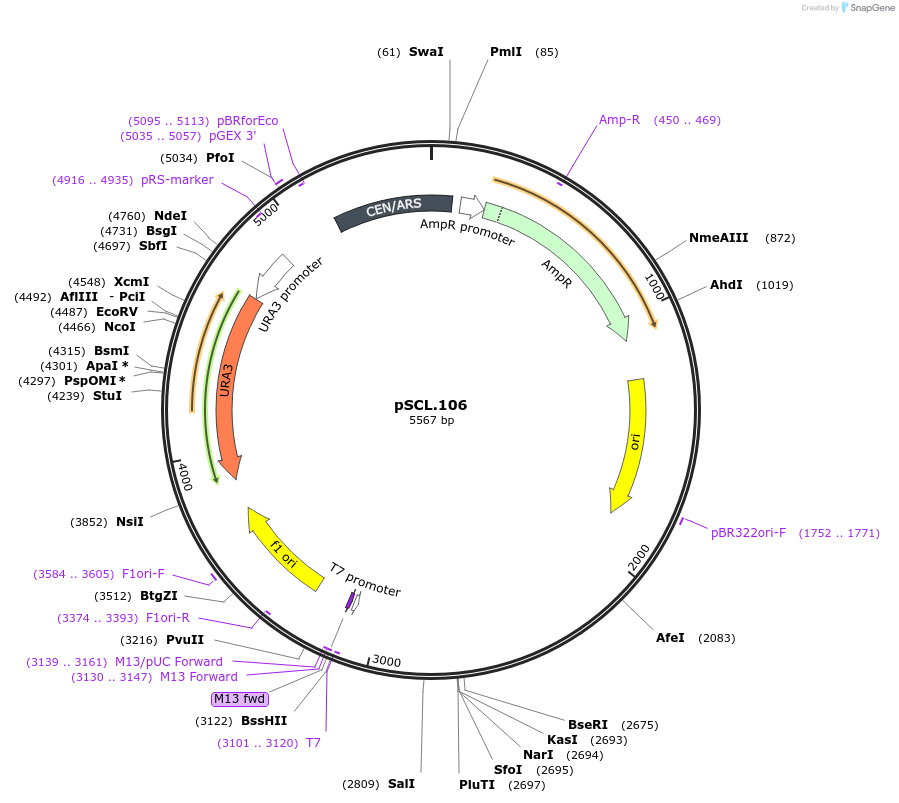
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
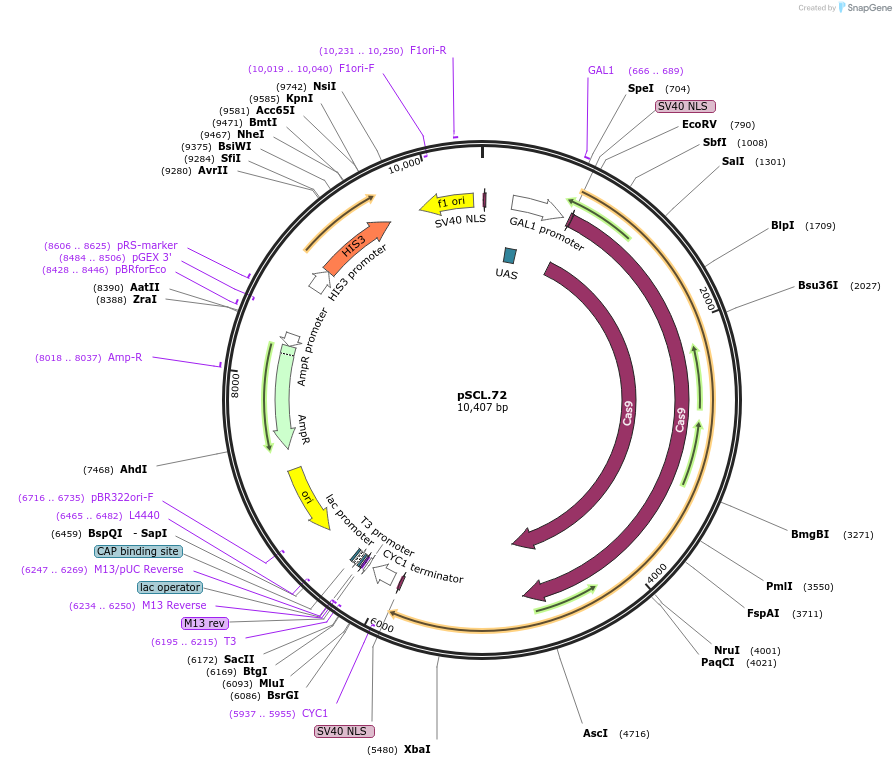
pSCL.72
Plasmid#184984PurposeExpress Cas9/-Eco1 RT fusion, using linker1DepositorInsertCas9-fusion_linker1-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
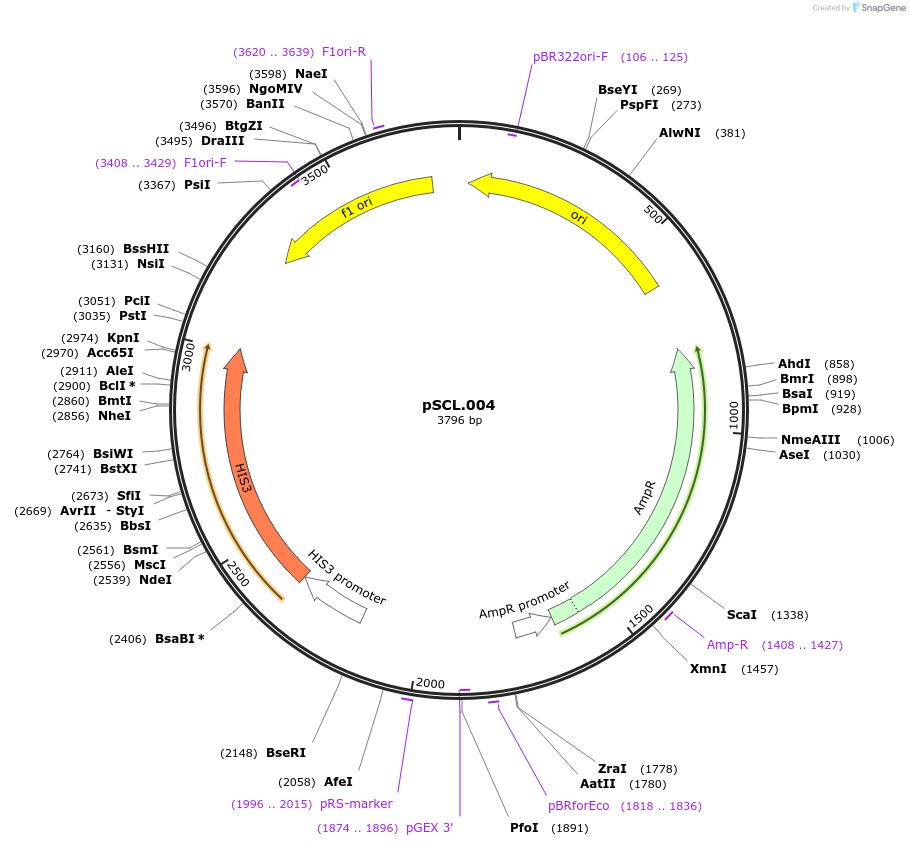
pSCL.004
Plasmid#184969PurposeNegative control, empty integrating inducible casetteDepositorTypeEmpty backboneExpressionYeastAvailable SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
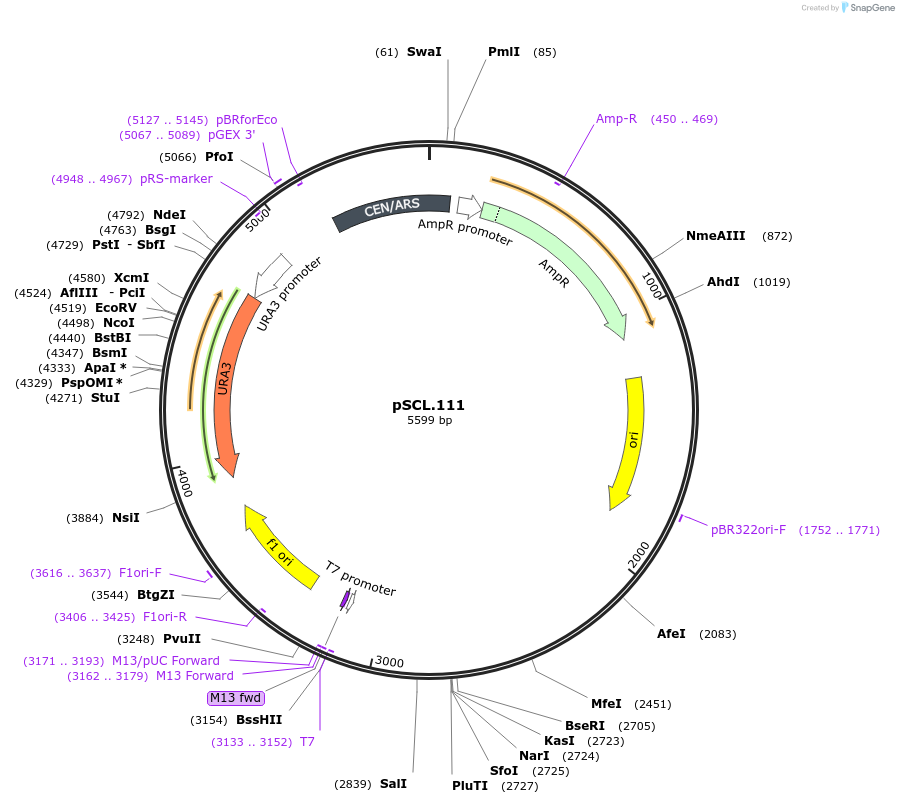
pSCL.111
Plasmid#184980PurposeTest effect of extending a1/a2 on TRP2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 27 v1
ExpressionYeastMutationTRP2 donor E64stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
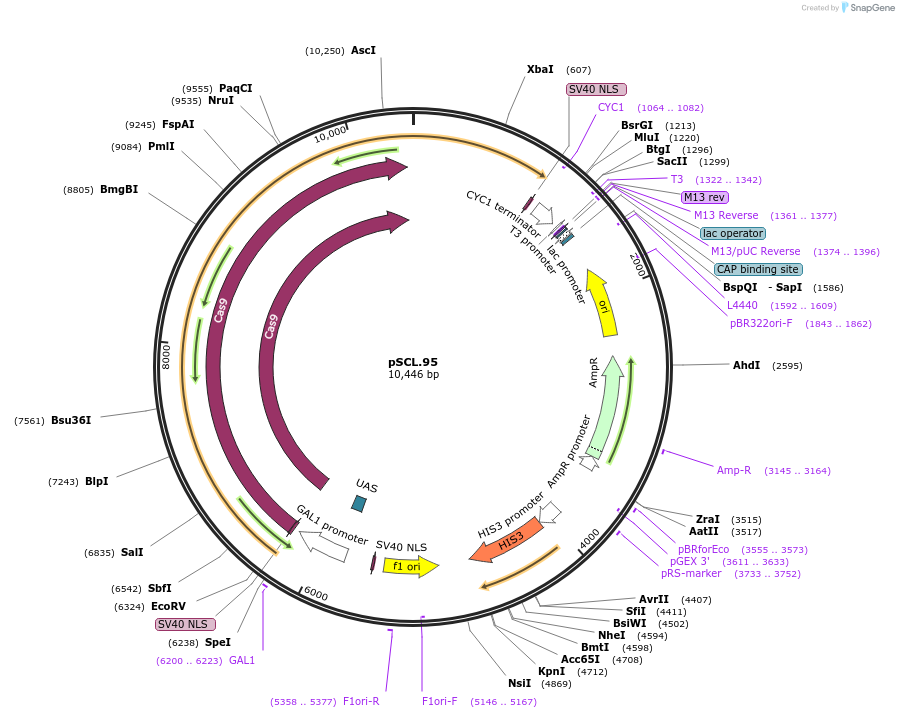
pSCL.95
Plasmid#184986PurposeExpress Cas9/-Eco1 RT fusion, using linker2DepositorInsertCas9-fusion_linker2-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
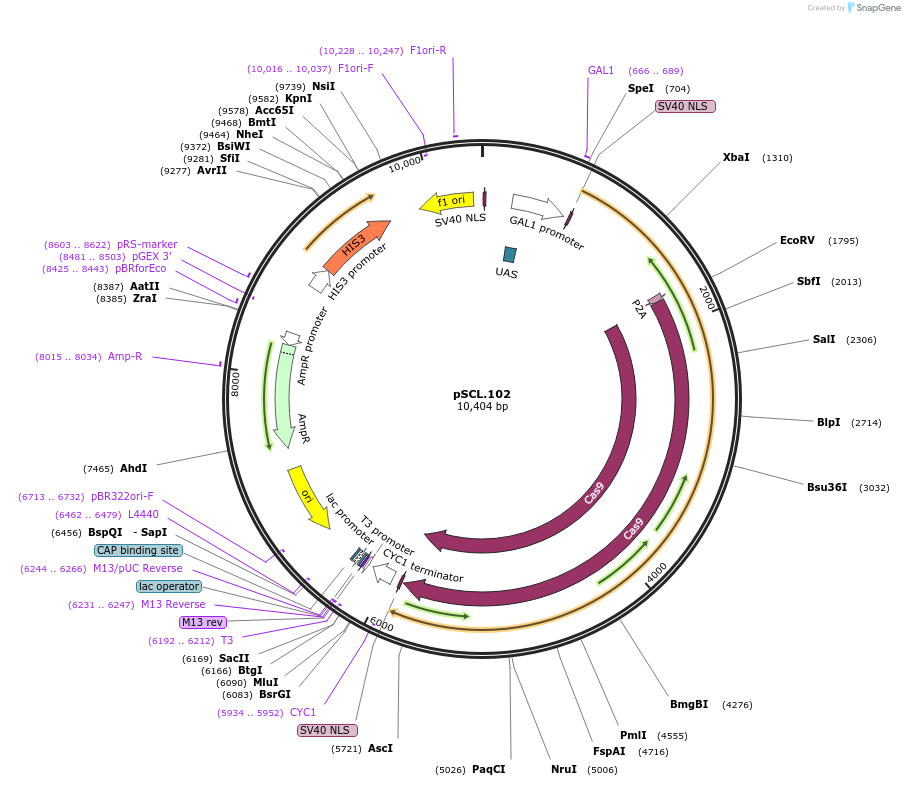
pSCL.102
Plasmid#184987PurposeExpress -Eco1 RT-P2A-Cas9DepositorInsertEco1RT-P2A-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
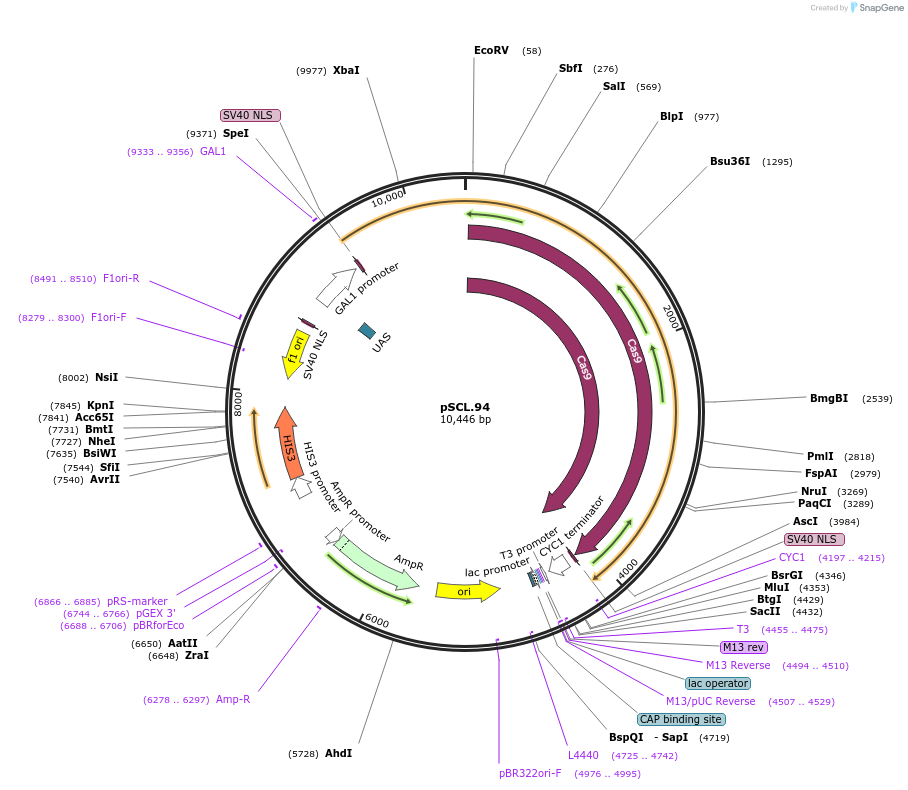
pSCL.94
Plasmid#184985PurposeExpress -Eco1 RT/Cas9 fusion, using linker2DepositorInsertEco1RT-fusion_linker2-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
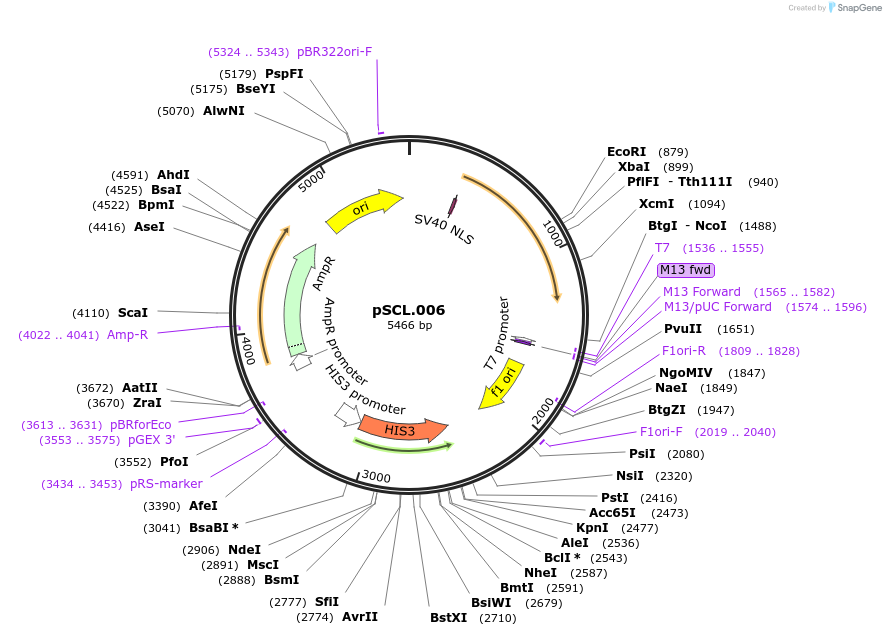
pSCL.006
Plasmid#184970PurposeExpress -Eco1 RT in yeastDepositorInsertIntegrating inducible cassette: Eco1 RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
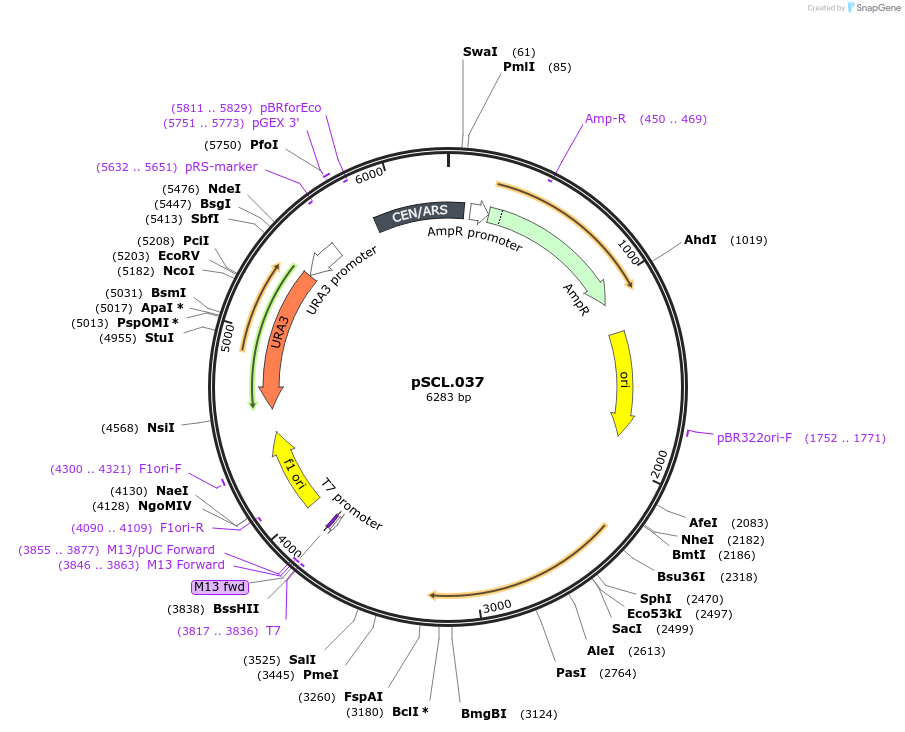
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
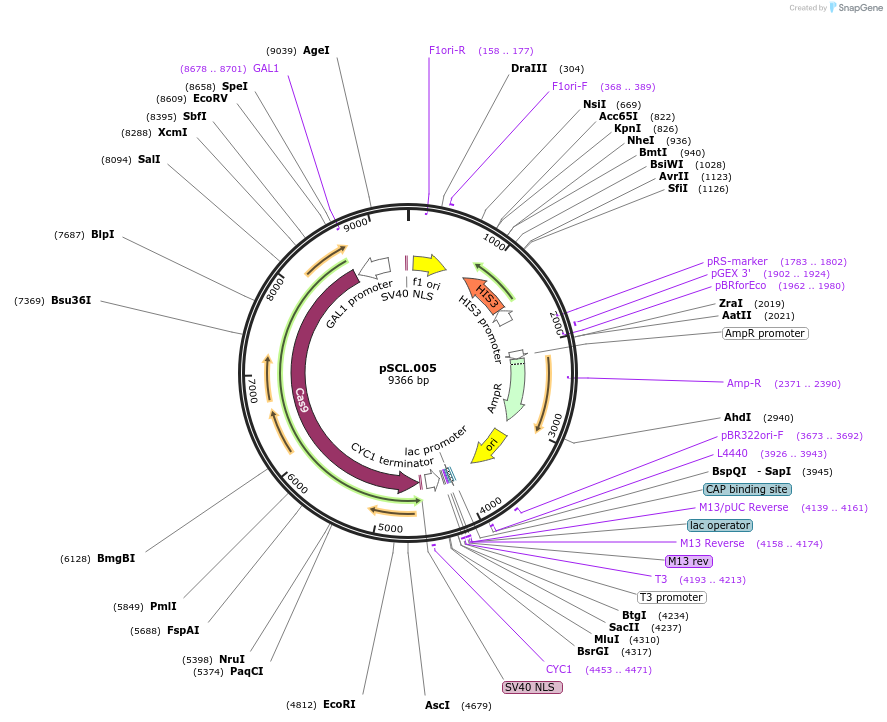
pSCL.005
Plasmid#184971PurposeExpress Cas9 in yeastDepositorInsertIntegrating inducible cassette: Cas9
TagsSV40NLSExpressionYeastPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
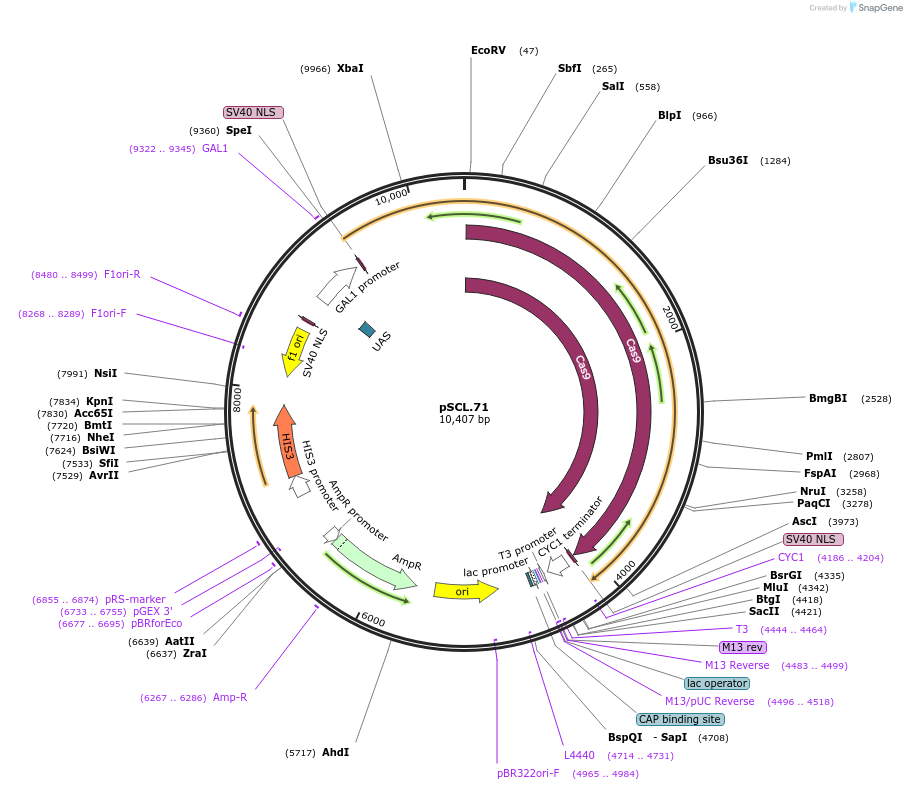
pSCL.71
Plasmid#184983PurposeExpress -Eco1 RT/Cas9 fusion, using linker1DepositorInsertEco1RT-fusion_linker1-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
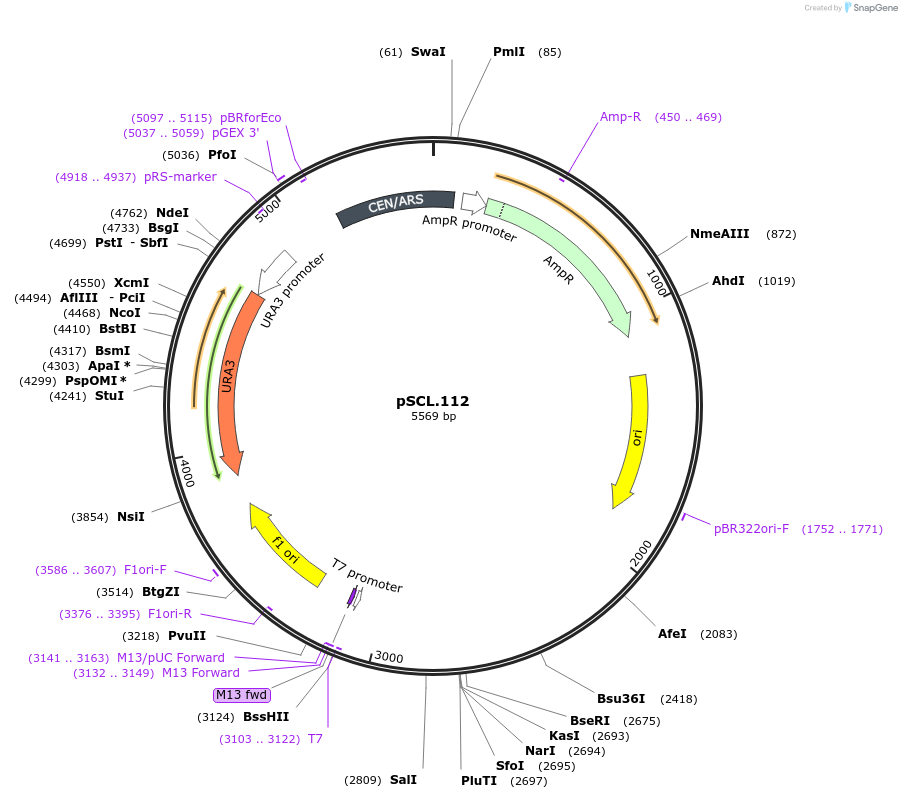
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
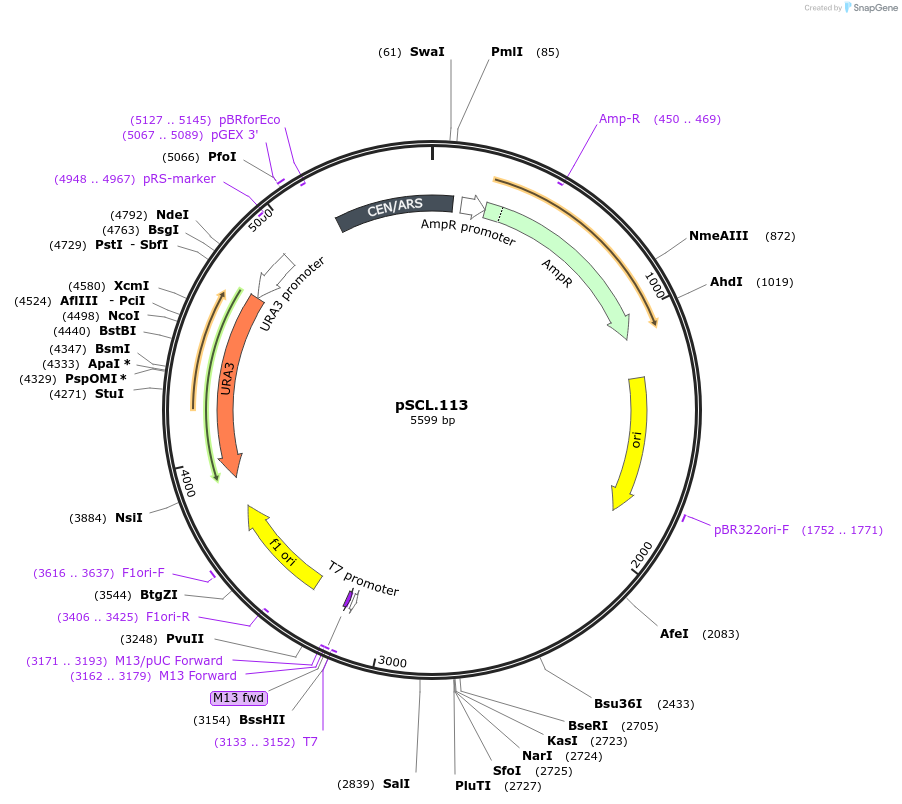
pSCL.113
Plasmid#184982PurposeTest effect of extending a1/a2 on FAA1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 27 v1
ExpressionYeastMutationFAA1 donor P233stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
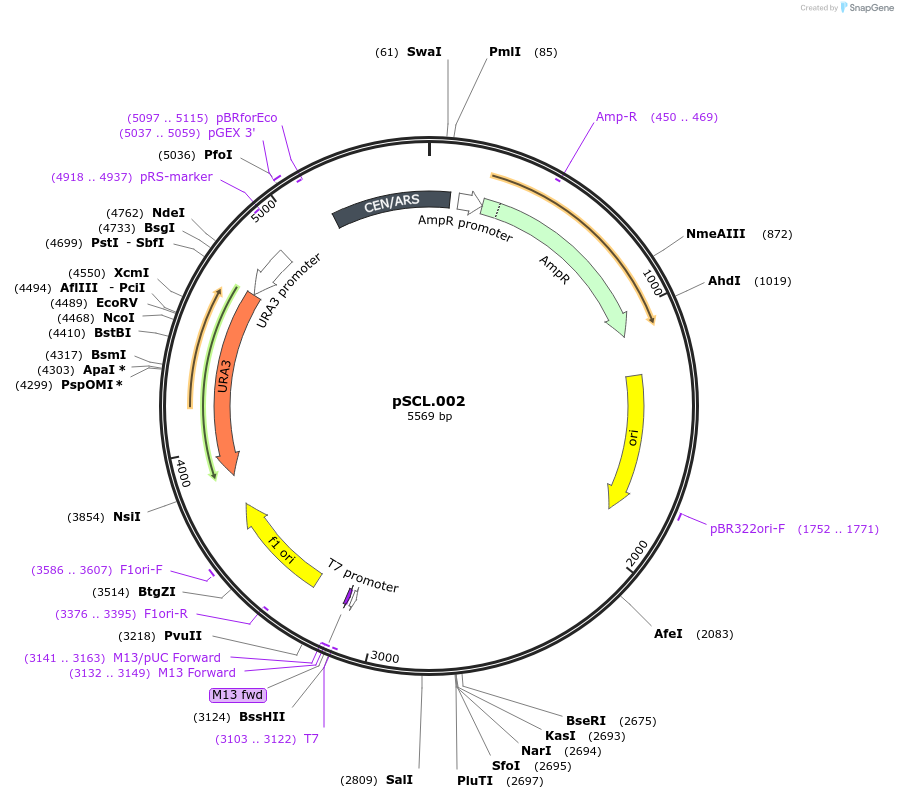
pSCL.002
Plasmid#184972PurposeExpress -Eco1 ADE2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 12
ExpressionYeastMutationADE2 donor P272stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
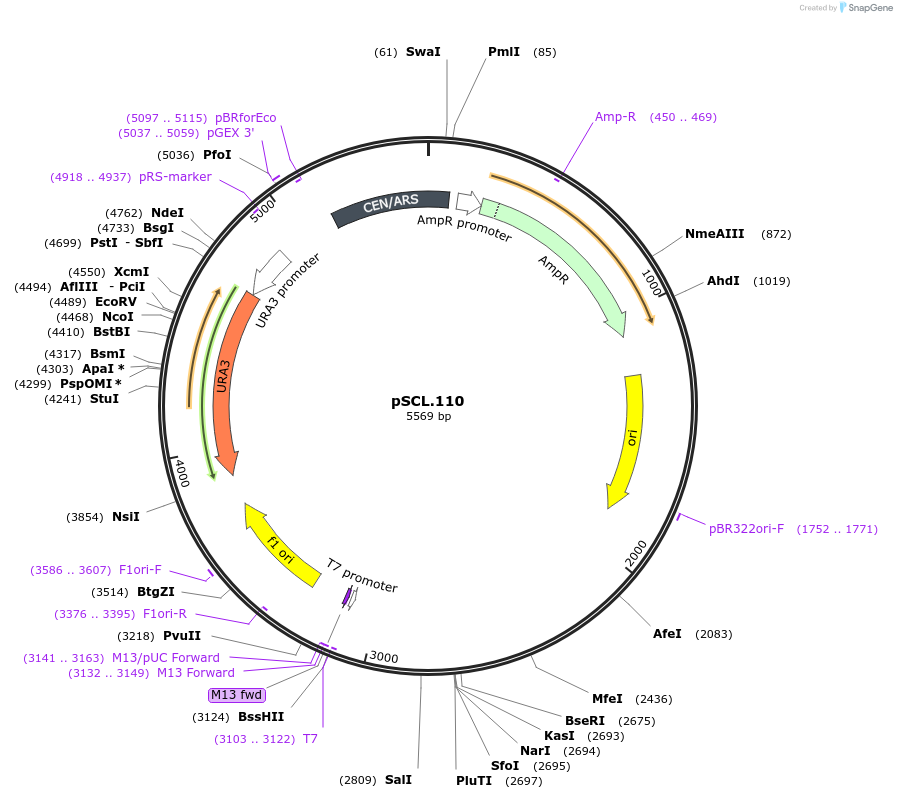
pSCL.110
Plasmid#184979PurposeExpress -Eco1 TRP2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 12
ExpressionYeastMutationTRP2 donor E64stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
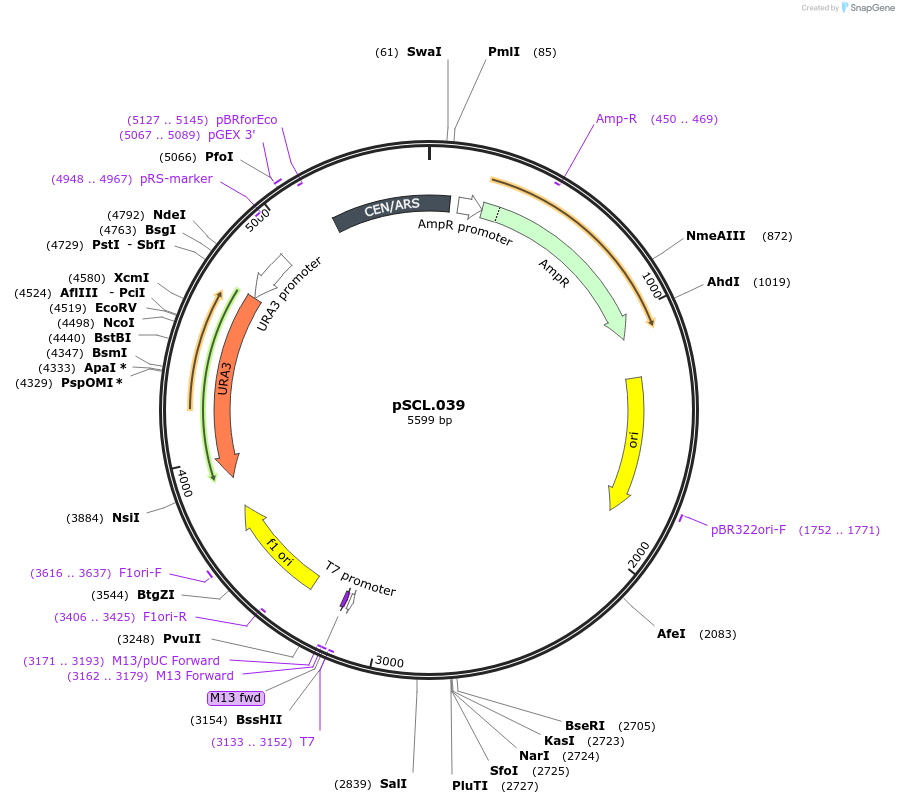
pSCL.039
Plasmid#184973PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v1
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
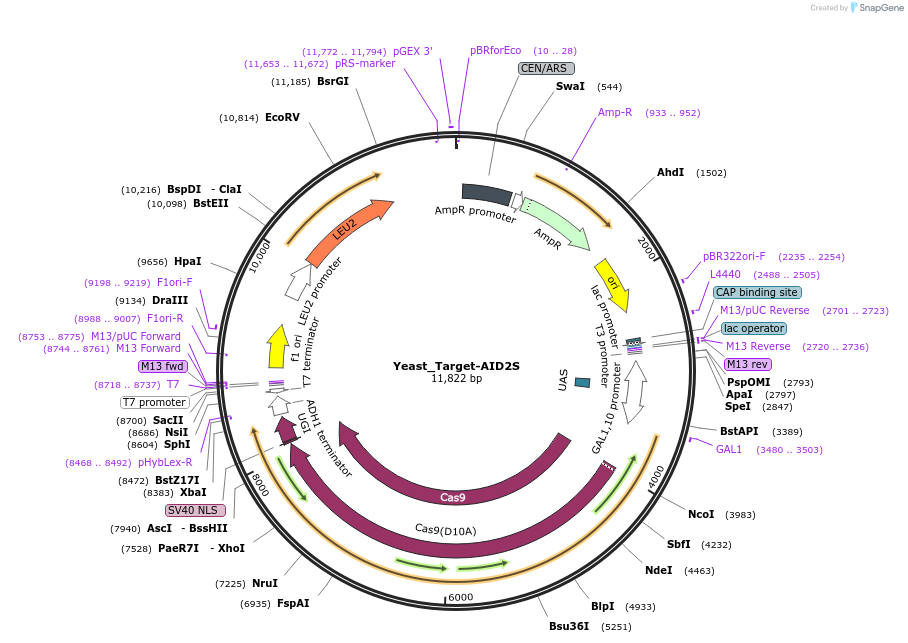
Yeast_Target-AID2S
Plasmid#188647PurposeExpressing Target-AID2S in yeastsDepositorInsertnCas9-AID2S-UGI
TagsUGIExpressionYeastMutationD10A for SpCas9PromoterpGal1Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
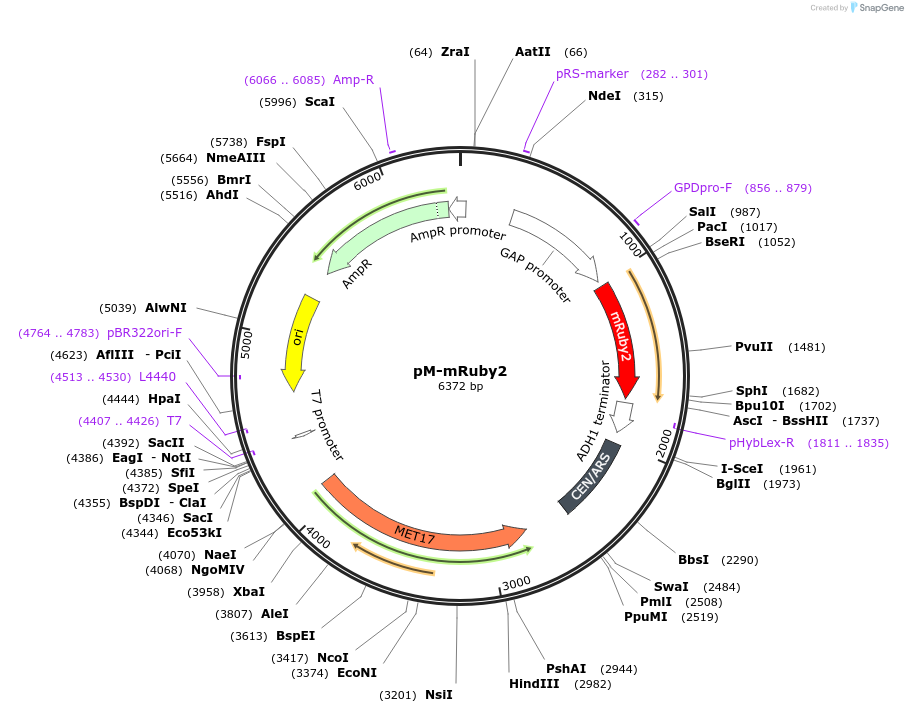
pM-mRuby2
Plasmid#176550PurposeExpression of mRuby2 for auxotrophic selection in the absence of methionineDepositorInsertyomRuby2
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
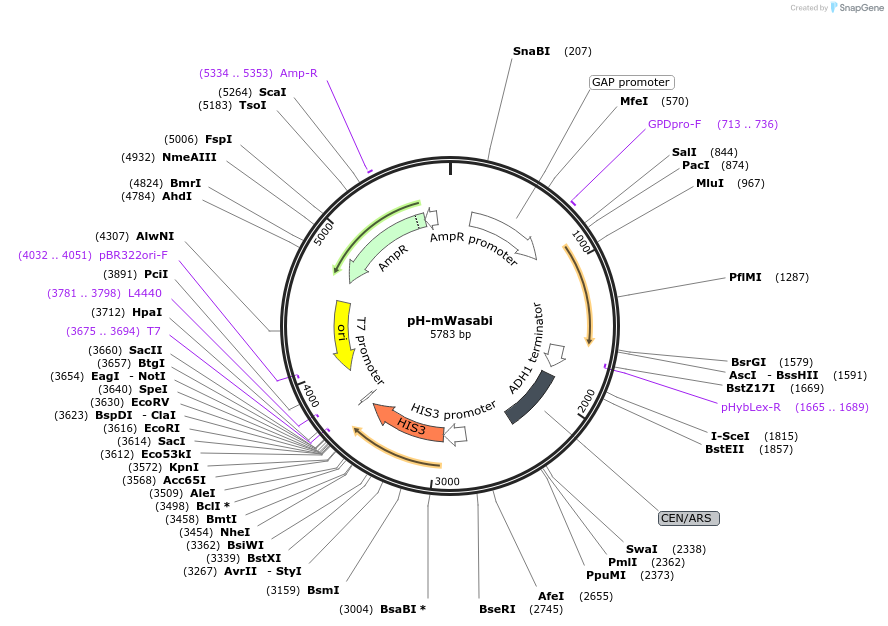
pH-mWasabi
Plasmid#176551PurposeExpression of mWasabi for auxotrophic selection in the absence of histidineDepositorInsertmWasabi
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
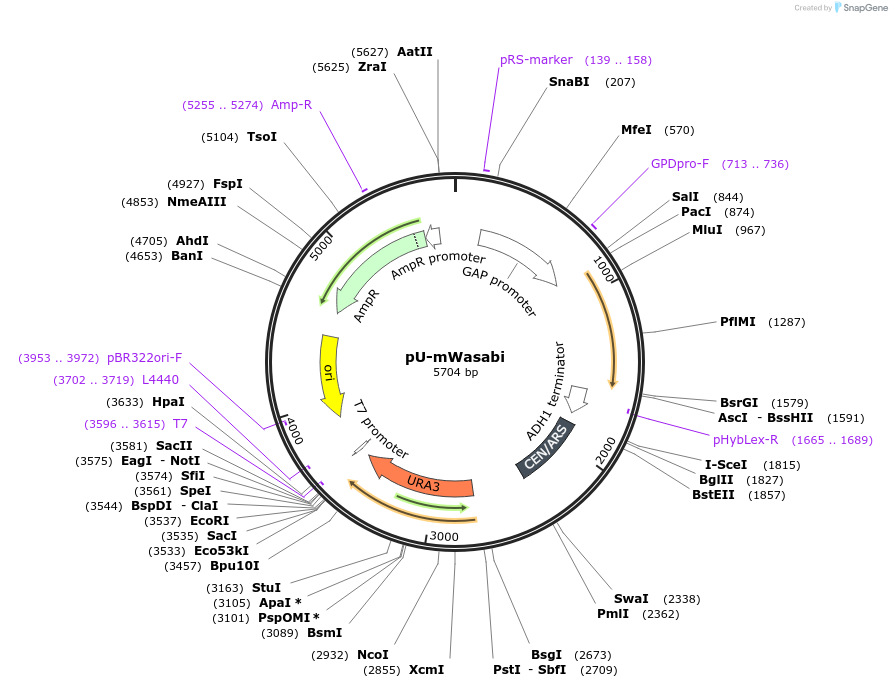
pU-mWasabi
Plasmid#176553PurposeExpression of mWasabi for auxotrophic selection in the absence of uracilDepositorInsertmWasabi
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
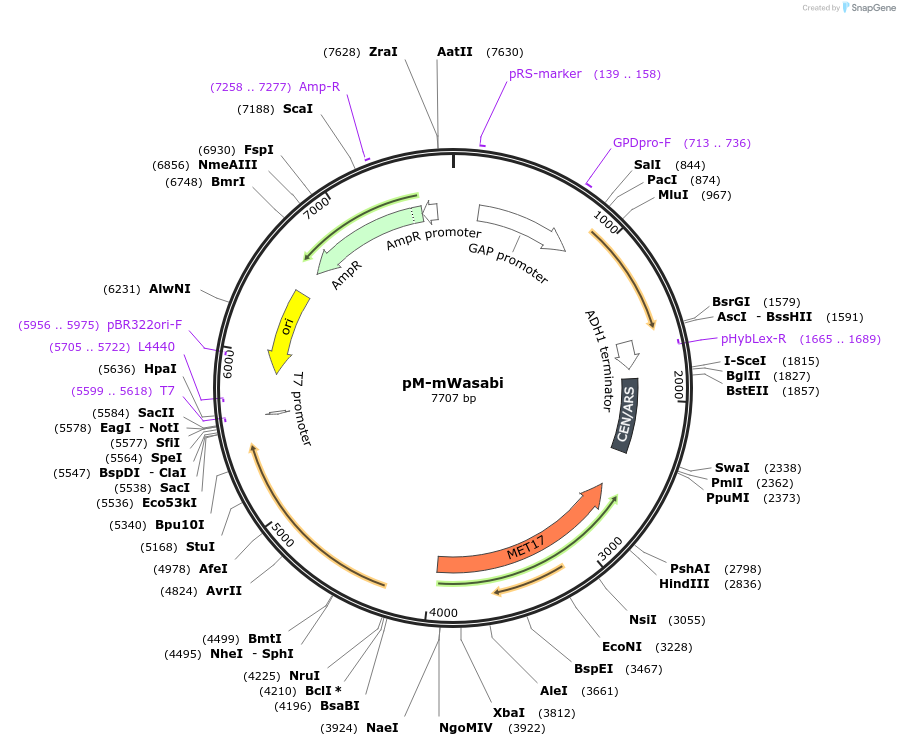
pM-mWasabi
Plasmid#176554PurposeExpression of mWasabi for auxotrophic selection in the absence of methionineDepositorInsertmWasabi
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
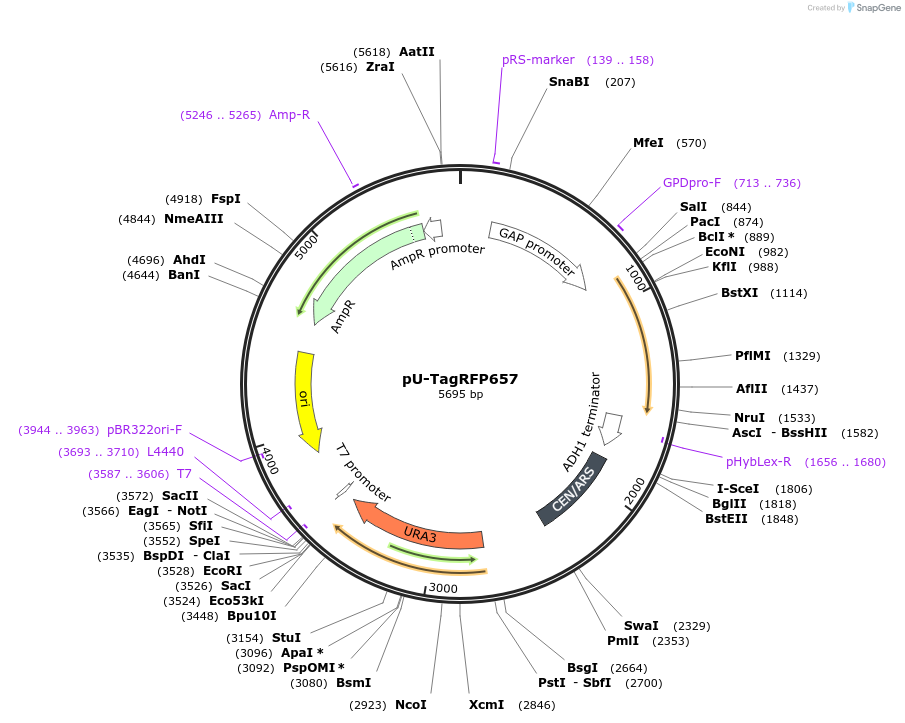
pU-TagRFP657
Plasmid#176562PurposeExpression of TagRFP657 for auxotrophic selection in the absence of uracilDepositorInsertTagRFP657
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
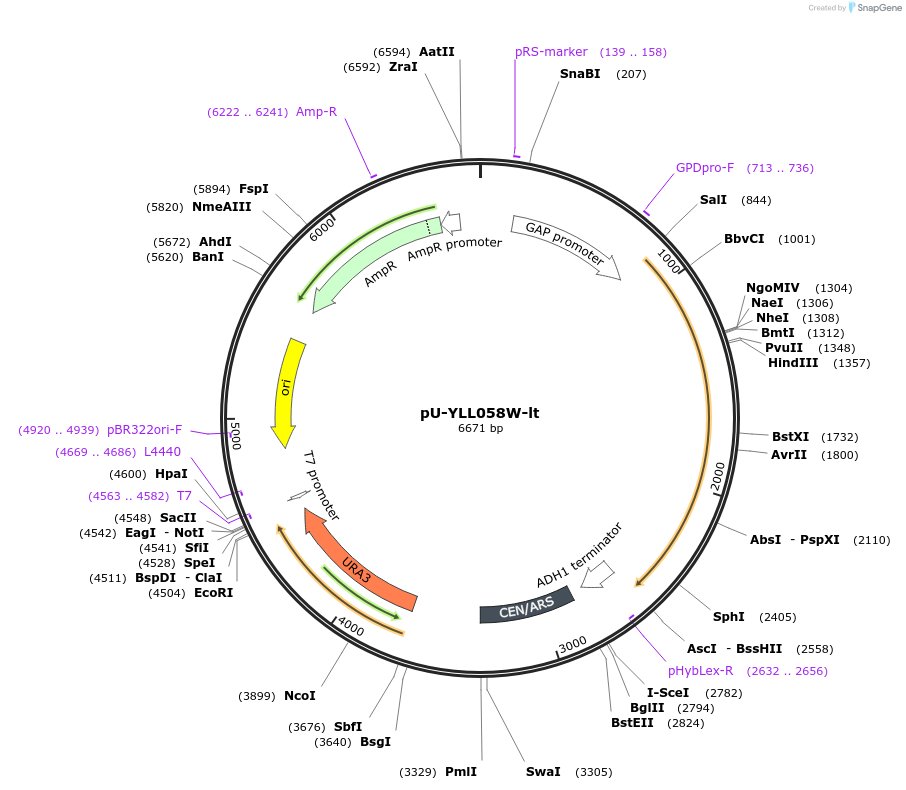
pU-YLL058W-lt
Plasmid#176564PurposeExpression of YLL058W for Lanchea thermotolerans for auxotrophic selection in the absence of uracilDepositorInsertYLL058W-lt
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
pU-YLL058W-zr
Plasmid#176565PurposeExpression of YLL058W from Zygosaccharomyces rouxii for auxotrophic selection in the absence of uracilDepositorInsertYLL058W-zr
ExpressionYeastMutationCodon optimised for expression in S.cerevisaePromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
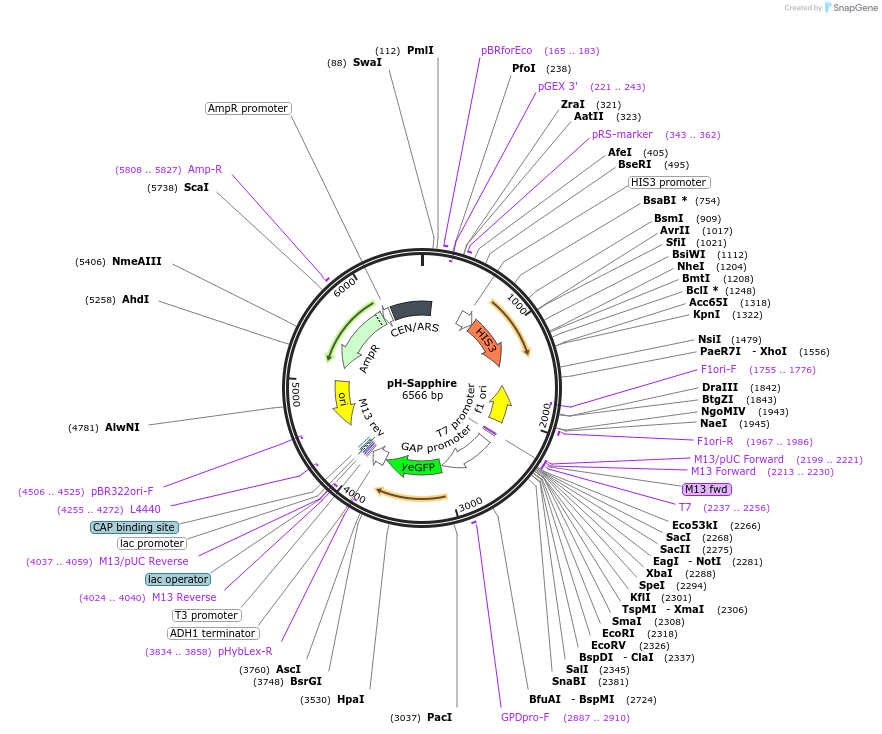
pH-Sapphire
Plasmid#176566PurposeExpression of Sapphire for auxotrophic selection in the absence of lysineDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
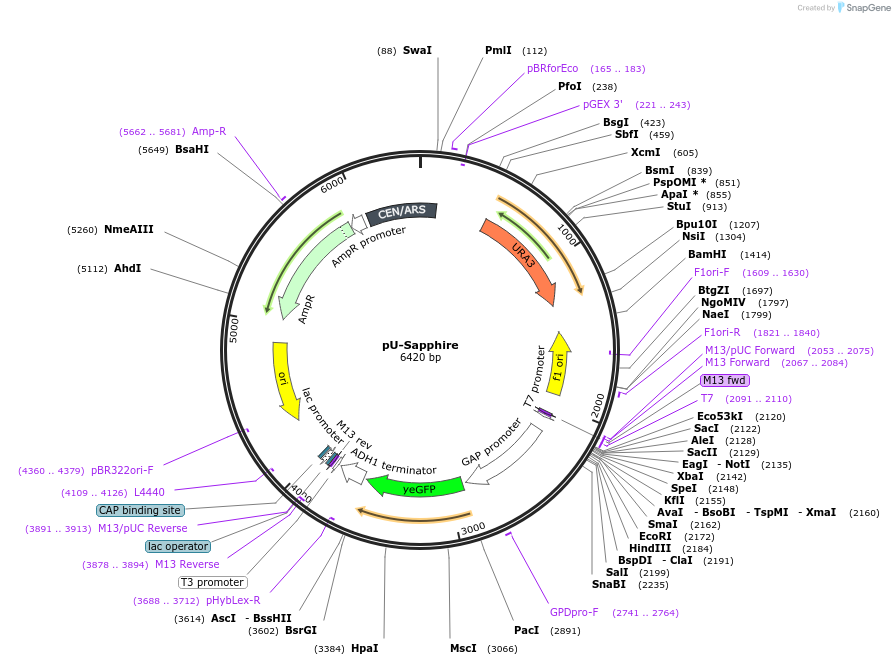
pU-Sapphire
Plasmid#176567PurposeExpression of Sapphire for auxotrophic selection in the absence of uracilDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
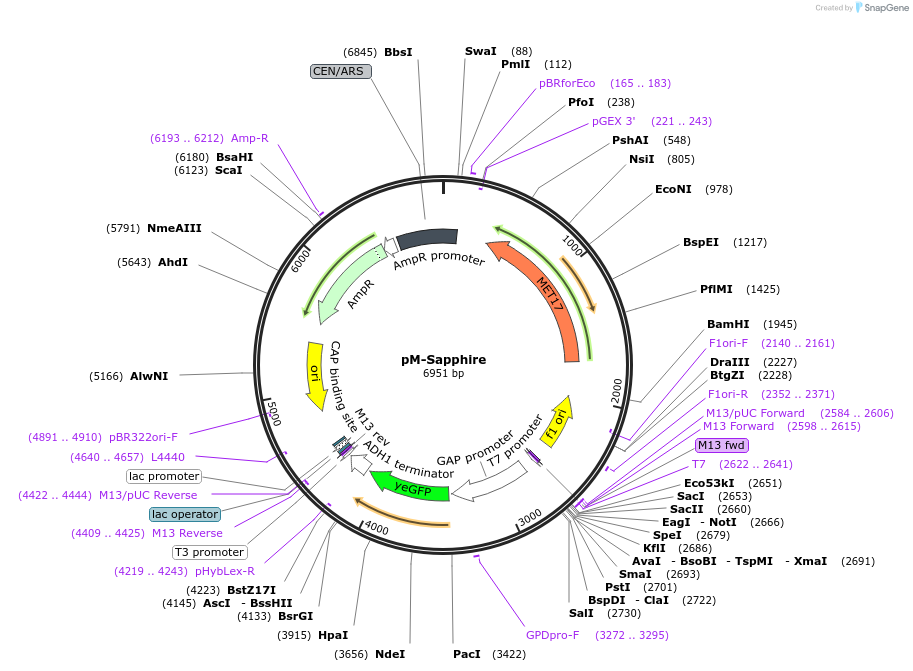
pM-Sapphire
Plasmid#176569PurposeExpression of Sapphire for auxotrophic selection in the absence of methionineDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
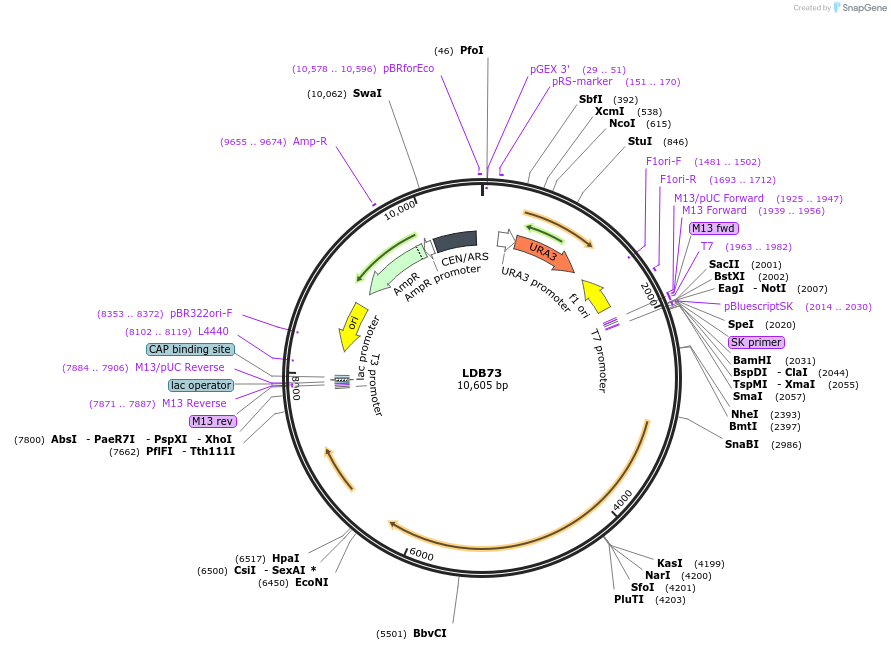
LDB73
Plasmid#185437PurposeNUP1 CEN URA3 vector for synthetic lethal sectoring assays and screeningDepositorInsertNUP1
ExpressionYeastAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
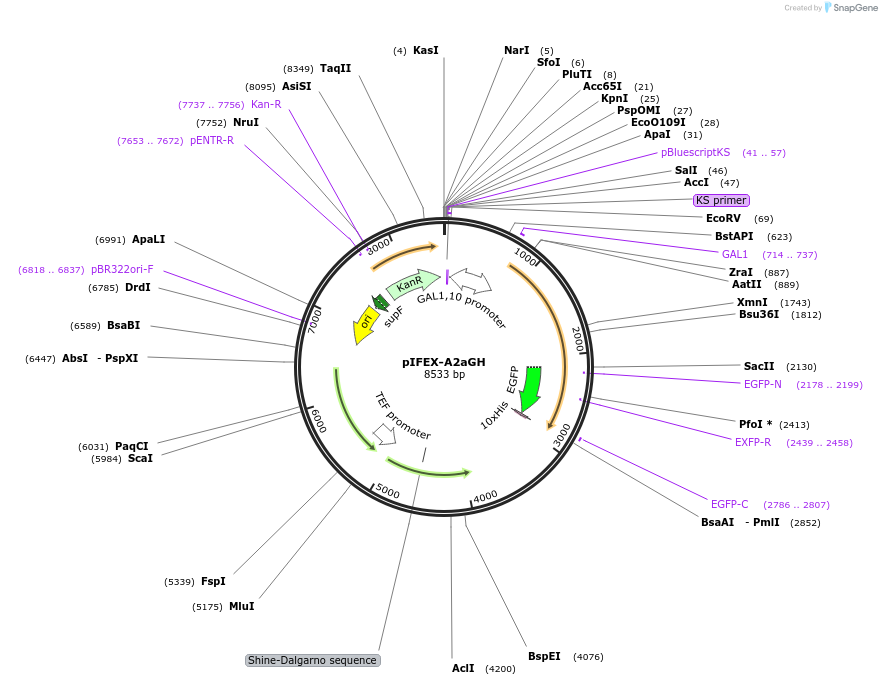
pIFEX-A2aGH
Plasmid#160542PurposepITY4 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
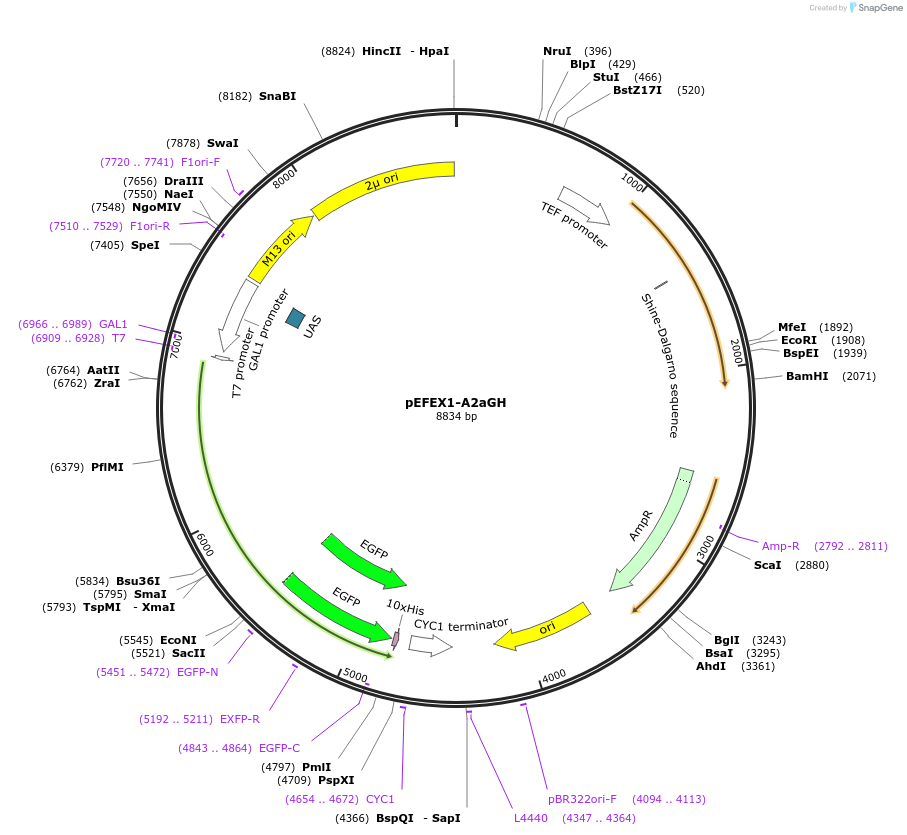
pEFEX1-A2aGH
Plasmid#160543PurposepYES2 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only